Abstract
Tumor protein D52 (TPD52) is overexpressed in different cancers, but its molecular functions are poorly defined. A large, low-stringency yeast two-hybrid screen using full-length TPD52 bait identified known partners (TPD52, TPD52L1, TPD52L2, MAL2) and four other preys that reproducibly bound TPD52 and TPD52L1 baits (PLP2, RAB5C, GOLGA5, YIF1A). PLP2 and RAB5 interactions with TPD52 were confirmed in pull down assays, with interaction domain mapping experiments indicating that both proteins interact with a novel binding region of TPD52. This study provides insights into TPD52 functions, and ways to maximise the efficiency of lowstringency yeast two-hybrid screens.
Keywords: TPD52, Yeast two-hybrid, Pull down, PLP2, RAB5
Introduction
Gene expression and genomic profiling analyses indicate that many human genes are overexpressed in human cancers, but the significance of most is poorly understood. An example is tumor protein D52 (TPD52), located at chromosome 8q21.13. Substantial evidence points to TPD52 representing a gene amplification target in common cancer types, and conferring or enhancing transformed phenotypes in cell culture models [1]. TPD52 is one of 4 paralogous mammalian genes whose translated coding sequences show approximately 50 % sequence identity [2]. Numerous protein–protein interaction studies have been undertaken to identify partners for TPD52 and related proteins [3–6], with most employing yeast two-hybrid (Y2H) screening [3–5]. While Y2H screens have repeatedly identified mutual interactions between TPD52-like proteins [3, 5, 7], fewer studies have identified heterologous partners, and particularly those partners that might help explain TPD52's oncogenic functions [4, 5]. Our knowledge of TPD52 molecular functions has therefore not kept pace with the growing recognition of TPD52 amplification and over-expression in human cancer.
Previous targeted Y2H screens using TPD52-like baits have been limited by technical factors, such as the numbers of prey constructs transformed, and/or the use of stringent screening environments [3–5]. These results contrasted those of a large, low-stringency Y2H screen using a MAL2 bait in our laboratory, which identified a number of informative novel partners [8, 9]. We therefore reasoned that a comparable Y2H screen could similarly extend our knowledge of TPD52 functions, through the identification of novel partners.
Materials and methods
Plasmid constructs
Plasmids encoding TPD52-like bait and prey proteins have been described previously, as have the corresponding plasmid constructs for pull-down assays [3, 10]. Plasmid constructs encoding TPD52, or fragments thereof, include sequences corresponding to TPD52 variant 3, NCBI Reference Sequence NM_005079.2. Plasmid constructs encoding Tpd52, or fragments thereof, include sequences corresponding to Tpd52 variant 5, NCBI Reference Sequence NM_009412.2, which is the Tpd52 variant most similar to TPD52 variant 3. Plasmid constructs encoding TPD52L1 include sequences corresponding to TPD52L1 variant 1, NCBI Reference Sequence NM_003287.2. Pull-down assays employed glutathione S-transferase (GST) and 6His-tagged recombinant proteins expressed from pGEX3X [3, 10], glutathione S-transferase-tagged recombinant proteins expressed from pGEX6P-1, and thioredoxin (Th) and 6His-tagged recombinant proteins expressed from the pET32a vector [10]. pGEX3X-Tpd52(1–152), and pGEX3X-Tpd52(40–185) plasmids were constructed as described for pGEX3X-Tpd52 [3]. The pGEX6P-1TPD52 construct was obtained by subcloning a BamH1-XhoI TPD52 fragment into the same sites of pGEX6P-1 expression vector. The pECFP-RAB5A, -RAB5B, and -RAB5C expression plasmids [11] were gifts from Professor Philip D. Stahl (Washington University, USA).
Yeast two-hybrid system and screening
Saccharomyces cerevisiae strain Hf7c cultures were grown at 30 °C in standard liquid or solid media [3]. A Y2H screen using full-length TPD52 bait was performed with a human breast carcinoma cDNA library over 14 days, as described [3]. For direct interaction testing, paired baits (pAS2-1 constructs) and preys (pACT2 or pAD-GAL4 constructs) were transformed into Hf7c cells, and plated on synthetic drop-out (SD) media lacking Leu and Trp (SD/-Leu–Trp). Positive and negative controls were TPD52 bait paired with TPD52L1 prey, and bait or prey paired with the opposing empty vector, respectively. Bait-prey interactions were scored according to time (in days) until visible colony growth on solid SD media lacking His, Leu, and Trp (SD/-His–Leu–Trp).
Breast cancer cell lines
Breast carcinoma (SK-BR-3, MCF7) cell lines were cultured in a humidified incubator at 37 °C with 5 % CO2 in RPMI media (Life Technologies) with 10 % fetal bovine serum (Life Technologies), 3 % L-glutamine (Life Technologies) and for MCF7 cells, 10 μg/ml insulin (Sigma-Aldrich). For transient transfections, SK-BR-3 cells were seeded into 100 mm tissue culture plates and grown until ~70 % confluency. Cells were transfected with 15 μg pECFP-RAB5 expression plasmids [11] for 8 h using TransIT-LT1 transfection reagent (Mirus) in serum-free RPMI media, as per the manufacturer's instructions. Transfection media were replaced with RPMI media and cells were incubated for 24 h before harvesting.
Pull-down assays
Recombinant Th-6-His-tagged truncated TPD52/Tpd52 proteins, Th-6-His protein, recombinant GST-TPD52, GST-Tpd52-6His and GST±6His proteins were produced in E. coli BL21 cells following induction of log-phase cultures with 0.1 mM isopropyl-ß-D-1-thiogalactopyrano-side (IPTG) for 3 h at 29 °C. GST- and 6-His fusion proteins were isolated from bacterial extracts as described previously [12] and purified by glutathione-Sepharose 4B beads (GE Healthcare) and Ni–NTA agarose (Qiagen), respectively. Protein product sizes were verified by electrophoresing pre- and post-induction protein samples on 10 % SDS–polyacrylamide gels, followed by Coomassie Brilliant Blue staining. Before pull-down assays, recombinant GST-fusion proteins were coupled to GSH-agarose beads. GST-fusion protein beads were transferred to Bio-Rad Bio-spin columns, centrifuged at 1,000 rpm for 5 min, and washed 4 times with 700 μl triethanolamine (TEA). Beads were resuspended in 500 μl 30 mM Dimethyl pimelimidate-2 HCl in TEA and incubated with rotation for 1 h at RT. Two ml quenching buffer (1M Tris–HCl pH 8.2) was added, columns were inverted for 10 min at RT, and centrifuged at 1,000 rpm for 5 min. Columns were washed 4 times with 700 μl freshly prepared 50 mM reduced glutathione in TBS pH 8.0 at RT. Ice-cold 700 μl pull-down wash buffer (150 mM NaCl, 20 mM Tris–HCl pH 7.4) was added, eluted under gravity, and columns were washed twice with cross-linking wash buffer (1.5 M NaCl, 25 mM Tris–HCl pH 7.6) and pull-down wash buffer.
MCF7 or SK-BR-3 cell lysates were pre-cleared 3 times with washed GSH or Ni–NTA beads. Recombinant proteins bound to beads were incubated with pre-cleared cell lysates for 1 h with rotation at 4 °C, followed by centrifugation at 500 ×g for 5 min at 4 °C. Bead pellets were transferred to pre-chilled MicroSpin columns (Bio-Rad) and washed 6 times (500 μl pull-down wash buffer for glutathione (GSH)-agarose beads, or PBS with 20 mM imidazole for Ni–NTA beads). Bound proteins were eluted in 20 μl SDS-loading buffer for 5 min at 85 °C, and analysed by electrophoresis on 15 % SDS–polyacrylamide gels.
Western blot analyses
Cultured cells were washed twice with ice-cold PBS and lysed in 100 mM NaCl, 50 mM Tris–HCl pH 7.4, 0.5 % Nonidet P-40, 5 mM EDTA, 1 mM PMSF, and protease inhibitors (Roche) for 30 min at 4 °C. Protein lysates were incubated for 15 min on ice and centrifuged at 13,000 rpm for 15 min at 4 °C prior to separation on 15 % SDS-PAGE gels. Proteins were electrotransferred to PVDF membranes (Millipore). Membranes were blocked with 5 % skim milk in TBS, washed twice with TBS, and incubated with one or more of the following primary antibodies for 2 h at RT: affinity-purified rabbit polyclonal TPD52 [13] (1/100), mouse monoclonal GAPDH (Ambion, 1/5,000) antisera, incubated in 0.1 % Tween 20 in TBS, and/or rabbit polyclonal RAB5C (Sigma, 1/300), rabbit polyclonal PLP2 (Sigma, 1/250) antisera, incubated in blocking buffer. Membranes were washed 4 times in TBS/0.1 % Tween 20, and incubated with horseradish peroxidase-conjugated donkey anti-rabbit or anti-mouse secondary antibody (GE Healthcare, 1/10,000 or 1/5,000, respectively) for 1 h at RT. Blots were washed 4 times, and antigen–antibody complexes were visualized by Western lightning chemiluminescent reagent (Perkin Elmer Life Science).
Statistical analysis
The SPSS for Windows package (version 19) was used to summarise prey categories using percentages within each group. Differences in proportions were compared using Pearson's Chi squared test.
Results
Putative novel TPD52 partners identified through yeast two-hybrid screening
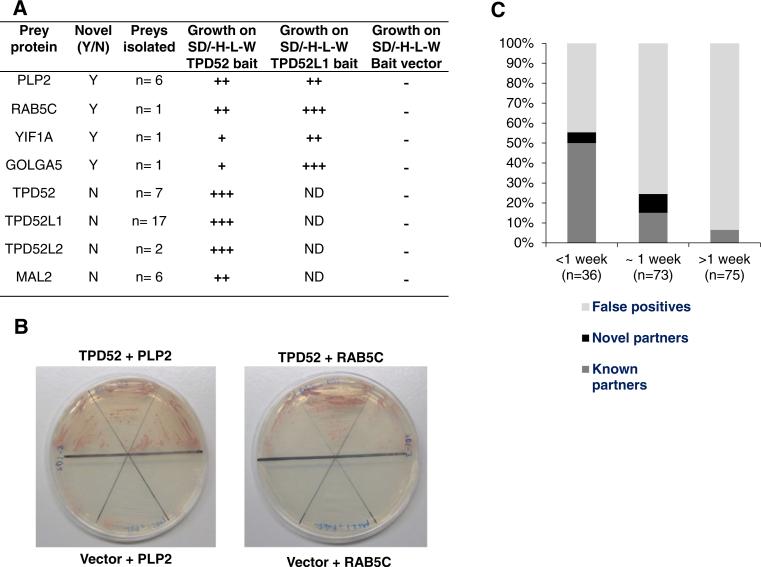
Approximately 3 × 106 colony forming units (cfu) of a human breast carcinoma cDNA expression library [3] were screened in Hf7c cells using a full-length TPD52 bait, and 204 His-positive colonies demonstrated reproducible growth when re-streaked on solid SD/-His–Leu–Trp media. Isolation and sequencing of plasmid DNA identified 184 prey constructs. Sequence analysis and direct interaction testing indicated that preys could be classified as known partners (n = 34, 18 %), novel putative partners (n = 9, 5 %), or false positives (n = 141, 77 %). The previously identified TPD52 partners TPD52L1, TPD52, TPD52L2 and MAL2 [3–5, 7] were all identified repeatedly (Fig. 1a). Most prey constructs encoding known partners (18/34, 53 %) were identified by 5 days of screening (<1 week), when they were the predominant prey category isolated (18/36 preys, 50 %) (Fig. 1c).
Fig. 1.
a Summary of known and novel candidate TPD52 partners identified. (+), growth on solid SD/-His–Leu–Trp media at 30 °C after 5–6 days; (++), after 3–4 days; (+++), after 1–2 days; (−), no visible growth after at least 7 days; ND not done. Results were obtained from three independent experiments in which each interaction was assessed in triplicate. b Hf7c cells co-transformed with PLP2 or RAB5C pAD-GAL4 prey vectors and pAS2-1TPD52 bait, or empty pAS2-1 vector. Triplicate colonies were transferred to solid SD/-His–Leu–Trp media, and incubated at 30 °C for 9 days, demonstrating growth of TPD52 and PLP2 or RAB5C co-transformants, but not of bait vector and PLP2 or RAB5C co-transformants. c Proportions of known partners (dark grey), candidate novel partners (black) and false positives (light grey) identified according to Y2H screening duration. Known partners were most frequently identified by 5 days of screening (<1 week), novel partners were most frequently identified at 1 week (1 week), and false positives were the most frequent prey category at 1 week, and between 1 and 2 weeks of screening (>1 week)
Nine additional prey constructs encoding 4 proteins showed reproducible interactions with TPD52 bait in direct testing analyses, namely proteolipid protein 2 (PLP2), Ras-associated binding protein 5C (RAB5C), Yip interacting factor homolog A (YIF1A) and Golgi autoantigen A5 (GOLGA5) (Fig. 1a, b). Full-length PLP2 coding sequences were isolated 6 times, whereas Full-length RAB5C and YIF1A coding sequences were isolated once (Fig. 1a). The single GOLGA5 prey contained a partial coding sequence, corresponding to GOLGA5 residues 490–731. Co-transfection of TPD52 bait and candidate preys produced detectable growth on selective media after 3–4 days (PLP2, RAB5C), or 5–6 days (YIF1A, GOLGA5) (Fig. 1a). Similarly, co-transfection of Full-length TPD52L1 bait produced detectable growth after 1–2 days (GOLGA5, RAB5C) or 3–4 days (PLP2, YIF1A) (Fig. 1a). Preys encoding novel partners were identified either before or at 1 week of screening (Fig. 1c). However at 1 week of screening, the preys identified were found to be predominantly false-positives (55/73 preys, 75 %), which increased to 70/75 (93 %) of prey constructs isolated between 1 and 2 weeks of screening (Fig. 1c). Chi squared analysis indicated significant differences in the proportions of prey categories isolated at different time-points during the screen (p < 0.001) (Fig. 1c).
Pull-down assays confirm PLP2 and RAB5 interactions with TPD52
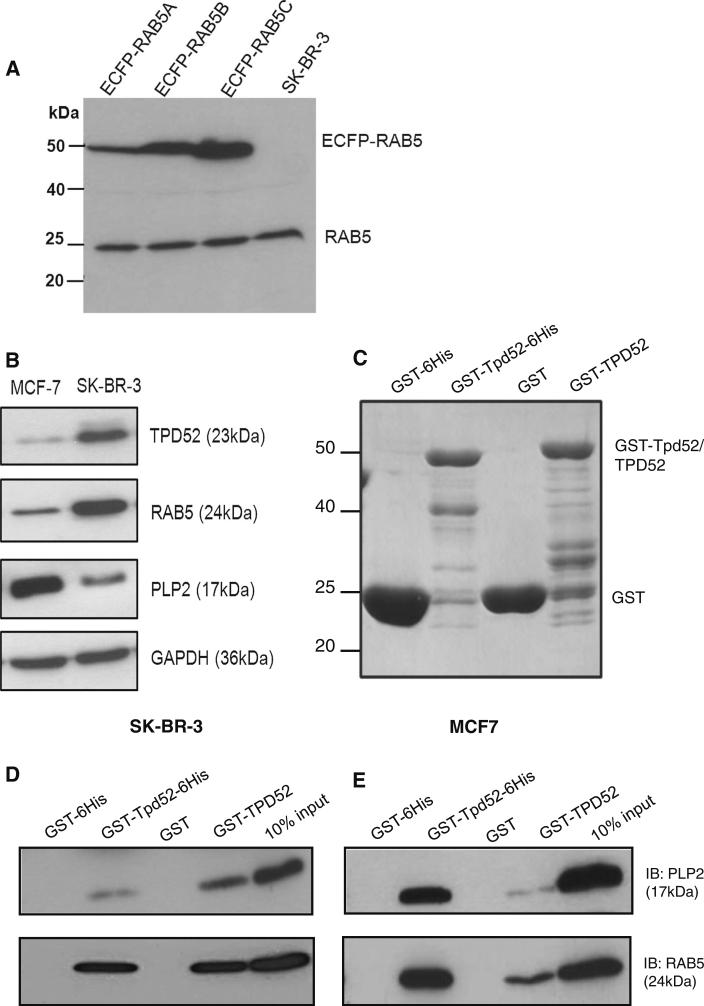
Of the novel candidate TPD52 partners, PLP2 and RAB5C were selected for further study. PLP2 was most frequently identified (Fig. 1a), and is related to the known TPD52 partner MAL2 [4, 7, 8], with both proteins featuring the MARVEL domain [14, 15]. RAB5C was also selected, as TPD52 has been reported to co-immunoprecipitate with RAB5 and other RAB proteins [16]. As RAB5 proteins are encoded by three highly related genes, we examined whether commercial RAB5C antisera specifically detected RAB5C. Western blot analyses showed that RAB5C antisera detected all 3 ECFP-tagged RAB5 isoforms [11], and were therefore employed as RAB5 antisera (Fig. 2a).
Fig. 2.
a Protein lysates from transfected (pECFP-RAB5A, -RAB5B, -RAB5C expression plasmids) and untransfected SK-BR-3 cells analysed by Western blotting using RAB5C antisera. Upper bands show positions of ECFP-tagged proteins (ECFP-RAB5) and the lower band indicates endogenous RAB5 (RAB5). Positions of molecular weight standards are shown on the left. b Western blot analysis of SKBR-3 and MCF7 protein lysates demonstrated detectable TPD52, PLP2 and RAB5 levels. GAPDH served as a loading control. Proteins detected and their molecular weights (in brackets) are shown on the right. c GST-Tpd52-6His, GSTTPD52 and GST tag proteins stained with Coomassie Brilliant Blue. Positions of molecular weight standards and expressed recombinant proteins are shown on the left and right, respectively. d, e Retention of PLP2 and RAB5 from SK-BR-3 (d) or MCF7 (e) protein lysates by GST-Tpd52-6His and GSTTPD52 but not GST tags. Proteins detected and their molecular weights (in brackets) are shown on the right of (e). Results shown are representative of those obtained in three independent experiments
Pull-down assays were used to test whether PLP2 and RAB5 bind TPD52 in vitro. Bound GST-TPD52 and GSTTpd52-6His or GST tags alone (Fig. 2c) were incubated with pre-cleared total cell lysates from SK-BR-3 or MCF7 cells, which detectably express both PLP2 and RAB5 (Fig. 2b). Both PLP2 and RAB5 from SK-BR-3 and MCF7 cells were retained on matrices to which GST-TPD52 and GST-Tpd52-6His were bound, but not on matrices to which GST tags had been bound (Fig. 2d, e).
TPD52 interaction domain mapping with PLP2 and RAB5
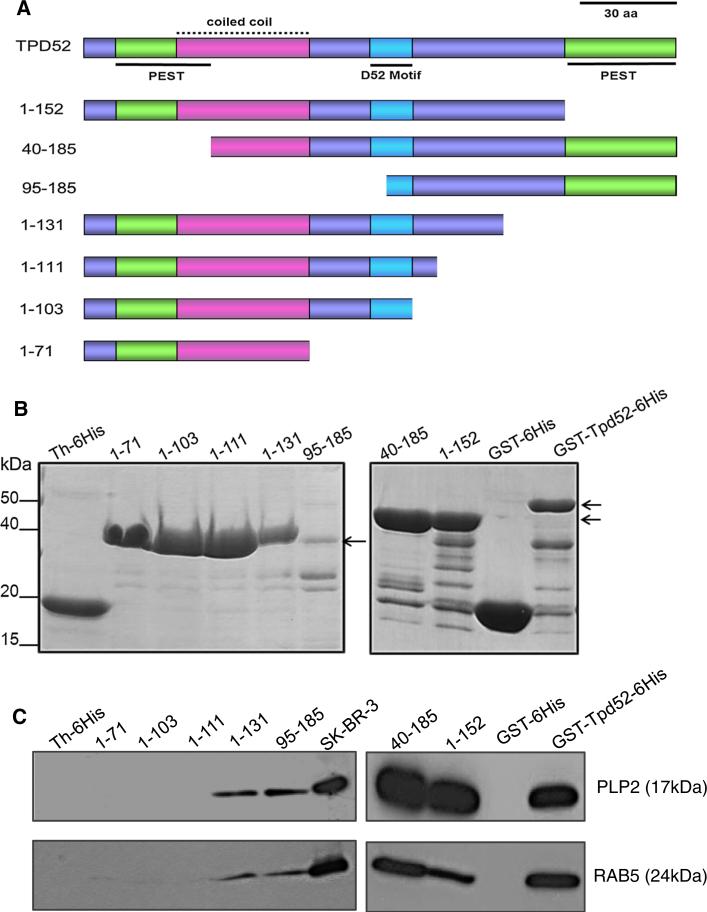
To determine the TPD52 regions required for interaction with PLP2 and RAB5, we used a series of TPD52/Tpd52 deletion mutants for pull-down assays (Fig. 3a). GSTTpd52 (40–185)-6His and GST-Tpd52 (1–152)-6His recombinant proteins lack N- or C-terminal PEST motifs, respectively (Fig. 3a). The TPD52 coiled-coil motif is encoded by all deleted constructs, except pET32a-Tpd52(95–185) [10]. Pull-down assays showed that both PLP2 and RAB5 from SK-BR-3 cell lysates (Fig. 3c) were retained by GSTTpd52 (1–152)-6His, GST-Tpd52 (40–185)-6His and Th-tagged Tpd52 (95–185)-6His and Tpd52 (1–131)-6His, but not by Th-6His or GST tag alone, or by Th-6His tagged Tpd52 (1–71), Tpd52 (1–103) or Tpd52 (1–111) (Fig. 3b, c). Comparable results were obtained in pull-down assays employing MCF7 cell lysates (data not shown).
Fig. 3.
a Schematic diagram of TPD52 sequence features included in deleted TPD52/ Tpd52 proteins used in pull-down assays. PEST motifs are shown in green, underlined, the coiled-coil motif is shown in pink, with a broken line, the D52 motif is shown in blue, underlined, with all other regions shown in purple. The N-terminal PEST and coiled-coil motifs partially overlap. Amino acid co-ordinates of deleted proteins are shown at the left. Scale bar = 30 aa. b Coomassie Brilliant Blue-stained recombinant TPD52/ Tpd52 (positions highlighted by arrows, right) and vector tag proteins. Th-6His-tagged recombinant proteins are shown in the left panel, whereas GST-6His-tagged recombinant proteins are shown in the right panel. c Retention of PLP2 (upper panel) or RAB5 (lower panel) by a subset of deleted TPD52/Tpd52 proteins and Full-length GST-Tpd52-6His, but not vector tag proteins. Proteins detected and their molecular weights (in brackets) are shown on the right. 10 % of the SKBR-3 lysate input (SK-BR-3) was included as a loading control. Results shown are representative of those obtained in three independent experiments. (Color figure online)
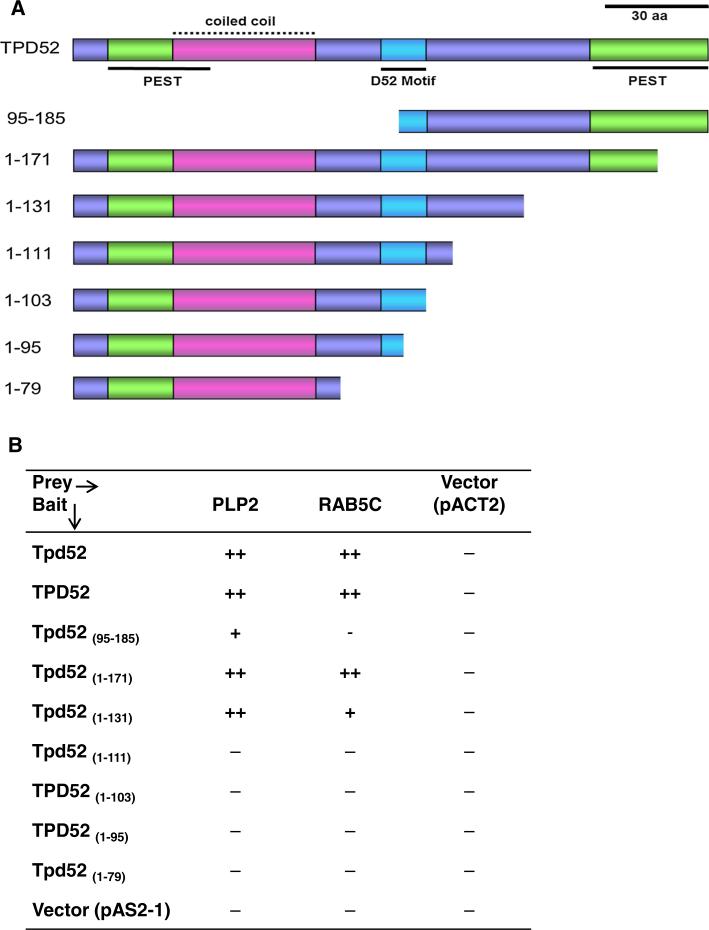
The Y2H system was used to replicate interactions between PLP2 and RAB5C preys and deleted TPD52/ Tpd52 baits (Fig. 4a). Both PLP2 and RAB5C preys detectably interacted with Tpd52 (1–171) and Tpd52 (1–131) but not with Tpd52 (1–111). Tpd52 (95–185) bait gave rise to weak growth on selective media when co-transfected with PLP2, but not RAB5C (Fig. 4b). The combined approach of pull-down and Y2H assays indicated that TPD52 residues Ser111–Ser131 were required for interactions between TPD52 and both PLP2 and RAB5.
Fig. 4.
a Deleted TPD52 baits used in Y2H assays, as per Fig. 4a. Scale bar = 30 aa. b Growth of H7fc cotransformants (deleted TPD52/ Tpd52 baits and PLP2, RAB5C preys, or empty pACT2 vector) on solid SD/-His–Leu–Trp media at 30 °C: (−) undetectable after 6–12 days; (+) detected after 5–6 days; (++) after 3–4 days. Results are from three independent Y2H experiments, assayed in triplicate. (Color figure online)
Discussion
TPD52 genes were first reported during the 1990s [1], but the identification of heterologous TPD52 binding partners has been slow. Whereas Y2H screens or direct interacting testing using paralogous TPD52-like baits have identified heterologous partners [4, 5, 13], a previous high-stringency TPD52 screen identified no known or novel partners, whereas a small screen in lower stringency Hf7c cells identified only known partners [3]. In the present study, a large Y2H screen in Hf7c cells identified 4 previously unreported prey proteins that showed reproducible interactions with both TPD52 and TPD52L1. Identification of novel partners was accompanied by larger numbers of preys encoding either known partners, or false-positives. The fact that novel partners were not detected beyond 1 week of screening indicates that extending screening time in Hf7c cells may not promote the identification of informative preys.
The 4 novel candidate partners identified are either integral membrane proteins (PLP2, GOLGA2, YIF1A) or membrane-associated proteins (RAB5C), which supports the identification of other trans-membrane or membrane-associated proteins as TPD52 partners [4, 6]. PLP2 is related to the known TPD52 partner MAL2 [4, 8], and was reported to enhance tumour-associated phenotypes in B16F10 melanoma cell lines [17]. In human breast cancer, PLP2 SAGE tags were found to contribute to an invasion-specific gene cluster [18], and PLP2 was also included in multi-gene classifiers associated with poorer outcomes in human astrocytic brain tumours [19] and glioblastoma [20]. PLP2 antisera suitable for in situ approaches will be required to pursue these transcript-based findings in breast and other cancers.
Direct interactions between RAB5 and TPD52 are supported by TPD52 co-immunoprecipitating with the bacterial product ExoS, as well as RAB5, RAB6, RAB9, and TIP47 [16]. This previous connection between TPD52 and RAB5 [16], combined with the identification of RAB5C by Y2H screening here and the high degree of sequence conservation between RAB5 isoforms [11], suggests that TPD52 may bind other RAB5 isoforms, and possibly other RAB proteins. RAB5 associating with TPD52L1 in Y2H assays and pull-down experiments (data not shown) is also consistent with endogenous and exogenously expressed Tpd52l1 co-localising with an early endosomal marker in PC12 and HeLa cells, respectively [5]. The significance of RAB5A immunostaining has been examined in human breast cancer specimens, where stronger RAB5A immunostaining was associated with higher histological grade and more axillary lymph node metastases [21]. RAB5A immunostaining was also detected in all breast cancer axillary lymph node metastases examined [21].
Both PLP2 and RAB5 were indicated to require TPD52 amino acids Ser111–Ser131 for binding, as neither bound deleted TPD52 recombinant proteins lacking Ser111–Ser131. This result contrasts those obtained for homomeric interactions between TPD52 proteins, where TPD52 was precipitated by recombinant TPD52 proteins that included the coiled coil motif, but not by a protein lacking the first 95 resides [10]. The present study therefore provides evidence for a second interaction interface within TPD52. Strikingly, the same region was required for interactions with PLP2 and RAB5, despite their different predicted structures [15, 22], suggesting that this TPD52 region may bind a range of partners. The fact that both PLP2 and RAB5 reproducibly bound TPD52L1 refines the likely binding region, as the alternatively-spliced 13 residue 14-3-3 binding site in TPD52L1 [13] inserts before TPD52L1 Asn143, which aligns with TPD52 residue Asn130. However, motif searches of residues within the TPD52 Ser111–Lys129 region that are identical in TPD52-like proteins (AAxSxxGxxIxxK) did not identify this motif in any other protein family (data not shown).
In summary, this study has reported novel candidate partners for TPD52 expressed in breast cancer tissue, and reinforced both membrane-associated functions for TPD52 [4, 6, 23], and a degree of functional redundancy between TPD52-like proteins [3–5, 7]. Approaches beyond the Y2H system may be required to identify isoform-specific partners for TPD52.
Acknowledgments
This work was supported by a project grant from the New South Wales Cancer Council (to JAB and BJS), a National Health and Medical Research Council of Australia postdoctoral fellowship (to YC), and Endeavour International- and International Postgraduate Research Scholarships (EIPRS and IPRS) (to HS), and by donations to the Children's Cancer Research Unit of the Children's Hospital at Westmead. The authors thank Dr Mona Shehata (CHW), Dr Erdahl Teber and A/Prof Jonathan Arthur (Children's Medical Research Institute, Australia) for discussions, and Prof Philip D. Stahl (Washington University, USA) for ECFPRAB5 expression plasmids.
Contributor Information
Hamideh Shahheydari, Children's Cancer Research Unit, Kids Research Institute, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia; The University of Sydney J. Discipline of Paediatrics and Child Health, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia.
Sarah Frost, Children's Cancer Research Unit, Kids Research Institute, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia; The University of Sydney J. Discipline of Paediatrics and Child Health, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia.
Brian J. Smith, Department of Chemistry, La Trobe Institute for Molecular Science, La Trobe University, Melbourne, VIC 3086, Australia
Guy E. Groblewski, Department of Nutritional Sciences, University of Wisconsin-Madison, Madison, WI, USA
Yuyan Chen, Children's Cancer Research Unit, Kids Research Institute, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia; The University of Sydney J. Discipline of Paediatrics and Child Health, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia.
Jennifer A. Byrne, Children's Cancer Research Unit, Kids Research Institute, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia The University of Sydney J. Discipline of Paediatrics and Child Health, The Children's Hospital at Westmead, Locked Bag 4001, Westmead, NSW 2145, Australia.
References
- 1.Shehata M, Weidenhofer J, Thamotharampillai K, Hardy JR, Byrne JA. Tumor protein D52 overexpression and gene amplification in cancers from a mosaic of microarrays. Crit Rev Oncog. 2008;14:33–55. doi: 10.1615/critrevoncog.v14.i1.30. [DOI] [PubMed] [Google Scholar]
- 2.Boutros R, Fanayan S, Shehata M, Byrne JA. The tumor protein D52 family: many pieces, many puzzles. Biochem Biophys Res Commun. 2004;325:1115–1121. doi: 10.1016/j.bbrc.2004.10.112. [DOI] [PubMed] [Google Scholar]
- 3.Byrne JA, Nourse CR, Basset P, Gunning P. Identification of homo- and heteromeric interactions between members of the breast carcinoma-associated D52 protein family using the yeast two-hybrid system. Oncogene. 1998;16:873–881. doi: 10.1038/sj.onc.1201604. [DOI] [PubMed] [Google Scholar]
- 4.Wilson SHD, Bailey AM, Nourse CR, Mattei MG, Byrne JA. Identification of MAL2, a novel member of the MAL proteolipid family, through interactions with TPD52-like proteins in the yeast two-hybrid system. Genomics. 2001;76:81–88. doi: 10.1006/geno.2001.6610. [DOI] [PubMed] [Google Scholar]
- 5.Proux-Gillardeaux V, Galli T, Callebaut I, Mikhailik A, Calothy G, Marx M. D53 is a novel endosomal SNARE-binding protein that enhances interaction of syntaxin 1 with the synaptobrevin 2 complex in vitro. Biochem J. 2003;370:213–221. doi: 10.1042/BJ20021309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas DD, Kaspar KM, Taft WB, Weng N, Rodenkirch LA, Groblewski GE. Identification of annexin VI as a Ca2+-sensitive CRHSP-28-binding protein in pancreatic acinar cells. J Biol Chem. 2002;277:35496–35502. doi: 10.1074/jbc.M110917200. [DOI] [PubMed] [Google Scholar]
- 7.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, Klitgord N, Simon C, Boxem M, Milstein S, Rosenberg J, Goldberg DS, Zhang LV, Wong SL, Franklin G, Li S, Albala JS, Lim J, Fraughton C, Llamosas E, Cevik S, Bex C, Lamesch P, Sikorski RS, Vandenhaute J, Zoghbi HY, Smolyar A, Bosak S, Sequerra R, Doucette-Stamm L, Cusick ME, Hill DE, Roth FP, Vidal M. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 8.Fanayan S, Shehata M, Agterof AP, McGuckin MA, Alonso MA, Byrne JA. Mucin 1 (MUC1) is a novel partner for MAL2 in breast carcinoma cells. BMC Cell Biol. 2009;10:7. doi: 10.1186/1471-2121-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Madrid R, Aranda JF, Rodríguez-Fraticelli AE, Ventimiglia L, Andrés-Delgado L, Shehata M, Fanayan S, Shahheydari H, Gómez S, Jiménez A, Martín-Belmonte F, Byrne JA, Alonso MA. The formin INF2 regulates basolateral-to-apical transcytosis and lumen formation in association with Cdc42 and MAL2. Dev Cell. 2010;18:814–827. doi: 10.1016/j.devcel.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Sathasivam P, Bailey AM, Crossley M, Byrne JA. The role of the coiled-coil motif in interactions mediated by TPD52. Biochem Biophys Res Comm. 2001;288:56–61. doi: 10.1006/bbrc.2001.5721. [DOI] [PubMed] [Google Scholar]
- 11.Chen PI, Kong C, Su X, Stahl PD. Rab5 isoforms differentially regulate the trafficking and degradation of epidermal growth factor receptors. J Biol Chem. 2009;284:30328–30338. doi: 10.1074/jbc.M109.034546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brymora A, Valova VA, Larsen MR, Roufogalis BD, Robinson PJ. The brain exocyst complex interacts with RalA in a GTP-dependent manner: identification of a novel mammalian Sec3 gene and a second Sec15 gene. J Biol Chem. 2001;276:29792–29797. doi: 10.1074/jbc.C100320200. [DOI] [PubMed] [Google Scholar]
- 13.Boutros R, Bailey AM, Wilson SH, Byrne JA. Alternative splicing as a mechanism for regulating 14-3-3 binding: interactions between hD53 (TPD52L1) and 14-3-3 proteins. J Mol Biol. 2003;332:675–687. doi: 10.1016/s0022-2836(03)00944-6. [DOI] [PubMed] [Google Scholar]
- 14.Sánchez-Pulido L, Martín-Belmonte F, Valencia A, Alonso MA. MARVEL: a conserved domain involved in membrane apposition events. Trends Biochem Sci. 2002;27:599–601. doi: 10.1016/s0968-0004(02)02229-6. [DOI] [PubMed] [Google Scholar]
- 15.Yaffe Y, Shepshelovitch J, Nevo-Yassaf I, Yeheskel A, Shmerling H, Kwiatek JM, Gaus K, Pasmanik-Chor M, Hirschberg K. The MARVEL transmembrane motif of occludin mediates oligomerization and targeting to the basolateral surface in epithelia. J Cell Sci. 2012;125:3545–3556. doi: 10.1242/jcs.100289. [DOI] [PubMed] [Google Scholar]
- 16.Zhang Y, Deng Q, Barbieri JT. Intracellular localization of type III-delivered Pseudomonas ExoS with endosome vesicles. J Biol Chem. 2007;282:13022–13032. doi: 10.1074/jbc.M606305200. [DOI] [PubMed] [Google Scholar]
- 17.Sonoda Y, Warita M, Suzuki T, Ozawa H, Fukuda Y, Funakoshi-Tago M, Kasahara T. Proteolipid protein 2 is associated with melanoma metastasis. Oncol Rep. 2010;23:371–376. [PubMed] [Google Scholar]
- 18.Iacobuzio-Donahue CA, Argani P, Hempen PM, Jones J, Kern SE. The desmoplastic response to infiltrating breast carcinoma: gene expression at the site of primary invasion and implications for comparisons between tumor types. Cancer Res. 2002;62:5351–5357. [PubMed] [Google Scholar]
- 19.Petalidis LP, Oulas A, Backlund M, Wayland MT, Liu L, Plant K, Happerfield L, Freeman TC, Poirazi P, Collins VP. Improved grading and survival prediction of human astrocytic brain tumors by artificial neural network analysis of gene expression microarray data. Mol Cancer Ther. 2008;7:1013–1024. doi: 10.1158/1535-7163.MCT-07-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colman H, Zhang L, Sulman EP, McDonald JM, Shooshtari NL, Rivera A, Popoff S, Nutt CL, Louis DN, Cairncross JG, Gilbert MR, Phillips HS, Mehta MP, Chakravarti A, Pelloski CE, Bhat K, Feuerstein BG, Jenkins RB, Aldape K. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12:49–57. doi: 10.1093/neuonc/nop007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang PS, Yin PH, Tseng LM, Yang CH, Hsu CY, Lee MY, Horng CF, Chi CW. Rab5A is associated with axillary lymph node metastasis in breast cancer patients. Cancer Sci. 2011;102:2172–2178. doi: 10.1111/j.1349-7006.2011.02089.x. [DOI] [PubMed] [Google Scholar]
- 22.Zhu G, Liu J, Terzyan S, Zhai P, Li G, Zhang XC. High resolution crystal structures of human Rab5a and five mutants with substitutions in the catalytically important phosphate-binding loop. J Biol Chem. 2003;278:2452–2460. doi: 10.1074/jbc.M211042200. [DOI] [PubMed] [Google Scholar]
- 23.Thomas DD, Martin CL, Weng N, Byrne JA, Groblewski GE. Tumor protein D52 expression and Ca2+-dependent phosphorylation modulates lysosomal membrane protein trafficking to the plasma membrane. Am J Physiol Cell Physiol. 2010;298:C725–C739. doi: 10.1152/ajpcell.00455.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]