Abstract
Genome sequencing efforts have identified many uncharacterized lipase/esterase enzymes that have potential to be drug targets for metabolic diseases such as obesity, diabetes, and atherosclerosis. However, sequence information and associated structural predictions provide only a loose framework for linking enzyme function to disease risk. We are now confronted with the challenge of functionally annotating a large number of uncharacterized lipases, with the goal of generating new therapies for metabolic diseases. This daunting challenge involves not gathering sequence-driven predictions, but more importantly structural, biochemical (substrates and products), and physiological data. At the center of such drug discovery efforts is the accurate identification of physiologically relevant substrates and products of individual lipases, and determining whether newly identified substrates/products can modulate disease in appropriate preclinical animal model systems. This review describes the importance of coupling in vivo metabolite profiling to in vitro enzymology as a powerful means to assign lipase function in disease specific contexts using animal models. In particular, we highlight recent examples using this multidisciplinary approach to functionally annotate genes within the α/β hydrolase fold domain (ABHD) family of enzymes. These new discoveries within the ABHD enzyme family serve as powerful examples of linking novel lipase function to human disease.
Keywords: ABHD3, ABHD6, ABHD12, Endocannabinoid, Enzymology, Lipase, Lysophospholipase, Mass Spectrometry
Introduction
In the post-genomic era we are presented with thousands of proteins whose functions have been predicted based only on computer modeling. However, the true function of most proteins identified by sequencing efforts remains elusive. In fact, whole families of proteins remain to be annotated and probed as potential drug targets to ameliorate disease. Among these are a plethora of lipid-metabolizing enzymes, which are very attractive candidates due to the profound role that lipids play in human physiology. Abnormal lipid metabolism and lipid signaling underlies many human diseases [1–3]. In fact, diabetes, obesity, liver disease, and atherosclerosis are predicted to plague us in the 21st century, and can be attributed almost exclusively to alterations in lipid enzymatic processes [4–7]. Furthermore, many forms of cancer, Alzheimer’s disease, and allergic diseases are now appreciated to be “metabolic” diseases driven by alterations in lipid metabolism and lipid signaling [8–11]. Therefore, it is imperative that we screen at the biochemical and physiological level various classes of lipid-metabolizing enzymes with the potential to become therapeutic targets. The functional annotation of uncharacterized lipid modifying enzymes holds great promise for identifying new drug targets for a number of key chronic diseases.
At the center of such drug discovery efforts is the accurate identification of physiologically relevant substrates and products of individual lipid modifying enzymes, and determining whether newly identified substrates/products can modulate disease in appropriate preclinical animal model systems. A certain knowledge base is needed as we move forward to effectively annotate uncharacterized lipases for disease prevention. Now that we have accurate sequence information and some structural predictions for most families of enzymes, the challenge now becomes to identify physiologically relevant substrates and products of lipid modifying enzymes, and create enzyme inhibitors and loss-of-function mouse models to interrogate the role of individual enzymes in appropriate disease-specific context.
In this review we describe synergistic biological and chemical methods to annotate novel lipase function. Using this multidisciplinary step-wise approach, a number of unannotated enzymes have rapidly become fully characterized. This review focuses on the importance of in vivo metabolite profiling as a powerful means to assign enzyme function in disease specific contexts. However, given that in vivo metabolite profiling methods have been eloquently described in other recent reviews [12,13], we instead focus on the approaches of integrating metabolic profiling data with synergistic biochemical and physiological approaches to gain a full picture of enzyme function. In particular, we highlight recent examples using this step-wise approach to functionally annotate genes within the α/β hydrolase fold domain (ABHD) family of enzymes, a newly described class of lysopholipases/phospholipases. These new discoveries within the ABHD enzyme family serve as powerful examples of linking novel lipase function to human disease, and provide a straightforward methodological approach to quickly annotating enzyme function.
In Vivo Metabolite Profiling Annotates ABHD3 as a Novel Enzymatic Regulator of Medium Chain Phospholipids
Using methods described essentially in Figure 1, the poorly characterized ABHD enzyme family member ABHD3 was recently functionally annotated by Long and colleagues [14]. Prior to this recent report there was essentially nothing known about the biochemical or physiological function of ABHD3, with only a handful of previous descriptive papers in the scientific literature [15–21]. The recent work by Long and colleagues [14] serves as a key example of how combining in vivo metabolite profiling with lipid enzymology can quickly provide new clues into enzyme function. ABHD3, also known as lung alpha/beta hydrolase 3 (LABH3) had previously been shown to be transcriptionally upregulated in some cancer models and downregulated in others [17–21], but the true biochemical function of ABHD3 remained elusive. Starting with an in vivo metabolic profiling approach, the authors demonstrated that lysates from cells overexpressing ABHD3 had elevated phospholipase activity (predominately PLA1 but some PLA2) toward myristate (C14)-containing phosphatidylcholine (C14-PCs) [14]. Importantly, cells overexpressing a catalytically dead serine mutant ABHD3 (ABHD3-S220A) did not display elevated phospholipase activity [14]. In agreement, tissue metabolomics of ABHD3 knockout (ABHD3−/−) mice showed elevated levels of C14-phosphatidylcholines (PCs) and other C14-phospholipids [14]. Support for this biochemical function has also been reported in human plasma with significant associations between ABHD3 polymorphisms and the molar percentage of phosphatidylcholine species [22]. In addition to its preference towards C14-PC and other shorter-chain PCs, ABHD3 can also hydrolyze PCs under conditions of oxidative stress [14]. This could be important in disease states such as atherosclerosis. Previous research has identified increased LPC levels within low density lipoproteins (LDL) during oxidation, which are associated with increased risk of atherosclerosis as well as acute and chronic inflammatory states [23–25]. Recent research also suggests that LPC can stimulate an increase in glucose-stimulated insulin secretion in pancreatic beta cells [26]. Therefore further characterization of the ABHD3−/− mice as well as development of ABHD3 inhibitors will be important to examine the signaling function of ABHD3 substrate lipids in appropriate disease specific contexts. Now that we know that short chain phosphatidylcholine species are physiologically relevant substrates of ABHD3 [14], we possess key information to move forward in defining the role that ABHD3 may play in atherosclerosis and other metabolic diseases.
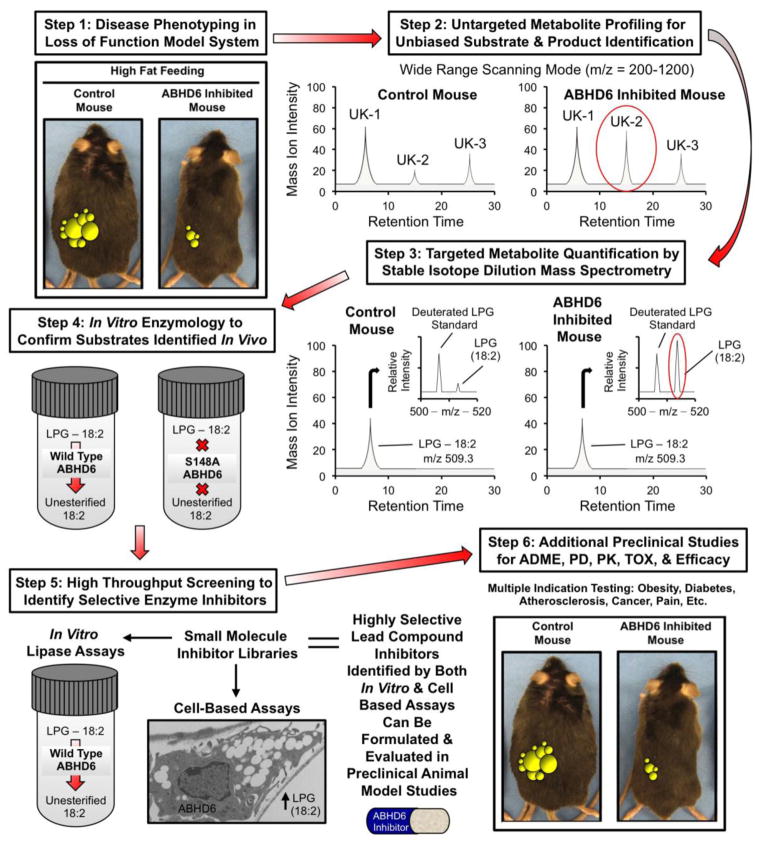
Fig. 1.
Synergistic biological and chemical methods to annotate novel lipase function. This diagram highlights the recent successful implementation of this stepwise process to annotate the previously uncharacterized lysophospholipase ABHD6. In step 1, a loss of function mouse model provides initial clues on whether enzyme inhibition would be therapeutically beneficial. For ABHD6 drug discovery, an antisense oligonucleotide (ASO) inhibition approach was taken, but knockout mouse models are the ideal screening tool. If experiments in step 1 provide support for further inquiry (ABHD6 inhibition protects against obesity, hepatic steatosis, and insulin resistance in high fat diet fed animals), steps 2 and 3 can be initiated using tissues or blood from the mice used in step 1. In step 2, lipid extracts from sources where the enzyme is abundantly expressed are unbiasedly analyzed using an untargeted LC-MS approach, where many unknown (UK-1, UK-2, and UK-3) metabolites are identified in a wide range (m/z 200–1200) scanning mode without the addition of an internal standards. The levels of unknown lipids are quantified by comparing the mass ion intensities between control vs. loss of function models. In step 3, lipid extracts from tissues or blood (where enzyme is abundantly expressed) are analyzed using a targeted quantitative approach where suspected lipid substrates are analyzed using stable isotope dilution LC-MS approaches. In both steps 2 and 3, potential enzyme substrates are expected to accumulate in loss of function mouse models, so these lipids (UK-2 or LPG-18:2) can move forward toward biochemical verification. In Step 4, the data generated in steps 2 and 3 provide clues for potential lipase substrates, and here potential substrate lipids are incubated in vitro with the purified enzyme to assay lipase activity in a reduced system. To ensure activity is intrinsic to the enzyme itself and not contaminants from the purified protein source, the same substrates are assayed with an enzyme that lacks key active site residues necessary for catalysis (S148A ABHD6). In step 5, once both in vitro and cell-based assays are developed to estimate enzyme activity; these can be used to screen small molecule inhibitor libraries to identify highly selective and biologically active lipase inhibitors. Finally, in step 6 lead small molecule inhibitors can be further evaluated for multiple indications in preclinical studies to examine ADME (absorption, distribution, metabolism, and excretion), PD (pharmacodynamics), PK (pharmacokinetics), TOX (toxicology), and efficacy.
ABHD6 has Dual Enzymatic Functions as a Monoacylglycerol Lipase and a Lysophospholipase
Another recent success story using methods described in Figure 1 to annotate enzyme function is illustrated by the recent identification of ABHD6 as a dual monoacylglycerol lipase and lysophospholipase promoting the metabolic syndrome [27]. Prior to this recent report there were only a few reports describing ABHD6 as a monoacylglycerol lipase hydrolyzing the key endocannabinoid lipid 2-arachidonoylglycerol in the brain [28–30]. However, ABHD6’s biochemical and physiological function in peripheral tissues had not been characterized. To address this, our group utilized targeted antisense oligonucleotides (ASOs) to selectively knock down ABHD6 in peripheral tissues in order to identify in vivo substrates and understand ABHD6’s role in energy metabolism (Fig. 1). Importantly, these studies demonstrated for the first time that selective knockdown of ABHD6 in metabolic tissues protects mice from high-fat-diet-induced obesity, hepatic steatosis, and systemic insulin resistance [27]. This protection against metabolic disease was attributed in part to decreased rates of de novo lipogenesis in the liver, increased energy expenditure, and increased physical activity levels [27]. Along with defining ABHD6’s role in metabolic disease, we were able to identify physiologically relevant substrates and products for ABHD6 using a metabolic profiling approach (Fig. 1). Based on the expectation that physiologically relevant substrates should accumulate when ABHD6 enzyme function is inhibited, we performed both unbiased and targeted lipidomics to identify potential substrate lipids. Interestingly, a number of phospholipids and lysophospholipids accumulated to varying degrees in ABHD6 ASO-treated livers, implicating them as potential physiologically relevant substrates [27]. ABHD6 knockdown significantly increased the hepatic levels of a number of phospholipids and lysophospholipids, with the most prominent accumulation seen in nearly all molecular species of phosphatidylglycerol and lysophosphatidylglycerol [27]. We subsequently tested whether ABHD6 can hydrolyze phospholipid and lysophospholipid substrates in vitro using recombinant ABHD6. Recombinant ABHD6 showed considerable lipase activity toward several lysophospholipids including lysophosphatidylglycerol, lysophosphatidylserine, lysophosphatidic acid, and lysophosphatidylethanolamine, but not lysophosphatidylcholine [27]. In contrast, ABHD6 exhibited no lipase activity against other major phospholipid classes [27]. This newly identified lysophospholipase activity for ABHD6 was indeed intrinsic to the recombinant enzyme, given that mutation of the active site serine abolished all lysophospholipase activity [27]. Taken together, these observations suggest that ABHD6 can act both as a monoacylglycerol lipase and lysophospholipase exhibiting a preference for LPG among the tested lysophospholipids. In further studies, we were able to demonstrate that a small molecule inhibitor of ABHD6 (WWL-70) likewise improved the metabolic disturbances driven by high fat feeding [27]. This indicates that further development of both small molecule inhibitors as well as ASO therapeutics hold promise as therapeutics for metabolic disease. However, given ABHD6’s dual role of hydrolyzing the endocannabinoid lipid 2-arachidonoylglycerol in the brain and hydrolyzing key lysophospholipids in the liver, tissue-selective inhibitors may be needed to avoid hyperactivation of the central endocannabinoind system while still providing metabolic benefit in the periphery. Collectively, these studies using combined in vivo lipidomic identification and in vitro enzymology approaches (Fig. 1) show that ABHD6 can hydrolyze both monoacylglycerols as well as lysophospholipids, positioning ABHD6 at the interface of glycerophospholipid metabolism and lipid signal transduction. Further, this work suggest that ABHD6 inhibition may be effective in preventing obesity, nonalcoholic fatty liver disease, and type II diabetes.
In Vivo Metabolite Profiling Annotates ABHD12 as a Dual Monoacylglycerol and Lysophosphatidylserine Lipase Regulating Microglial Activation and PHARC Progression
In another powerful example of coupling in vivo lipidomic identification and in vitro enzymology approaches (Fig. 1), the uncharacterized lipase ABHD12 was recently biochemically and physiologically annotated. Like ABHD6, ABHD12 had previously been shown to hydrolyze the endocannabinoid lipid 2-arachidonylglycerol in vitro [28–30], but true physiologically relevant substrates had not been identified until the elegant recent work of Blankman and colleagues [31]. Prior to this important study, mutations in ABHD12 had been shown to be the cause of a neurodegenerative disease called polyneuropathy, hearing loss, ataxia, retinitis pigmentosa and cataract (PHARC) in humans [32–34]. It was originally thought ABHD12’s enzymatic ability to hydrolyze 2-arachidonylglycerol may contribute to the development of PHARC, but a recent metabolic profiling study has identified a novel role for ABHD12 acting as a lysophospholipase to regulate the progression of PHARC [31]. By coupling both untargeted and targeted lipidomic approaches in ABHD12 knockout (ABHD12−/−) mice, Blankman and colleagues uncovered a massive accumulation of very long chain lysophosphatidylserine lipids in the brain [31]. Using a complimentary biochemical approach, the authors showed that recombinant ABHD12 has robust intrinsic lysophosphatidylserine lipase activity [31]. Interestingly, the accumulation of lysophosphatidylserine species in the brains of ABHD12−/− mice preceded the development of age-dependent microglial activation and behavior phenotypes that resemble phenotypes seen in PHARC patients [31]. Collectively, this important set of studies demonstrates that ABHD12’s ability to hydrolyze lysophosphatidylserine lipids in brain lies at the center of the human disease PHARC [31]. Collectively, the accumulation of lysophosphatidylserine seen with ABHD12 mutations subsequently causes hyperactivation of proinflammatory signals [31]. This hyperactivation drives chronic microglial activation and associated behavioral abnormalities [31]. ABHD12 may also be involved in other disease states in addition to PHARC. Notably, a genome-wide association study looking at concentrations of liver enzymes in plasma, which is a widely used indicator of liver disease, found ABHD12 to be an associated candidate gene [35]. A deeper look into ABHD12’s shared activity with ABHD6 as a 2-AG hydrolase in the brain and in peripheral tissues may uncover the contributions of 2-AG or alternative substrates of ABHD12 to metabolic disease.. Looking forward, we believe that utilizing a similar in vivo loss-of-function approach may prove useful for mapping natural enzyme-substrate relationships for other uncharacterized enzymes of lipid metabolism.
Looking Ahead: Functional Annotation of Other Uncharacterized Lipases
Along with the ABHD family, there are more than 200 serine hydrolases present in the human genome. Many of these members function as proteases whereas others are considered “metabolic” and can function as esterases, amide hydrolases, or thioesterases [36]. Within the groups of small-molecule hydrolases, extracellular neutral lipid lipases, phospholipase A2 enzymes, peptidases, protein- and glycan-modifying hydrolases, and other phospholipases several remain understudied [36]. Members of these groups may share similar abilities to hydrolyze or synthesize specific substrates that play important roles in human diseases. Within the ABHD family, several members whose substrates have been identified seem to share the ability to regulate glycerophospholipid metabolism, and may also play a role in lipid and energy metabolism [37]. The methods used to study the ABHD family shown in Figure 1 have been highly successful in annotating the activity of several members, while also identifying substrates and linking them to human diseases of altered lipid metabolism. We believe that use of the multidisciplinary approach described here (Fig. 1) will prove useful for mapping natural enzyme-substrate relationships for the other uncharacterized enzymes. However, as shown in Figure 1, it remains critical that we develop good enzyme inhibitors or knockout mouse models to support the process of interrogation. Therefore, continued efforts to generate a complete mouse knockout repository [38,39] and enzyme specific inhibitors [40–41] will be extremely important as we move forward in functionally annotating new enzymes. It is truly an exciting time in drug discovery where we have access to the complete human genome, and have the knowledge and tools to interrogate the complexity of gene and protein function. Over the last few decades we have significantly improved methodological approaches, especially in the field of mass spectrometry that will allow us to now forge ahead to functionally annotate new enzymes relevant to human disease.
Highlights.
Uncharacterized lipases have potential to be drug targets for metabolic diseases.
In vivo metabolite profiling studies are key to functionally annotating new lipases.
Recent annotation of α/β-hydrolase domain (ABHD) provide a methodological framework.
Acknowledgments
This work was supported in part by the National Heart, Lung, and Blood Institute (R00-HL096166 to J.M.B and T32-HL091797 to G.T.) and the American Heart Association (11BGIA-7840072 to J.M.B and 13GRNT17050074 to J.M.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Zechner R, Zimmermann R, Eichmann TO, Kohlwein SD, Haemmerle G, Lass A, Madeo F. FAT SIGNALS – lipases and lipolysis in lipid metabolism and signaling. Cell Metab. 2012;15:279–291. doi: 10.1016/j.cmet.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wymann MP, Schneiter Lipid signaling in disease. Nat Rev Mol Cell Biol. 2008;9:162–176. doi: 10.1038/nrm2335. [DOI] [PubMed] [Google Scholar]
- 3.Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J Biol Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- 4.Unger RH, Scherer PE. Gluttony, sloth and the metabolic syndrome: a roadmap to lipotoxicity. Trends Endocrinol Metab. 2010;21:345–352. doi: 10.1016/j.tem.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cohen JC, Horton JD, Hobbs HH. Human fatty liver disease: old questions and new insights. Science. 2011;332:1519–1523. doi: 10.1126/science.1204265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samuel VT, Shulman GI. Mechanisms for insulin resistance: common threads and missing links. Cell. 2012;148:852–871. doi: 10.1016/j.cell.2012.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steinberg D. Thematic review series: the pathogenesis of atherosclerosis. An interpretive history of the cholesterol controversy. Part V: the discovery of statins and the end of the controversy. J Lipid Res. 2006;47:1339–1351. doi: 10.1194/jlr.R600009-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Currie E, Schulze A, Zechner R, Walther TC, Farese RV., Jr Cellular fatty acid metabolism and cancer. Cell Metab. 2013;18:153–161. doi: 10.1016/j.cmet.2013.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBerardinis RJ, Lum JJ, Hatzivassilou G, Thompson CB. The biology of cacner: metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Liu CC, Kanekiyo T, Xu H, Bu G. Apolipoprotein E and Alzheimer disease: risk, mechanisms and therapy. Nat Rev Neurol. 2013;9:106–118. doi: 10.1038/nrneurol.2012.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fanning LB, Boyce JA. Lipid mediators and allergic disease. Ann Allergy Asthma Immunol. 2013;111:155–162. doi: 10.1016/j.anai.2013.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saghatelian A, Cravatt BF. Assignment of protein function in the postgenomic era. Nat Chem Biol. 2005;1:130–142. doi: 10.1038/nchembio0805-130. [DOI] [PubMed] [Google Scholar]
- 13.Want EJ, Cravatt BF, Siuzdak G. The expanding role of mass spectrometry in metabolite profiling and characterization. Chembiochem. 2005;6:1941–1951. doi: 10.1002/cbic.200500151. [DOI] [PubMed] [Google Scholar]
- 14.Long JZ, Cisar JS, Milliken D, Niessen S, Wang C, Trauger SA, Siuzdak G, Cravatt BF. Metabolomics annotates ABHD3 as a physiologic regulator of medium-chain phospholipids. Nat Chem Biol. 2012;7:763–765. doi: 10.1038/nchembio.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernandez-Becker NQ, Moss AC. In silico analysis of T-bet activity in peripheral blood mononuclear cells in patients with inflammatory bowel disease (IBD) In Silico Biol. 2009;9:355–363. doi: 10.3233/ISB-2009-0410. [DOI] [PubMed] [Google Scholar]
- 16.Edgar AJ, Polak JM. Cloning and tissue distribution of three murine alpha/beta hydrolase fold protein cDNAs. Biochem Biophys Res Commun. 2002;292:617–625. doi: 10.1006/bbrc.2002.6692. [DOI] [PubMed] [Google Scholar]
- 17.L'Esperance S, Bachvarova M, Tetu B, Mes-Masson AM, Bachvarov D. Global gene expression analysis of early response to chemotherapy treatment in ovarian cancer spheroids. BMC Genomics. 2008;9:99. doi: 10.1186/1471-2164-9-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin B, Huntley D, Abuali G, Langley SR, Sindelar G, Petretto E, Butcher S, Grimm S. Determining signalling nodes for apoptosis by a genetic high-throughput screen. PLoS One. 2011;6:e25023. doi: 10.1371/journal.pone.0025023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Rechem C, Rood BR, Touka M, Pinte S, Jenal M, Guerardel C, Ramsey K, Monte D, Begue A, Tschan MP, Stephan DA, Leprince D. Scavenger chemokine (CXC motif) receptor 7 (CXCR7) is a direct target gene of HIC1 (hypermethylated in cancer 1) J Biol Chem. 2009;284:20927–20935. doi: 10.1074/jbc.M109.022350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez-Becker NQ, Moss AC. In silico Analysis of T-bet Activity in Peripheral Blood Mononuclear Cells in Patients with Inflammatory Bowel Disease (IBD) In Silico Biol. 2009;9:355–363. doi: 10.3233/ISB-2009-0410. [DOI] [PubMed] [Google Scholar]
- 21.Johnson EC, Doser TA, Cepurna WO, Dyck JA, Jia L, Guo Y, Lambert WS, Morrison JC. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52:504–518. doi: 10.1167/iovs.10-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson A, Rudan I, Aulchenko YS, Kirichenko AV, Janssens AC, Jansen RC, Gnewuch C, Domingues FS, Pattaro C, Wild SH, Jonasson I, Polasek O, Zorkoltseva IV, Hofman A, Karssen LC, Struchalin M, Floyd J, Igl W, Biloglav Z, Broer L, Pfeufer A, Pichler I, Campbell S, Zaboli G, Kolcic I, Rivadeneira F, Huffman J, Hastie ND, Uitterlinden A, Franke L, Franklin CS, Vitart V, Nelson CP, Preuss M, Bis JC, O'Donnell CJ, Franceschini N, Witteman JC, Axenovich T, Oostra BA, Meitinger T, Hicks AA, Hayward C, Wright AF, Gyllensten U, Campbell H, Schmitz G. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmitz G, Ruebsaamen K. Metabolism and atherogenic disease association of lysophosphatidylcholine. Atherosclerosis. 2010;208:10–18. doi: 10.1016/j.atherosclerosis.2009.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Makide K, Kitamura H, Sato Y, Okutani M, Aoki J. Emerging lysophospholipid mediators, lysophosphatidylserine, lysophosphatidylthreonine, lysophosphatidylethanolamine and lysophosphatidylglycerol. Prostaglandins Other Lipid Mediat. 2009;89:135–139. doi: 10.1016/j.prostaglandins.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Grzelczyk A, Dengaszewska-Darmach E. Novel bioactive glycerol-based lysophospholipids: New data – New insight into their function. Biochimie. 2013;95:667–679. doi: 10.1016/j.biochi.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Soga T, Ohishi T, Matsui T, Saito T, Matsumoto M, Takasaki J, Matsumoto S, Kamohara M, Hiyama H, Yoshida S, Momose K, Ueda Y, Matsushime H, Kobori M, Furuichi K. Lysophosphatidylcholine enhances glucose-dependent insulin secretion via an orphan G protein-coupled receptor. Biochem Biophys Res Commum. 2005;326:744–751. doi: 10.1016/j.bbrc.2004.11.120. [DOI] [PubMed] [Google Scholar]
- 27.Thomas G, Betters JL, Lord CC, Brown AL, Marshall S, Ferguson D, Sawyer J, Davis MA, Melchoir J, Blume LC, Howlett AC, Ivanova PT, Milne SB, Myers DS, Mrak I, Leber V, Heier C, Taschler U, Blankman J, Cravatt BF, Lee RG, Crooke RM, Graham MJ, Zimmermann R, Brown HA, Brown JM. The Serine Hydrolase ABHD6 is a Critical Regulator of the Metabolic Syndrome. Cell Rep. 2013;5:508–520. doi: 10.1016/j.celrep.2013.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blankman JL, Simon GM, Cravatt BF. A Comprehensive Profile of Brain Enzymes that Hydrolyze the Endocannabinoid 2-Arachidonoylglycerol. Chem Biol. 2007;14:1347–1356. doi: 10.1016/j.chembiol.2007.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marrs WR, Blankman JL, Horne EA, Thomazeau A, Lin YH, Coy J, Bodor AL, Muccioli GG, Hu SSJ, Woodruff G, Fung S, Lafourcade M, Alexander JP, Long JZ, Li W, Xu C, Möller T, Mackie K, Manzoni OJ, Cravatt BF, Stella N. The serine hydrolase ABHD6 controls the accumulation and efficacy of 2-AG at cannabinoid receptors. Nat Neurosci. 2010;13:951–957. doi: 10.1038/nn.2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Navia-Paldanius D, Savinainen JR, Laitinen JT. Biochemical and pharmacological characterization of human α/β-hydrolase domain containing 6 (ABHD6) and 12 (ABHD12) J Lipid Res. 2012;53:2413–2424. doi: 10.1194/jlr.M030411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Blankman JL, Long JZ, Trauger SA, Siuzdak G, Cravatt BF. ABHD12 controls brain lysophosphatidylserine pathways that are deregulated in a murine model of the neurodegenerative disease PHARC. PNAS. 2013;110:1500–1505. doi: 10.1073/pnas.1217121110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fiskerstrand T, H’mida-Ben Brahim D, Johansson S, M’zahem A, Haukanes BI, Drouot N, Zimmermann J, Cole AJ, Vedeler C, Bredrup C, Assoum M, Tazir M, Klockgether T, Hamri A, Steen VM, Boman H, Bindoff LA, Koenig M, Knappskog PM. Mutations in ABHD12 Cause the Neurodegenerative Disease PHARC: An Inborn Error of Endocannabinoid Metabolism. American Journal of Human Genetics. 2010;87:410–417. doi: 10.1016/j.ajhg.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eisenberger T, Slim R, Mansour A, Nauck M, Numberg G, Decker C, Dafinger C, Ebermann I, Bergmann C, Boltz HJ. Targeted next-generation sequencing identifies a homozygous nonsense mutations in ABHD12, the gene underlying PHARC in a family clincally diagnosed with Usher syndrome type 3. Orphanet J Rare Dis. 2012;7:59. doi: 10.1186/1750-1172-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen DH, Naydenov A, Blankman JL, Mefford HC, Davis M, Sul Y, Barloon AS, Bonkowski E, Wolff J, Matsushita M, Smith C, Cravatt BF, Mackie K, Raskind WH, Stella N, Bird TD. Two novel mutations in ABHD12: expansion of the mutation spectrum in PHARC and Assessment of their functional effects. Hum Mutat. doi: 10.1002/humu.22437. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers JC, Zhang W, Sehmi J, Li X, Wass MN, Van der Harst P, Hold H, Sanna S, Kavousi M, Baumeister SE, Coin LJ, Deng G, Geiger C, Heard-Costa NL, Hottenga JJ, Kühnel B, Kumar V, Lagou V, Liang L, Luan J, Vidal PM, Leach IM, O’Reilly PF, Peden JF, Rahmioglu N, Soininen P, Speliotes EK, Yuan X, Thorleifsson G, Alizadeh BZ, Atwood LD, Borecki IB, Brown MJ, Charoen P, Cucca F, Das D, de Geus EJC, Dixon AL, Döring A, Ehret G, Eyjolfsson GI, Farrall M, Forouhi NG, Friedrich N, Goessling W, Gudbjartsson DF, Harris TB, Hartikainen AL, Heath S, Hirschfield GM, Hofman A, Homuth G, Hyppönen E, Jansses HLA, Johnson T, Kangas AJ, Kema IP, Kühn JP, Lai S, Lathrop M, Lerch MM, Li Y, Liang TJ, Lin J, Floos RJ, Martin NG, Moffatt MF, Montgomery GW, Monroe PB, Musunuru K, Nakamura Y, O’Donnell CJ, Olafsson I, Penninx BW, Pouta A, Prins BP, Prokopenko I, Puls R, Ruokonen A, Savolainen MJ, Schlessinger D, Schouten JNL, Seedorf U, Sen-Chowdry S, Siminovitch KA, Schmidt JH, Spector TD, Tan W, Teslovich TM, Tukianen T, Uitterlinden AG, Van der Klauw MM, Vasan RS, Wallace C, Wallaschofski H, Wichmann HE, Willemsen G, ürtz PW, Xu C, Yerges-Armstrong LM, Abecasis GR, Ahmadi KR, Boomsma DI, Caulfield M, Cookson WO, van Duijn CM, Froguel P, Matsuda K, McCarthy MI, Meisinger C, Mooser V, Pietiläinen KH, Schumann G, Snieder H, Sternberg MJE, Stolk RP, Thomas HC, Thorsteinsdottir U, da MU, Waeber G, Wareham NJ, Waterworth DM, Watkins H, Whitfield JB, Witteman JCM, Wolffenbuttel BHR, Fox CS, Ala-Korpela M, Stefansson K, Vollenweider P, Völzke H, Schadt EE, Scott J, Järvelin MR, Elliott P, Skooner J Alcohol Genome-wide Association (AlcGen) Consortium Diabetes Genetics Replication and Meta-analyses (DIAGRAM+) Study Genetic Investigation of Anthropometric Trains (GIANT) Consortium Global Lipids Genetics Consortium Genetics of Liver Disease (GOLD) Consortium International Consortium for Blood Pressure (ICBP-GWAS) Meta-analyses of Glucose and Insulin-Related Traits Consortium (MAGIC) Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43:1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Long JZ, Cravatt BF. The Metabolic Serine Hydrolases and Their Functions in Mammalian Physiology and Disease. Chem Rev. 2011;111:6022–6063. doi: 10.1021/cr200075y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lord CC, Thomas G, Brown JM. Mammalian alpha beta hydrolase domain (ABHD) proteins: Lipid metabolizing enzymes at the interface of cell signaling and energy metabolism. Biochem Biophys Acta. 2013;1831:792–802. doi: 10.1016/j.bbalip.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guan C, Ye C, Yang X, Gao J. A review of current large-scale mouse knockout efforts. Genesis. 2010;48:73–85. doi: 10.1002/dvg.20594. [DOI] [PubMed] [Google Scholar]
- 39.Schofield PN, Hoehndorf R, Gkoutos GV. Mouse genetic and phenotypic resources for human genetics. Hum Mutat. 2012;33:826–836. doi: 10.1002/humu.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hsu KL, Tsuboi K, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Optimization and characterization of triazole urea inhibitors for abhydrolase domain containing protein 6 (ABHD6) Probe Reports from the NIH Molecular Libraries Program (2010–2012) [PubMed] [Google Scholar]
- 41.Adibekian A, Hsu KL, Speers AE, Brown SJ, Spicer T, Fernandez-Vega V, Ferguson J, Cravatt BF, Hodder P, Rosen H. Optimization and characterization of a triazole urea inhibitor for alpha/beta hydrolase domain-containing protein 11 (ABHD11): anti-probe for LYPLA1/LYPLA2 dual inhibitor ML211. Probe Reports from the NIH Molecular Libraries Program (2010–2012) [PubMed] [Google Scholar]