Abstract
Background
Conceptually, allergic responses may involve cross-reactivity by antibodies or T-cells. While IgE cross-reactivity amongst grass pollen allergens has been observed, cross-reactivity at the allergen-specific T-cell level has been less documented. Identification of the patterns of cross-reactivity may improve our understanding, allowing optimization of better immunotherapy strategies.
Objectives
We use Phleum pratense as model for the studying of cross-reactivity at the allergen-specific CD4+ T cell level amongst DR04:01 restricted Pooideae grass pollen T-cell epitopes.
Methods
After In vitro culture of blood mononucleated cells from Grass-pollen allergic subjects with specific Pooideae antigenic epitopes, dual tetramer staining with APC-labeled DR04:01/Phleum pratense tetramers and PE-labeled DR04:01/Pooideae grass homolog tetramers was assessed to identify cross-reactivity amongst allergen-specific DR04:01-restricted T-cells in 6 subjects. Direct ex vivo staining enabled the comparison of frequency and phenotype of different Pooideae grass pollen reactive T-cells. Intracellular cytokine staining (ICS) assays were also used to examine phenotypes of these T-cells.
Results
T-cells with various degree of cross reactive profiles could be detected. Poa p 1 97-116, Lol p 1 221-240, Lol p 5a 199-218, and Poa p 5a 199-218 were identified as minimally-cross-reactive T-cell epitopes that do not show cross reactivity to Phl p 1 and Phl p 5a epitopes. Ex vivo tetramer staining assays demonstrated T-cells that recognized these minimally-cross reactive T-cell epitopes are present in Grass-pollen allergic subjects.
Conclusions
Our results suggest that not all Pooideae grass epitopes with sequence homology are cross-reactive. Non-cross reactive T-cells with comparable frequency, phenotype and functionality to Phl p-specific T-cells, suggest that a multiple allergen system should be considered for immunotherapy instead of a mono allergen system.
Keywords: Cross-reactivity, T-cells, MHC class II tetramers, epitopes, grass-pollen, allergy, Pooideae, CD4+
INTRODUCTION
In atopic individuals exposure to allergens from taxonomically related species plays an important role in eliciting and maintaining clinical symptoms [1;2]. Structural similarities among proteins derived from these species allow humoral and cell mediated immunity to target homologous regions that originally did not serve as the source of sensitization [1;2]. An example is found amongst grass-pollens from the Pooideae subfamily. Pooideae-grasses coexist geographically and share pollination periods, therefore, allergic subjects in temperate zones are poly-exposed and poly-sensitized to multiple pollens from this subfamily [3-6]. Due to their taxonomical relatedness, high amino acid sequence homology is found within clinically relevant major allergens in different Pooideae species: 90% for Group 1 (Beta-expansins) and about 55-85% for Group 5 (Ribonucleases) allergens [2;7-10]. Thus, cross-reactivity for both humoral and T-cell responses can be expected amongst allergens of grass-pollens [2;7-10]. Phleum pratense (Timothy grass), has been accounted as an index species in this group because it exhibits the most dominant epitope profile [3;9;11]. Several investigators have suggested that immunotherapy with this species alone is sufficient to cover other species due to observed cross-reactivity at the IgE level [3;9;11]. On the other hand, it is now firmly established that allergen-specific T-cells play an important role in allergic inflammation [12] and that induction of antigen specific Treg or elimination of allergen-specific TH2 cells might be a prerequisite for the induction of specific tolerance [13]. Yet, evaluation of cross-reactivity at the T-cell level has been less documented. Some studies advocate that there are cross-reacting and non-cross-reacting T-cell epitopes for both major allergens [14;15]. In this study, we determined the patterns of cross-reactivity of CD4+ T-cells specific for homologous Pooideae-grass-pollen epitopes derived from Timothy grass against Kentucky, Orchard, Rye, Velvet, Barley and Canary grass. We determined whether grass-pollen allergic subjects that were diagnosed based upon IgE reactivity to Timothy grass pollen (TGP) extract were also sensitized to other related grass species at the T-cell level. The implications of our findings and the choices of using a single extract verses multiple extracts in immunotherapy will be discussed.
MATERIALS AND METHODS
Human Subjects
Subjects were recruited from the Virginia Mason Medical Center Allergy Clinic and Benaroya Research Institute. All subjects were recruited with informed consent and institutional review board approval (IRB title “Allergen and T cell reagent resources for the study of allergic diseases,” Approval number IRB7109.) A total of 6 DR04:01, 2 DR07:01 and 2 DRB5*01:01 grass-pollen (GP) allergic patients, diagnosed upon an ImmunoCAP score for TGP extract of ≥3 (Phadia AB, Uppsala, Sweden) were recruited. DNA samples were HLA-typed using Dynal Unitray™ SSP Kits (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The attributes of these human subjects are summarized in Supplementary Table 1.
Basophil stimulation tests
Basophil activation was measured as previously described [16]. Briefly, heparinized whole blood from TGP allergic subjects was incubated with pollen extract from different grass-species (2 μg/mL): Timothy grass (Phleum pratense, Phl p), Velvet grass (Holcus lanatus,Hol l), Canary grass (Phalaris aquatica,Pha a), Barley grass (Hordeum vulgare,Hor v), Rye grass (Lolium perenne,Lol p), Orchard grass (Dactylis glomerata,Dac g) and Kentucky grass (Poa pratensis,Poa p) (Greer Laboratories, USA, Pollen extracts are sterile solutions containing the extractables of pollens (20,000 BAU/mL), 0.5% Sodium Chloride, 0.275% Sodium Bicarbonate, and 50% Glycerin by volume as preservative. The content of group 1 and group 5 of the extracts can be found in [17], the extract contains isoforms for both groups), and simultaneously stained with anti-CD3(eBiosciences), anti-CD203(Beckman Coulter), anti-CRTH2(BD Biosciences) for 25 minutes at 37°C. Basophils were identified as CD3- CRTH2+ and activation status was assessed following the detection of CD203c in the presence of the allergens tested [16]. Whole blood stimulated with buffer without allergen and a mixture of grass-pollen extract were used as negative and positive controls, respectively.
Peptide binding assays
Peptide binding assays were measured as previously described [18]. Briefly, Non-biotinylated target peptides were incubated with DR04:01 protein at final concentrations ranging from 0.01 μM to 10 μM for 1 h at 37°C, followed by additional 16 h incubation in the presence of 0.01 μM biotinylated reference peptide (Influenza HA306-318, PKYVKQNTLKLAT). The binding reaction was stopped by adding an equal volume of 50 mM Tris-Cl buffer (pH 8.0). The DR04:01 molecules were then immobilized on 96-well plates coated with anti-HLA-DR monoclonal antibody (L243). The amount of biotinylated reference peptide-bound to DR04:01 was quantified using a europium streptavidin detection system on a Victor 2 microtiter plate reader (Perkin-Elmer, Waltham, MA). The concentrations of target peptides required to inhibit 50% of maximal biotinylated reference peptide binding were retrieved from regression curves fitted by a sigmoidal dose-response equation provided by Prism software (GraphPad, San Diego, CA). Relative binding was calculated using Phl p peptides as reference.
Dual tetramer staining
CD4+ T-cells (5 ×106) were stimulated for 2 weeks in vitro with homologous grass-pollen antigenic epitopes (20-mer for Group 1 or 13-mer for Group 5a), cultures were then co-stained with allophycocyanin (APC) conjugated pMHC II tetramers loaded with TGP-derived peptides(Phl p 1 or Phl p 5a peptides)and phycoerythrin (PE) labeled tetramer with homologous grass-pollen peptides at 37°C for 1 h. FITC-conjugated anti-CD4 (eBioscience) was then added to the cell suspension for a 20 minute incubation at 4°C. Cells were analyzed by flow cytometry. Data were analyzed utilizing FlowJo (Tree Star, Ashland, Ore); cells were gated on CD4+ and PE-tetramer+ subsets. The average of cross-reactive T-cells was calculated utilizing the percentage of co-stained T-cell populations divided by the total of tetramer+ stained T-cells. Tetramer+ T-cells showed three different cross-staining patterns: co-staining of greater than 85% was arbitrarily defined as full-scale cross-reactivity; T-cells that have 25 to 85% co-staining were designated as partially cross-reactive T-cells; and T-cells that showed co-staining of lower than 25% were designated as minimally cross-reactive T-cells. We defined epitope regions that elicit full-scale cross-reactivity as full-scale cross-reactive T-cell epitopes, partially cross-reactive T-cells as partially cross-reactive T-cell epitopes and minimally cross-reactive T-cells as minimally cross-reactive T-cell epitopes.
Ex vivo tetramer staining to determine the frequency of Phl p- and grass homolog-specific CD4+ T-cells
The frequency of Phl p 1- and Phl p 5a-specific T-cells was measured as previously described [12]. Briefly, 30 million PBMC in 200 μL T-cell culture medium were stained with 20μg/mL PE-labeled tetramers and/or APC-labeled tetramers (for ex vivo dual tetramer staining) for 100 minutes. Cells were then washed and incubated with anti-PE or anti-APC magnetic beads (Miltenyi Biotec) for 20 minutes at 4°C and a 1/100 fraction was saved for analysis; the other fraction was passed through a magnetic column (Miltenyi Biotec, Bergisch Gladbach, Germany). Bound PE- and/or APC-labeled cells were flushed and collected. Cells in the bound and precolumn fractions were stained with a panel of antibodies of interest for 20 minutes at room temperature. After staining, cells were stained with Via-probe+ (BD Biosciences) for 10 minutes at 4°C before flow-cytometry. Data were analyzed utilizing FlowJo (Tree Star, Ashland, Ore) gating on forward scatter/side scatter and excluding CD14+, CD19+ and Viaprobe populations. Frequency was calculated as previously described [12]. For phenotyping studies, antibodies were used against markers of interest; CCR4 (R&D systems), CD45RA (eBioscience) and CD38 (eBioscience).
Intracellular cytokine staining
For intracellular staining of IFN-ϒ and IL-4, PBMC were stimulated for 2 weeks with specific peptides and stained with the corresponding PE-labeled tetramers for 30 minutes at 37°C. Cells were restimulated with 50 ng/mL phorbol 12-myristate-13-acetate and 1 mg/mL ionomycin in the presence of 1x brefeldin-A (eBiosciences) for 5 hours at 37°C, 5% CO2. After restimulation, cells were stained with anti-CD4 (BD Biosciences) and anti-CD3 (eBioscience). After 10 minutes at room temperature, cells were then fixed with fixation buffer (eBioscience) and washed twice with a permeabilization buffer (eBioscience). Cells were then stained with a panel of antibodies directed against cytokines of interest, IFN-ϒ (Biolegend), IL-4 (eBioscience) for 20 minutes at room temperature; cells were washed and immediately analyzed by flow cytometry. Cells were gated on CD4 and PE-tetramer subsets.
RESULTS
GP allergic subjects have IgE sensitivity to different grass-species
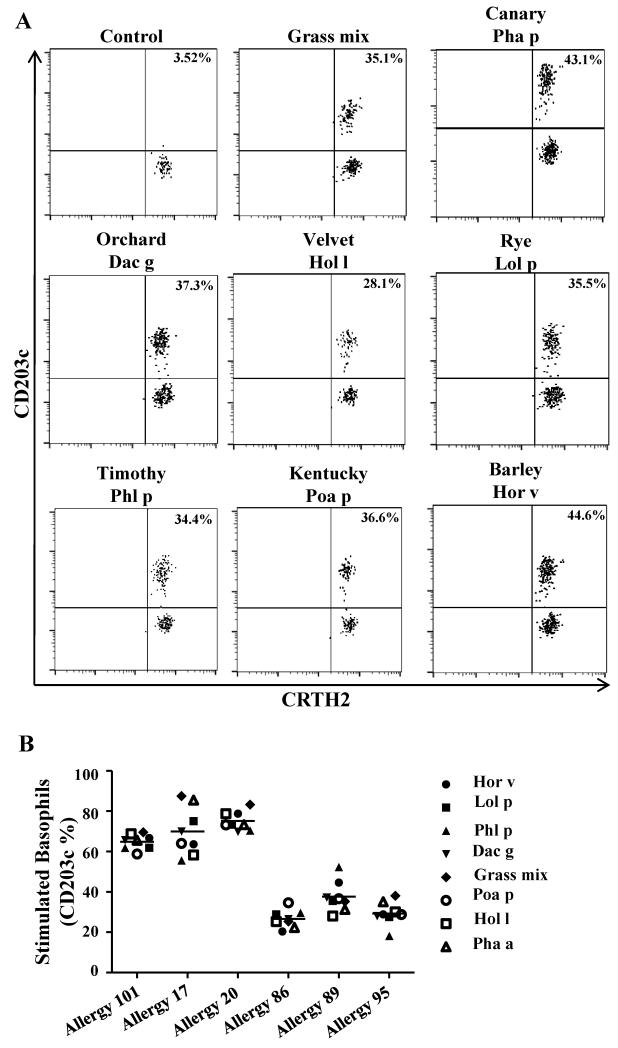
A total of 6 DR04:01 GP allergic subjects with an ImmunoCAP score of 3 or greater for TGP extract were recruited for this study (Supplementary Table 1). DR04:01 was selected since it is the most frequent MHC class II allele found in our cohort of GP allergic subjects. Subjects were subsequently evaluated for IgE reactivity to five additional Pooideae grass-pollen extracts by using a basophil activation assay. All GP allergic individuals tested exhibited reactivity to all five additional grass species (Figure 1). This confirms that allergic subjects sensitized to Phl p are also sensitized to other Pooideae species. However, due to significant homology at the conformational level among Pooideae allergens[2], the involvement of a specific grass species in symptom manifestation cannot be established.
Figure 1.
Subjects allergic to TGP have IgE sensitivity to different grass-species. Basophils were stimulated with pollen extracts from 6 different Pooideae species or a mix containing grass-pollen from all species (Greer Laboratories, USA) and were stained simultaneously with a panel of antibodies of interest. Upregulation of CD203c indicated activation of basophils. A. Representative result for Allergy #89. B. Percentages of CD203c on stimulated basophils are summarized for each DR04:01 allergic subject.
Pooideae grass-pollen specific CD4+ T-cells exhibit varying degrees of cross-reactivity: High sequence homology does not imply cross-reactivity.
Previous studies in our laboratory have identified Phl p 197-226 and Phl p 1221-240 (Phl p 1 is a Group 1 allergen) and Phl p 5a167-186 and Phl p 5a199-218 (Phl p 5a is a Group 5a allergen)as dominant DR04:01-restricted TGP TH2 T-cell epitopes [19]. Since major allergens from different Poodieae species share high amino acid sequence homology (90% homology for Group 1 and 55-85% for Group 5 allergens) [2], T-cell cross-reactivity amongst these different allergens may be present. Potential epitopes for other Pooideae species were identified by selecting sequences homologous to the previously reported TGP-derived epitopes using Blast alignments [20] (Table 1). As shown in Table 1, the majority of Group 1 and Group 5a homologs from the different Pooideae species bound DR04:01 with binding affinities similar to the previously verified TGP-derived epitopes. However, we observed that, Group 1 homolog Dac g 1 221-240 along with Group 5 homologs, Hol l 5a 199-218, Dac g 5a 199-218, and Poa p 5a 199-218 bound with much higher affinity than their Phl p counterpart (10 to 15 fold higher). This data suggests that all homologous peptides could be presented by DR04:01 as plausible DR04:01 restricted T-cell epitopes.
Table 1.
Phl p and grass homolog CD4+ T-cell epitopes
| Group allergen a | Grass species b | Aminoacid sequence c | IC50 (μM) d | RBA e |
|---|---|---|---|---|
| Group 1 0102 97-116 | Phl p 1 | EEPIAPYHFDLSGHAFGAMA | 0.274590811 | 1 |
| Lol p 1 | EEPIAPYHFDLSGHAFGSMA | 0.435355464 | 0.63072784 | |
| Pha a 1 | EEPIAPYHFDLSGHAFGSMA | 0.435355464 | 0.63072784 | |
| Poa p 1 | EEPIAAYHFDLSGKAFGAMA | 0.826658242 | 0.33216969 | |
| Group 1 0102 221-240 | Phl p 1 | TEAEDVIPEGWKADTSYESK | 2.921366199 | 1 |
| Lol p 1 | SEFEDVIPEGWKADTSYSAK | 2.921366199 | 1 | |
| Poa p 1 | GEAEDVIPEGWKADTAYASK | 0.453158364 | 6.44667832 | |
| Dac g 1 | SEVEDVIPEGWKADTSYEAK | 0.176697335 | 16.533165 | |
| Group 5 01 01 167-186 | Phl p 5a | AAFKVAATAANAA | 0.215901172 | 1 |
| Hor v 5a | AAFRTAATAADAA | 0.970389302 | 0.22248923 | |
| Hol l 5a | AAFRIAATAANAA | 0.116004811 | 1.86113982 | |
| Lol p 5a | AAYRTAATAANAA | 0.531949004 | 0.40586818 | |
| Pha a 5a | AAFKIAATAANSA | 0.38603706 | 0.55927576 | |
| Poa p 5a | AAFKVAATAANAA | 0.05 | 4.31802348 | |
| Dac g 5a | AAYKIAATAANAA | 0.05 | 4.31802348 | |
| Group 5 0101 199-218 | Phl p 5a | ESYKFIPALEAAV | 0.970389302 | 1 |
| Hor v 5a | ESYKFIPALEAAV | 0.990031471 | 0.98016006 | |
| Hol l 5a | EAYKFIPSLETAV | 0.104945543 | 9.24659849 | |
| Lol p 5a | DSYKFIPTLVAAV | 0.409955767 | 2.36705855 | |
| Pha a 5a | ETYKFIPSLEAAV | 0.051012076 | 19.0227368 | |
| Poa p 5a | DTYKSIPSLEAAV | 0.061093499 | 15.8836753 | |
| Dac g 5a | ESYKFIPTLEAAV | 0.074648485 | 12.9994507 |
Two distinct regions were used for Group 1 (20-mer) and Group 5a (13-mer) major allergens; Major allergen Phl p 5 consists of two isoforms, Phl p 5a and 5b.
Peptides for different grass species were used.
Homologous sequences of amino acids for each epitope region. Bold letters variant residues. Underlined amino acids depict DRB1*0401 motifs P1, P4, P6, P7 and P9.
IC50, concentration required by a target peptide to inhibit 50 % binding of the reference peptide.
RBA, relative binding affinity (relative to that of the Phl p peptide).
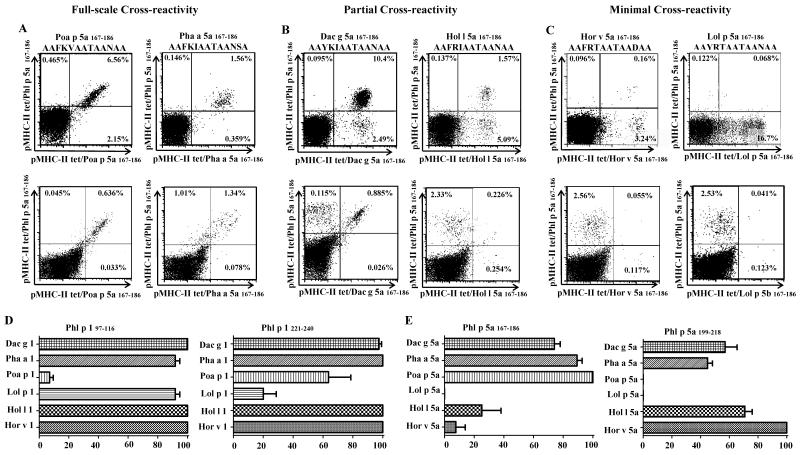
PBMC from GP allergic subjects were then stimulated with the Pooideae homologs (other than Phl p peptides). T-cell cross-reactivity was evaluated by co-staining assays utilizing pMHC II tetramers loaded with TGP-derived peptides (Phl p 1 or Phl p 5a peptides) and tetramers with homologous grass-pollen peptides corresponding to the stimulating epitope (Figure 2A, B and C top panels). Three different cross-staining patterns were observed: (i) Full-scale cross reactivity (Figure 2A); (ii) Partial cross-reactivity (Figure 2B; and (iii) Minimal cross-reactivity (Figure 2C). Even though Group 1 allergens share high sequence similarity, homologous antigenic-epitopes (Figure 2D), Poa p 1 97-116 and Lol p 1 221-240 were identified as minimally cross-reactive T-cell epitopes. The majority of T-cells elicited by these epitopes did not cross recognize the Phl p 1 epitopes in the corresponding region. On the other hand, along Group 5 epitopes the patterns of cross-reactivity were more diversified (Figure 2E). Lol p 5a 167-186, Hor v 5a 167-186, Hol l 5a 167-186, Poa a 5a 199-218 and Lol p 5a 199-218-reactive T-cell populations co-stained less than 25%. The patterns of cross-reactivity were also evaluated by stimulating PBMC with Phl p epitopes, and subsequently staining with both Phl p tetramers and tetramers loaded with other homologous GP peptides (Figure 2A, B and C bottom panels). The results were similar to those obtained by stimulating PBMC with homologous grass-peptides (Figure 2A, B and C top panels). The cross-reactive patterns for each epitope region are summarized in Table 2. This data suggests that the patterns of cross-reactivity among Pooideae GP homolog epitopes are diversified and that minimally cross-reactive T-cell epitopes are present amongst Group 1 and Group 5 allergens, meaning that high sequence homology does not imply cross-reactivity.
Figure 2.
Grass-specific CD4+ T-cells exhibit varying degrees of cross-reactivity. After in vitro stimulation with Phl p homologous grass peptides (top panels of A, B and C, amino-acid sequence of the stimulating peptide is specified on top of each panel) or Phl p 167-186 AAFKVAATAANAA, bottom panels of A, B and C), cells were co-stained with APC-labeled DR04:01/Phl p grass peptide-loaded tetramers (Y axis) and with PE-labeled DR04:01/homologous grass peptide-loaded tetramers corresponding to the stimulating epitopes (X axis). The plots in A, B and C show representative results for different cross-reactivity profiles for the TGP-derived epitopes in the Group 5a 167-186 region. A. Full-scale cross-reactivity was observed for Poa p 5a 167-186, and Pha a 5a 167-186. B. Partial cross-reactivity was observed for Dac g 5a 167-186 and Hol 5a 167-186. C. Minimal cross-reactivity was observed for Lol p 5a 167-186 and Hor v 5a 167-186. D. Comparison of percentages of cross-reactive T-cells for Group 1 homologs. E. Comparison of percentages of cross-reactive T-cells for Group 5a homologs. Summarized results from (n=6) allergic subjects are presented for distinct antigenic epitope regions. Each bar depicts the average of dual-tetramer stained T-cells (cross-reactive) observed per epitope.
Table 2.
Cross-reactivity between Phl p and grass homologous epitopes
| Group allergen * | Cross-staining I | Full-scale cross-reactivity II | Partial cross-reactivity II | ’ Minimal cross-reactivity II |
|---|---|---|---|---|
| Group 1 97-116 | Pha a 1 and Phl p 1 | X | ||
| Poa p 1 and Phl p 1 | X | |||
| Lol p 1 and Phl p 1 | X | |||
| Group 1 221-240 | Dac g 1 and Phl p 1 | X | ||
| Poa p 1 and Phl p 1 | X | |||
| Lol p 1 and Phl p 1 | X | |||
| Group 5 167-186 | Dac g 5a and Phl p 5a | X | ||
| Pha a 5a and Phl p 5a | X | |||
| Poa p 5a and Phl p 5a | X | |||
| Lol p 5a and Phl p 5a | X | |||
| Hol l 5a and Phl p 5a | X | |||
| Hor v 5a and Phl p 5a | X | |||
| Group 5 199-218 | Dac g 5a and Phl p 5a | X | ||
| Pha a 5a and Phl p 5a | X | |||
| Poa p 5a and Phl p 5a | X | |||
| Lol p 5a and Phl p 5a | X | |||
| Hol l 5a and Phl p 5a | X | |||
| Hor v 5a and Phl p 5a | X |
Group 1 refers to isoform 0102; Group 5a refers to isoform 0101 and 5b to isoform 0102.
The first epitope listed is the stimulatory epitope; the second epitope listed is the cross-stained epitope.
Cross-reactivity assessed on the basis of tetramer co-staining experiments.
Minimally cross-reactive CD4+ T-cells can be detected in GP allergic subjects
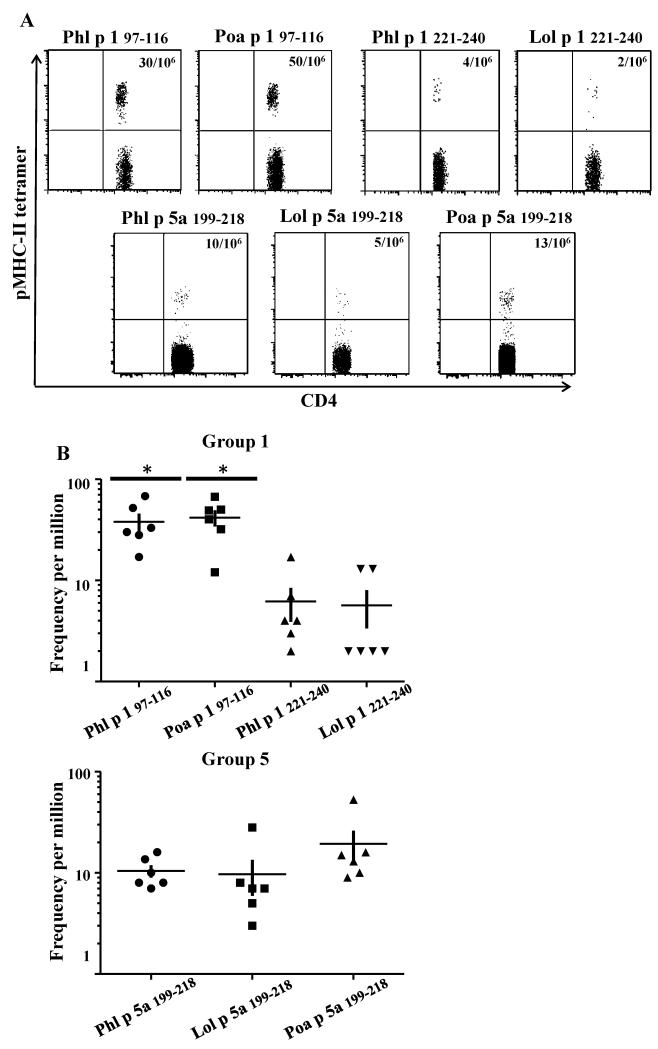
On the basis of these findings, we asked whether T-cells that recognize the minimally cross-reactive T-cell epitopes of the different Pooideae species can be detected directly ex-vivo in PBMC of DR04:01 GP allergic subjects without an amplification step. Ex-vivo frequencies of minimally cross-reactive T-cell epitopes of Poa p 1 and Lol p 1 were compared to its counterpart Phl p 1. Interestingly for TGP-derived epitope region 97-116 in Group 1 (Figure 3), both Phl p 1 and Poa p 1 were identified as the immunodominant epitopes (frequency ranged from 40 to 65 T-cells per million), while for TGP-derived region 221-240, low frequency was observed for both Phl p 1 and Lol p 1 (frequency ranged from 2 to 6 T-cells per million). For Group 5a allergens, (Figure), T-cell frequencies for TGP-derived region 199-218 for Phl p 5a, Lol p 5a and Poa p 5a were similar and ranged from 5 to 14 cells per million). Thus T-cells from TGP-derived region 97-116 in Group 1 are found in higher frequencies (P<0.05) than epitope-specific T-cells from TGP-derived regions 221-240 in Group 1 and TGP-derived region 199-218 in Group 5a allergens. These experiments show that subjects with GP allergy, diagnosed upon IgE reactivity to TGP extract are also sensitized to pollen from other Pooideae species at the T-cell level.
Figure 3.
Minimally cross-reactive CD4+ T-cells can be detected ex vivo in TGP allergic subjects. Determination of frequencies of Phl p- and grass homolog-reacting CD4+ T-cells for different regions of Group 1 and 5a allergens. A. Example of ex-vivo tetramer staining for TGP–derived Group 1 97-116, Group 1 221-240 and Group 5a 97-116-Phl p-specific and homologous minimally cross-reactive CD4+ T-cells from a grass pollen allergic subject with an ImmunoCAP score of 5 for Phl p specific-IgE. B. Frequencies for TGP-derived Group 1 97-116 and Group 1 221-240, and Group 5a 199-218 grass homolog-reactive CD4+ T-cells. Each data point denotes the frequency of epitope specific CD4+ T-cells for a different individual. A t student test was used in the statistical analysis.*P < 0.05 to compare populations from different epitopes.
Minimally cross-reactive CD4+ T-cells have a TH2 phenotype and are functionally active during pollen season
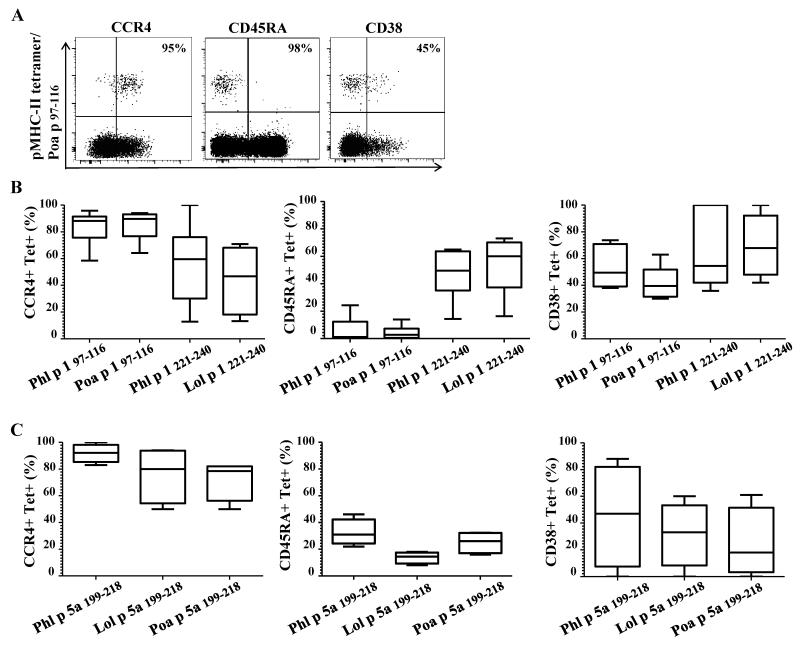
To further characterize the phenotype of minimally cross-reactive T-cells, antibodies directed against CCR4 (TH2 marker), CD45RA (a naïve T-cell marker) and CD38 (an activation marker) were utilized with ex vivo tetramer staining. The majority of epitope-specific T-cells from Group 1 97-116 region and Group 5a 199-218 region displayed a CCR4+ and CD45RA− 218 phenotype (Figure 4) indicating these are memory T-cells with a TH2 phenotype. However, a higher percentage of epitope-specific T-cells from Group 1 221-240 region displayed a mixed CCR4+ and CD45RA+ phenotype (Figure 4B). The activation status of these cells was also examined during pollen season by analyzing CD38 expression on CD454RA-T-cell populations. These stainings show that substantial activation could be seen for all these T-cells (Figures 4B and 4C) and that there is no difference in CD38 expression among the different immunodominant TGP-derived epitopes.
Figure 4.
Minimally cross-reactive CD4+ T-cells are memory T-cells with a TH2 phenotype and are activated during the pollen-season. A. Anti-PE enrichment permitted the analysis of surface marker expression; a naïve marker (CD45RA), a TH2 marker (CCR4) and an activation marker (CD38) were analyzed to compare Phl p grass-specific CD4+ T-cells with homologous grass-specific CD4+ T-cells. PBMC from a TPG allergic subject were stained with Poa p corresponding for TGP–derived region Group 1 97-116 and are presented as an example. B. Each bar represents surface expression of different markers for Group 1 reactive T-cells for 6 different allergic subjects. C. Each bar represents surface expression of different markers for Group 5 reactive T-cells for 6 different subjects.
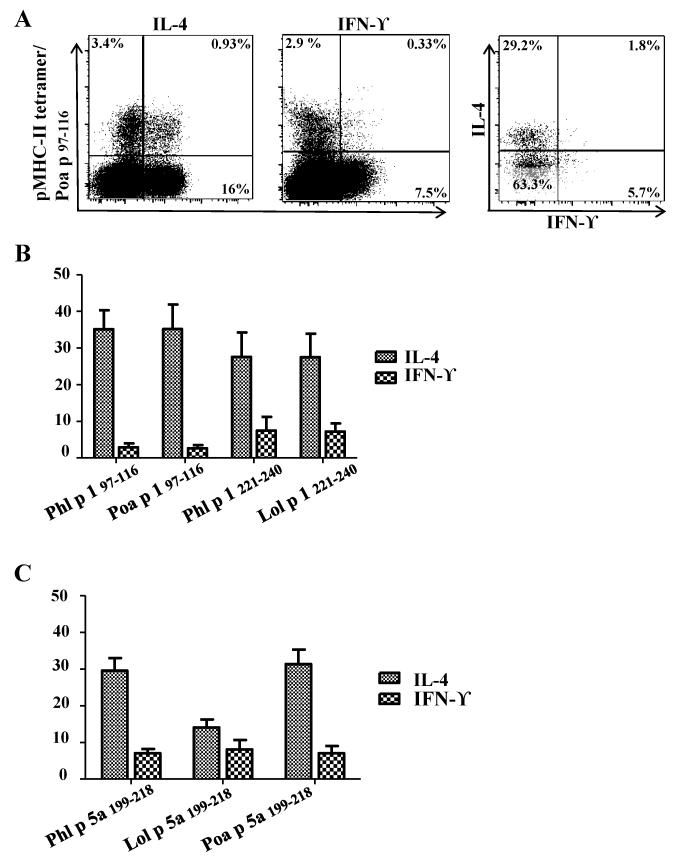
Functional assays were performed to confirm that minimally cross-reactive epitope-specific T-cells belong to the TH2-lineage (CCR4+); cells that had been stimulated in vitro with peptides and cultured for 2 weeks were stained with tetramer and co-stained for cytokine expression. A TH2-cytokine (IL-4) and a TH1 cytokine (IFN-ϒ) were used to discern amongst both lineages. As presented in Fig 5, tetramer-stained T-cells predominantly produced IL-4 with minimal production of IFN-ϒ. Overall these data suggest that minimal cross-reactive T-cells are memory TH2 cells and functionally, are equally potent as its Phl p counterparts. This data suggest that allergic subjects may experience symptoms through exposure to Phl p or other Pooideae species during the pollen season.
Figure 5.
Minimally cross-reactive CD4+ T-cells functional profiles correlate with a TH2 dominated response. An example of intracellular cytokine staining for Phl p- and grass homolog-reactive T-cells is provided. A. Representative result for ICS. PBMC from a TGP allergic subject were stimulated with Poa p peptide corresponding to region Group 1 97-116 and cultured for 2 weeks, cells were than stained with the corresponding PE-labeled DR04:01/Poa p grass peptide-loaded tetramers. Tetramer+ cells were gated and dual cytokine analysis is presented. B. Summarized results of ICS for TGP-derived Group 1 97-116, Group 1 221-240 and Group 5 199-218 epitopes, each bar depicts percentage of tetramer-positive cytokine producing cells in each individual.
Pooideae grass-specific CD4+ T-cells have varying degrees of cross-reactivity that depend on the HLA-allele and epitope they recognize
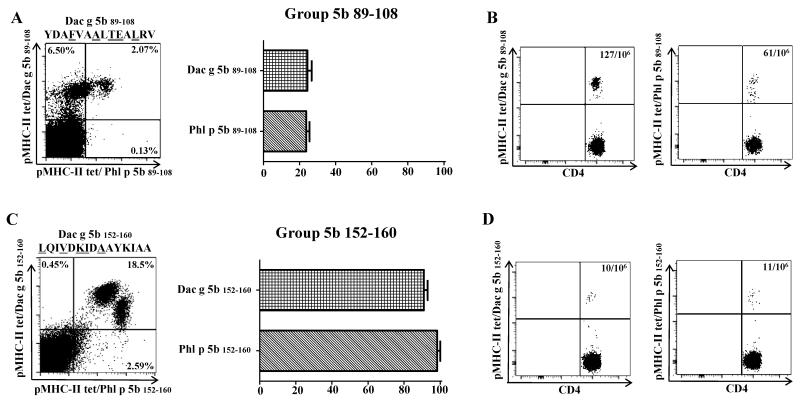
To generalize our findings, T-cell cross-reactivity in other HLA-DR alleles was also investigated. Homologous epitopes for Dac g 5b were identified by selecting sequences homologous to previously immunodominant DR07:01 and DRB5*01:01 Phl p-specific epitopes identified in our laboratory [19].For DR07:01, minimal cross-reactivity was observed between Dac g 5b 89-108 or Phl p 5b 89-108 T-cells(Figure 6A). However, Dac g 5b-specific T-cell frequency was at least two fold higher in DR07:01 subjects in comparison to Phl p-specific T-cell frequency (Figure 6B). In DRB5 subjects, full-scale cross reactivity was observed between both homologous TGP-derived region 152-160 in Group 5b epitopes and the T-cell frequency was approximately 10 cells per million (Figure 6C and 6D).
Figure 6.
Grass-pollen-specific CD4+ T-cells have varying degrees of cross-reactivity that vary on the HLA and epitope they recognize. A. PBMC (n=2) from DR07:01 allergic subjects were stimulated in vitro with Dac g peptide corresponding to region Group 5b 89-108. Cells were then co-stained with APC-labeled DR07:01/Phl p grass peptide-loaded tetramers (Y axis) as well as PE-labeled DR0701/Dac g 5b peptide-loaded tetramers (X axis). The plot shows a representative result for Group 5 89-108 grass homologs, sequence of stimulating peptide is shown on top of plot. T-cells were gated on Sidescatter and CD4+. The graph represents percentage of T-cells that were able to double stain; for this particular HLA-restriction minimal cross-reactivity was detected stimulating with either Dac g 5b or Phl p 5b peptides (about 20% of T-cells double stained) B. Ex vivo frequency of allergen-specific CD4+ T-cells for Group 5b 89-108 homologs. C. PBMC (n=2) from DRB5 allergic subjects were stimulated in vitro with either Dac g peptide corresponding to region Group 5b 152-160. Cells where then co-stained with APC-labeled DRB5/Phl p grass peptide-loaded tetramers (Y axis) as well as PE-labeled DRB5/Dac g 5b peptide-loaded tetramers (X axis). The plot shows a representative result for Group 5b 152-160 grass homologs, sequence of stimulating peptide is shown on top of plot. T-cells were gated on Side scatter and CD4+. The graph represents percentage of double stained T-cells, for this particular HLA-restriction full scale cross-reactivity was detected stimulating with either Dac g 5b or Phl p 5b peptides (≥ 90% of T-cells double stained) D. Ex vivo frequency of allergen-specific CD4+ T-cells for Group 5 152-160 grass homologs.
DISCUSSION
Although the efficacy and safety of single-grass-allergen (Phl p) immunotherapy has been documented [21;22], the use of single or multiple allergen products for Pooideae-grass pollen immunotherapy is still debated [11;23-25]. In the present study we utilized a multiplex pMHCII tetramer staining approach to determine the patterns of cross-reactivity between TGP-derived epitopes and epitopes of pollens from multiple grass-species within the Pooideae family.
Prior studies showed that Pooideae pollen extracts demonstrate significant IgE cross-reactivity [3-6;9;10;26;27]. In contrast, cross-reactivity at the T-cell level has not been thoroughly documented. Heijl et al showed both Phl p 1- and Phl p 5-specific T-cell lines (TCL) responded robustly to extract from other Pooideae species, suggesting cross-reactivity at the T-cell level [11]. However, TCL being used were generated by pollen extract, and T-cells specific for minimally cross-reactive T-cell epitopes could be overlooked. In this present study, Phl p-specific CD4+ T cells in allergic individuals had a range of patterns of cross-reactivity against allergens from other Pooideae species. The cross-reactivity profile depended on the HLA-restriction and epitope involved. Consistent with previous studies [14;15], our approach identified full-scale, partially and minimally-cross-reactive T-cell epitopes for both Group 1 and 5 major allergens. The majority of T-cells specific for minimally-cross-reactive epitopes did not cross-recognize the Phl p 1 derived epitopes in the corresponding regions. However, a minor population of T-cells, capable of cross-recognizing these epitopes was detected in some subjects, indicating that TCR diversity can be present in Pooideae grass-pollen specific T-cells. This suggests that although high sequence homology is found amongst grass-pollen allergens from the Pooideae subfamily, a single species (Timothy grass) does not cover the broad epitopic repertoire at the T-cell level. Interestingly, there is discordance in known cross-reactivity at the IgE level as compared to the T-cell level [3-6;9;10;26;27]. IgE epitopes are conformational and Pooideae grass-pollen allergens contain IgE-binding sites that are capable of generating IgE responses to allergens from other Pooideae species [1;2]. On the other hand, T-cell epitopes are linear and selectively target allergen-specific T-cells. A recent study by Focke-Tejkl et al showed discrepancy among the IgE- and T-cell-reactive domains in Phl p 5 major allergens, showing the importance of studying B-cell and T-cell epitopes [28]. However, a caveat is noteworthy in our study, as the result of the cross-reactivity patterns might be influenced by the size of the cohort and the variety of HLA-alleles. It is possible that minimally cross-reactive epitopes may be absent in other HLA types.
Class II MHC/peptide structures have been thoroughly characterized [29-31;31], and it is established that peptide side chains at position p1, p4, p6, and p9 project into pockets along the MHC peptide-binding groove acting as anchor motifs [32]. Conversely, peptide side-chains at position p-1, p2, p3, p5, p8, p10 project directly upward from the MHC-molecule surface and impact T-cell recognition [30-32]. Interestingly, anchor positions for most of the allergenic epitopes are well conserved among Pooideae species. For example, anchor motifs in TGP derived epitopes from region 97-116 correspond to Y, D, S and G, and these residues are well conserved amongst Phl p, Lol p, Pha a and Poa p (Table 1). In contrast, most of the minimally cross-reactive T-cell epitopes feature variability at p-1, p2, p3 and/or p8. As shown in Table 1, Poa p 1 97-116 variant residues are present in p-1 (A) and p8 (K). For this reason, homologous peptides from diverse grass species can bind to MHC class II but have varying degrees of recognition by different TCRs.
The current study demonstrated that different grass-species contain species-specific T-cell epitopes. Considering the overlap of pollination calendars and geographical distribution for Pooideae grass species, allergic subjects can be exposed to multiple pollens in the same geographical region [3;6;23;24]. As minimally cross-reactive T-cell epitopes can also bind to MHC-II, exposure to pollens from different grass species may generate T-cell population’s specific to those minimally cross-reactive T-cell epitopes in allergic subjects. Indeed, homologous grass-pollen epitope-specific CD4+ T-cells specific for minimally cross-reactive T-cell epitopes of other Pooideae species could be readily detected in PBMC from grass-pollen allergic subjects. For example, frequency of Poa p 1 97-116 in PBMC was found in similar range as the immunodominant Phl p epitope in DR04:01 subjects, suggesting that poly-sensitization occurs at the T-cell level. Side-by-side comparison of surface phenotype and functional profiles reveals that these minimally cross-reactive CD4+ T-cells have a TH2 dominated response and equal allergenic properties as Phl p-specific CD4+ T-cells. In addition, these homologous grass-pollen epitope-specific CD4+ T-cells are also activated during pollen season. The presence of the minimally cross-reactive T-cells for different Pooideae species in a group recruited on the basis of IgE reactivity to TGP implies that GP allergic subjects are usually poly-sensitized to other Pooideae species at the T-cell level. A recent study suggested that depletion of allergen specific T-cells would be essential for successful antigen specific immunotherapy [13]. In addition, it has also been established that the strength of the TCR signal induced by an epitope can affect the outcome in inducing tolerance [33]. Conversely, lack of cross-staining and the featured variability at sites of T-cell contact suggest that minimally cross-reactive CD4+ T-cells specific for other Pooideae species will not be targeted by TGP specific mono immunotherapy as most of GP allergic subjects are sensitized to various Pooideae species.
On the other hand, the presence of full-scale and partially cross-reactive T-cell populations raises the possibility that single species immunotherapy might be effective for these cross-reactive populations with similar TH2 phenotype (data not shown). Campbell et al previously showed that peptide immunotherapy with selected epitopes from a single allergen could render T-cell tolerance to one epitope, enabling the suppression of the function of T-cells specific for other epitopes within the same allergen [34;35]. However, this linked epitope suppression is confined to a single allergen. Nonetheless, further investigations are required to confirm that Phl p-specific T-cells can mediate suppression of immune responses elicited by Phl p-nonspecific homologous T-cell populations.
In summary, class II tetramer staining experiments show that the patterns of cross-reactivity vary for 4 different DR04:01-, 1 DR07:01- and 1 DRB5*01:01-restricted TGP derived epitope regions tested. Indeed, T-cells with various degrees of cross reactivity profiles could be detected with tetramers both in vitro and ex-vivo settings. On the basis of these findings, we confirmed that both Phl p-specific as well as their Phl p-homologous CD4+ TH2 populations exist in vivo, suggesting GP allergic patients diagnosed upon IgE reactivity to Phleum pratense pollen extract were sensitized to various grass species at the T-cell level. This finding implies that mono-allergen immunotherapy with Phl p allergen would fail to elicit Treg or delete those species-specific T-cells that show minimum cross reactivity to Phl p. Although a direct clinical comparison between immunotherapy with single grass-extract verses multiple species-extracts utilizing a variety of HLAs would be obligatory, the current study suggests multiple-grass-pollen-species immunotherapy should be more beneficial than single species immunotherapy.
Supplementary Material
ACKNOWLEDGEMENTS AND CONFLICTS OF INTEREST
We thank Jennifer Heaton for help with subject recruitment. We also thank Diana Sorus for assistance in preparing the article. This work is supported by National Institute of Health contract HHSN272200700046C and NIH grant 5 R01 AI095074.
List of nonstandard abbreviations used
- HLA
Human histocompatibility leukocyte antigen
- MHC
Major histocompatibility complex
- PBMC
Peripheral blood mononuclear cell
- PE
Phycoerythrin
- APC
Allophycocyanin
- Phl p
Phleum pratense
- Hol l
Holcus lanatus
- Pha a
Phalaris aquatica
- Hor v
Hordeum vulgare
- Lol p
Lolium perenne
- Dac g
Dactylis glomerata
- Poa p
Poa pratensis
- pMHCII
Peptide/MHC class II
- TH
T helper
- TGP
Timothy grass pollen
- GP
Grass pollen
- TCL
T-cell line
References
- 1.Weber RW. Patterns of pollen cross-allergenicity. J Allergy Clin Immunol. 2003;112:229–39. doi: 10.1067/mai.2003.1683. [DOI] [PubMed] [Google Scholar]
- 2.Weber RW. Cross-reactivity of pollen allergens. Curr Allergy Asthma Rep. 2004;4:401–8. doi: 10.1007/s11882-004-0091-4. [DOI] [PubMed] [Google Scholar]
- 3.White JF, Bernstein DI. Key pollen allergens in North America. Ann Allergy Asthma Immunol. 2003;91:425–35. doi: 10.1016/S1081-1206(10)61509-8. [DOI] [PubMed] [Google Scholar]
- 4.Weber RW, Nelson HS. Pollen allergens and their interrelationships. Clin Rev Allergy. 1985;3:291–318. doi: 10.1007/BF02992997. [DOI] [PubMed] [Google Scholar]
- 5.Marcucci F, Sensi L, Di CG, Incorvaia C, Puccinelli P, Scurati S, Frati F. Which allergen extract for grass pollen immunotherapy? An in vitro study. Immunol Invest. 2010;39:635–44. doi: 10.3109/08820131003796876. [DOI] [PubMed] [Google Scholar]
- 6.Jager S. Exposure to grass pollen in Europe. 2008;8:2–6. [Google Scholar]
- 7.Hrabina M, Peltre G, Van Ree R, Moingeon P. Grass pollen allergens. Clinical and Experimental Allergy Reviews. 2008;8:7–11. [Google Scholar]
- 8.Andersson K, Lidholm J. Characteristics and immunobiology of grass pollen allergens. Int Arch Allergy Immunol. 2003;130:87–107. doi: 10.1159/000069013. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Cocera C, Sastre J, Cimarra M, Quirce S, Fernandez-Rivas M, Enriquez-Matas A, Rodriguez-Alvarez M, Martin S. Immunotherapy with a Phleum pratense allergen extract induces an immune response to a grass-mix allergen extract. J Investig Allergol Clin Immunol. 2010;20:13–9. [PubMed] [Google Scholar]
- 10.Weber RW. Cross-reactivity of plant and animal allergens. Clin Rev Allergy Immunol. 2001;21:153–202. doi: 10.1385/CRIAI:21:2-3:153. [DOI] [PubMed] [Google Scholar]
- 11.Hejl C, Wurtzen PA, Kleine-Tebbe J, Johansen N, Broge L, Ipsen H. Phleum pratense alone is sufficient for allergen-specific immunotherapy against allergy to Pooideae grass pollens. Clin Exp Allergy. 2009;39:752–9. doi: 10.1111/j.1365-2222.2008.03195.x. [DOI] [PubMed] [Google Scholar]
- 12.Kwok WW, Roti M, Delong JH, Tan V, Wambre E, James EA, Robinson D. Direct ex vivo analysis of allergen-specific CD4+ T cells. J Allergy Clin Immunol. 2010;125:1407–9. doi: 10.1016/j.jaci.2010.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wambre E, Delong JH, James EA, LaFond RE, Robinson D, Kwok WW. Differentiation stage determines pathologic and protective allergen-specific CD4+ T-cell outcomes during specific immunotherapy. J Allergy Clin Immunol. 2012;129:544–51. doi: 10.1016/j.jaci.2011.08.034. 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schenk S, Breiteneder H, Susani M, Najafian N, Laffer S, Duchene M, Valenta R, Fischer G, Scheiner O, Kraft D. T-cell epitopes of Phl p 1, major pollen allergen of timothy grass (Phleum pratense): evidence for crossreacting and non-crossreacting T-cell epitopes within grass group I allergens. J Allergy Clin Immunol. 1995;96:986–96. doi: 10.1016/s0091-6749(95)70237-7. [DOI] [PubMed] [Google Scholar]
- 15.Muller WD, Karamfilov T, Kahlert H, Stuwe HT, Fahlbusch B, Cromwell O, Fiebig H, Jager L. Mapping of T-cell epitopes of Phl p 5: evidence for crossreacting and non-crossreacting T-cell epitopes within Phl p 5 isoallergens. Clin Exp Allergy. 1998;28:1538–48. doi: 10.1046/j.1365-2222.1998.00432.x. [DOI] [PubMed] [Google Scholar]
- 16.Burtin D, Chabre H, Olagnier B, Didierlaurent A, Couret MN, Comeau D, Wambre E, Laparra H, Van OL, Montandon F, Batard T, Jonval V, Lorphelin A, Merle C, Berrouet C, Parry L, Gomord V, Van RR, Moingeon P. Production of native and modified recombinant Der p 1 molecules in tobacco plants. Clin Exp Allergy. 2009;39:760–70. doi: 10.1111/j.1365-2222.2009.03201.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt H, Gelhaus C, Nebendahl M, Janssen O, Petersen A. Characterization of Phleum pratense pollen extracts by 2-D DIGE and allergen immunoreactivity. Proteomics. 2010;10:4352–62. doi: 10.1002/pmic.201000451. [DOI] [PubMed] [Google Scholar]
- 18.Ge X, Tan V, Bollyky PL, Standifer NE, James EA, Kwok WW. Assessment of seasonal influenza A virus-specific CD4 T-cell responses to 2009 pandemic H1N1 swine-origin influenza A virus. J Virol. 2010;84:3312–9. doi: 10.1128/JVI.02226-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wambre E, Delong JH, James EA, Torres-Chinn N, Pfutzner W, Mobs C, Durham S, Till SJ, Robinson D, Kwok WW. Specific immunotherapy modifies allergen-specific CD4+ T cell responses in an epitope-dependent manner. J Allergy Clin Immunol. 2013 doi: 10.1016/j.jaci.2013.10.054. Article in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Dahl R, Kapp A, Colombo G, de Monchy JG, Rak S, Emminger W, Rivas MF, Ribel M, Durham SR. Efficacy and safety of sublingual immunotherapy with grass allergen tablets for seasonal allergic rhinoconjunctivitis. J Allergy Clin Immunol. 2006;118:434–40. doi: 10.1016/j.jaci.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 22.Durham SR, Yang WH, Pedersen MR, Johansen N, Rak S. Sublingual immunotherapy with once-daily grass allergen tablets: a randomized controlled trial in seasonal allergic rhinoconjunctivitis. J Allergy Clin Immunol. 2006;117:802–9. doi: 10.1016/j.jaci.2005.12.1358. [DOI] [PubMed] [Google Scholar]
- 23.Moingeon P, Hrabina M, Bergmann KC, Jaeger S, Frati F, Bordas V, Peltre G. Specific immunotherapy for common grass pollen allergies: pertinence of a five grass pollen vaccine. Int Arch Allergy Immunol. 2008;146:338–42. doi: 10.1159/000121468. [DOI] [PubMed] [Google Scholar]
- 24.Chabre H, Gouyon B, Huet A, Baron-Bodo V, Nony E, Hrabina M, Fenaille F, Lautrette A, Bonvalet M, Maillere B, Bordas-Le F,V, Van OL, Jain K, Ezan E, Batard T, Moingeon P. Molecular variability of group 1 and 5 grass pollen allergens between Pooideae species: implications for immunotherapy. Clin Exp Allergy. 2010;40:505–19. doi: 10.1111/j.1365-2222.2009.03380.x. [DOI] [PubMed] [Google Scholar]
- 25.Didier A, Malling HJ, Worm M, Horak F, Jager S, Montagut A, Andre C, de Beaumont O, Melac M. Optimal dose, efficacy, and safety of once-daily sublingual immunotherapy with a 5-grass pollen tablet for seasonal allergic rhinitis. J Allergy Clin Immunol. 2007;120:1338–45. doi: 10.1016/j.jaci.2007.07.046. [DOI] [PubMed] [Google Scholar]
- 26.Hrabina M, Jain K, Gouyon B. Cross-reactivity between pollen allergens from common Pooideae grasses and rye. Clinical and Experimental Allergy Reviews. 2008;8:18–20. [Google Scholar]
- 27.Leiferman KM, Gleich GJ. The cross-reactivity of IgE antibodies with pollen allergens. I. Analyses of various species of grass pollens. J Allergy Clin Immunol. 1976;58:129–39. doi: 10.1016/0091-6749(76)90148-2. [DOI] [PubMed] [Google Scholar]
- 28.Focke-Tejkl M, Campana R, Reininger R, Lupinek C, Blatt K, Valent P, Pavkov-Keller T, Keller W, Valenta R. Dissection of the IgE and T-cell recognition of the major group 5 grass pollen allergen Phl p 5. J Allergy Clin Immunol. 2014 doi: 10.1016/j.jaci.2013.08.038. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dessen A, Lawrence CM, Cupo S, Zaller DM, Wiley DC. X-ray crystal structure of HLA-DR4 (DRA*0101, DRB1*0401) complexed with a peptide from human collagen II. Immunity. 1997;7:473–81. doi: 10.1016/s1074-7613(00)80369-6. [DOI] [PubMed] [Google Scholar]
- 30.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–73. [PubMed] [Google Scholar]
- 31.Yassai M, Afsari A, Garlie J, Gorski J. C-terminal anchoring of a peptide to class II MHC via the P10 residue is compatible with a peptide bulge. J Immunol. 2002;168:1281–5. doi: 10.4049/jimmunol.168.3.1281. [DOI] [PubMed] [Google Scholar]
- 32.Sette A, Sidney J, Oseroff C, del Guercio MF, Southwood S, Arrhenius T, Powell MF, Colon SM, Gaeta FC, Grey HM. HLA DR4w4-binding motifs illustrate the biochemical basis of degeneracy and specificity in peptide-DR interactions. J Immunol. 1993;151:3163–70. [PubMed] [Google Scholar]
- 33.Gabrysova L, Wraith DC. Antigenic strength controls the generation of antigen-specific IL-10-secreting T regulatory cells. Eur J Immunol. 2010;40:1386–95. doi: 10.1002/eji.200940151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell JD, Buckland KF, McMillan SJ, Kearley J, Oldfield WL, Stern LJ, Gronlund H, van HM, Reynolds CJ, Boyton RJ, Cobbold SP, Kay AB, Altmann DM, Lloyd CM, Larche M. Peptide immunotherapy in allergic asthma generates IL-10-dependent immunological tolerance associated with linked epitope suppression. J Exp Med. 2009;206:1535–47. doi: 10.1084/jem.20082901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holan V, Mitchison NA. Haplotype-specific suppressor T cells mediating linked suppression of immune responses elicited by third-party H-2 alloantigens. Eur J Immunol. 1983;13:652–7. doi: 10.1002/eji.1830130809. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.