Abstract
The role of the habenular nuclei in modulating fear and reward pathways has sparked a renewed interest in this conserved forebrain region. The bilaterally paired habenular nuclei, each consisting of a medial/dorsal and lateral/ventral nucleus, can be further divided into discrete subdomains whose neuronal populations, precise connectivity and specific functions are not well understood. An added complexity is that the left and right habenulae show pronounced morphological differences in many non-mammalian species. Notably, the dorsal habenulae of larval zebrafish provide a vertebrate genetic model to probe the development and functional significance of brain asymmetry. Previous reports have described a number of genes that are expressed in the zebrafish habenulae, either in bilaterally symmetric patterns or more extensively on one side of the brain than the other. The goal of our study was to generate a comprehensive map of the zebrafish dorsal habenular nuclei, by delineating the relationship between gene expression domains, comparing the extent of left-right asymmetry at larval and adult stages, and identifying potentially functional subnuclear regions as defined by neurotransmitter phenotype. While many aspects of habenular organization appear conserved with rodents, the zebrafish habenulae also possess unique properties that may underlie lateralization of their functions.
Keywords: Epithalamus, Left-right asymmetry, habenula, interpeduncular nucleus, somatostatin, ano2, mbnl3, gng8
INTRODUCTION
The habenular nucleus influences a wide range of behaviors, from sleep and reward to fear and anxiety (Bianco and Wilson, 2009; Hikosaka, 2010; Hikosaka et al., 2008; Klemm, 2004; Lecourtier and Kelly, 2007; Okamoto et al., 2012; Sutherland, 1982), and has been a recent focus in research on addiction and depression (Aizawa et al., 2013; Hikosaka, 2010; Hikosaka et al., 2008; Viswanath et al., 2014). Despite increasing functional studies, our knowledge of its organization and neuronal properties is limited. Moreover, many vertebrates, including zebrafish, display prominent left-right (L-R) differences in this dorsal diencephalic structure (Concha and Wilson, 2001), further complicating our understanding of habenular organization, connectivity and function.
The habenulae of mammals consist of bilaterally paired medial and lateral nuclei (Gurdjian, 1925) that have different afferent input (Herkenham and Nauta, 1977) and efferent connections (Herkenham and Nauta, 1979). The lateral nuclei project to a variety of areas in the midbrain and hindbrain, whereas the medial nuclei chiefly innervate the unpaired interpeduncular nucleus (IPN) in the ventral midbrain (Herkenham and Nauta, 1979). Axons from both nuclei contribute to the fasciculus retroflexus (FR) nerve bundles to form one of the most highly conserved conduction systems in the vertebrate brain (Herrick, 1948). While it was initially suggested that zebrafish only possess a habenular region analogous to the medial nucleus of mammals (Concha and Wilson, 2001), further characterization of molecular and anatomical properties confirmed the presence of both a dorsal and ventral nucleus (Amo et al., 2010), which, respectively, correspond to the medial and lateral habenulae of mammals.
In several vertebrate species, the medial/dorsal and lateral/ventral nuclei have been further subdivided into discrete neuronal populations or subnuclei largely based on morphological criteria (Contestabile et al., 1987; Iwahori, 1977; Marburg, 1944; Quina et al., 2009; Wagner et al., 2014). Four subnuclei were initially recognized in the medial nucleus of the well-studied rat habenula by neuronal shape and packing, neuropil density and neurotransmitter immunoreactivity (Andres et al., 1999; Contestabile et al., 1987; Geisler et al., 2003). Antibody labeling demonstrated that neurons located more dorsally produce substance P (Cuello et al., 1978), while those in more ventral regions are cholinergic (Eckenrode et al., 1987). More recent analyses of gene expression corroborate this major sub-division, as well as demonstrate that genes from the vesicular glutamate transporter (vglut) family are expressed throughout the habenulae (Aizawa et al., 2012; Quina et al., 2009). The dorsal region can be further divided into glutamatergic-only and substance P-expressing/glutamatergic neuronal populations by the localization of transcripts for vglut1and 2 and the substance P precursor, tachykinin 1 (tac1) (Aizawa et al., 2012; Quina et al., 2009). Immunolabeling indicates that the cholinergic neurons largely project to the central core of the IPN, whereas substance P-expressing neurons innervate peripheral subnuclei (Contestabile et al., 1987). Additional combinatorial patterns of gene expression reveal 5 distinct subnuclear regions in the medial habenula of the adult rat (Aizawa et al., 2012), and a recent study suggests a similar organization for the mouse (Wagner et al., 2014).
Discrete subnuclei identified in the dorsal habenulae of amphibians and fish show prominent differences in their size and organization between the left and right sides of the brain (refer to Concha and Wilson, 2001). For example, in Rana esculenta, the right dorsal habenula is a single nucleus, while the left contains lateral and medial subnuclei, which innervate the IPN via different routes (Braitenberg and Kemali, 1970; Kemali and Guglielmotti, 1984). Substance P neurons are present in the left and right dorsal habenulae; however, but only in the lateral subnucleus on the left side (Kemali and Guglielmotti, 1984).
In zebrafish, where the dorsal habenulae have emerged as a powerful genetic model to study the development and function of L-R asymmetry in the brain, subnuclear organization has been defined on the basis of patterns of gene and transgene expression. For example, combinatorial expression of the potassium channel tetramerization domain containing (kctd) genes kctd12.1, ktcd12.2, and ktcd8 revealed 6 molecularly distinct domains at the larval stage, with 3 differing in size between the left and right dorsal habenula, 2 unique to the left, and 1 unique to the right (Gamse et al., 2005). The transgenic line Tg(brn3a-hsp70:GFP), which was generated using regulatory sequences from the POU domain, class 4, transcription factor 1 (pou4f1) gene, labels neurons in the adult dorsal habenula in a mostly non-overlapping pattern with kctd12.1 expression (Aizawa et al., 2005). The brn3a and kctd12.1 domains have been proposed to correspond to the medial (dHbM) and lateral (dHbL) subnuclei of the dorsal habenulae, respectively (Aizawa et al., 2005). Integration into the neuronal activity regulated pentraxin (nptx2) locus generated the Tg(nptx2:Gal4-VP16) driver line, which also activates reporter gene expression in a largely complementary pattern to Tg(brn3a-hsp70:GFP) in the dHbL of the adult brain (Agetsuma et al., 2010). Habenular axons emanating from the dHbL are thought to innervate the dorsal, intermediate and most dorsal part of the ventral IPN, while those originating from dHbM neurons project to the intermediate and ventral IPN (Agetsuma et al., 2010; Aizawa et al., 2005; Gamse et al., 2005).
Transgenic lines have also been used to assess habenular L-R asymmetry. The Tg(brn3a-hsp70:GFP) labeled neuronal population is larger in the right habenula while, conversely, in nptx2:Gal4; UAS:DsRed2 double heterozygous fish more DsRed2-positive neurons are located in the left habenula. The size asymmetry between the dHbM and dHbL subnuclei has been attributed to a higher number of early born dHbL neurons in the left dorsal nucleus and later born dHbM neurons enriched on the right (Aizawa et al., 2007). Prolonging the early period of neurogenesis may also increase the dHbL subnuclei and decrease the dHbM, resulting in bilaterally symmetric dorsal habenulae (Doll et al., 2011).
The goal of the present study was to determine how distinct neuronal populations defined by neurotransmitter phenotype correspond to the previously designated subnuclear regions of the dorsal habenulae. Through double labeling in the larval and adult brain, we provide a consolidated molecular map of the dorsal habenular subnuclei and demonstrate that L-R asymmetric cholinergic and peptidergic subregions only partially overlap with the previously proposed dHbL and dHbM subnuclear organization. By comparing the results in zebrafish with published studies on the medial habenulae of the adult rodent brain, we find that specific neurotransmitter-expressing domains and their spatial relationships are mostly conserved, even though L-R differences are not. Precise definition of subnuclei is important when monitoring neuronal activity and interpreting behavioral responses in efforts to understand how the habenular region of the brain regulates its diverse functions. Moreover, performing such analyses using the zebrafish model could shed light on the significance of directional asymmetry in an essential neural pathway.
MATERIALS AND METHODS
Zebrafish
Zebrafish were raised and housed at 27°C on a 14/10 h light dark cycle. We used the wild-type AB strain (Walker, 1999) and the transgenic lines TgBAC(gng8:Eco.NfsB- 2A-CAAX-GFP)c375 (deCarvalho et al., 2013), Tg(vglut2a: loxP-DsRed-loxP-GFP)nns9 (Miyasaka et al., 2009), Tg(brn3a-hsp70:GFP)rw0110b (also known as Tg(pou4f1-hsp70l:GFP); Aizawa et al., 2005), Tg(nptx2:Gal4-VP16)rw0143a (also known as Tg(narp:Gal4-VP16); Agetsuma et al., 2010), Tg(UAS:DsRed2)rw0135 (Agetsuma et al., 2010), Tg(4xnrUAS:GFP)c354 (Akitake et al., 2008) and Tg(14xUAS:BGi-NLS-emGFP)y262 (H. Burgess, personal communication). For simplicity, we refer to these transgenic lines as Tg(gng8:GFP), Tg(vglut:DsRed), Tg(brn3a:GFP), Tg(nptx2:Gal4), Tg(UAS:DsRed2), Tg(4xUAS:GFP) and Tg(14xUAS:GFP). Maintenance of zebrafish and experimental procedures on larvae and adults were carried out in accordance with the protocol approved by Carnegie Institutional Animal Care and Use Committee.
RNA hybridization and immunofluorescence
Larvae and dissected adult brains were fixed in 4% paraformaldehyde (in PBS) at 4°C overnight. Single and double label colorimetric RNA in situ hybridization experiments were performed as described previously for whole larvae (Gamse et al., 2003) and modified for adult brain tissue (Gorelick et al., 2008). Single and double fluorescent RNA in situ hybridization (FISH) or FISH coupled with immunolabeling for green fluorescent protein (anti-GFP rabbit antibody; Torrey Pines TP401) was carried out as in a prior study (deCarvalho et al., 2013).
To synthesize antisense RNA probe for amine oxidase, copper containing 1 (aoc1), a 1019 base-pair fragment was PCR-amplified from the cDNA clone MGC154101 (Thermo Scientific) using primers GTCACTGAATACATCGTTGGCCC and TTGTAGACTGTAGATGTAGTTCTGATC and sub-cloned into the pCRII-TOPO vector using the TOPO TA Cloning kit (Invitrogen). pCRII-TOPO-aoc1 was linearized with ApaI and RNA transcribed with SP6 RNA polymerase. For anoctamin 2 (ano2), an EcoRI to NotI fragment from the cDNA clone MGC171498 (Open Biosystems) was subcloned into the pBluescript SK(−) vector, linearized with EcoRI and transcribed with T3 RNA polymerase. A zebrafish homologue to Drosophila muscleblind-like 3 (mbnl3) was obtained from a zebrafish kidney cDNA library and cloned into the EcoRI and XhoI sites of the pBK-CMV vector. pBK-CMV-mbnl3 was linearized with SalI and RNA transcribed with T7 RNA polymerase.
Methods for cloning and probe synthesis have been described previously for guanine nucleotide binding protein (G protein), gamma 8 (gng8; Thisse and Thisse, 2004), kctd12.1 (previously known as leftover; Gamse et al., 2003), kctd8 (previously known as dexter; Gamse et al., 2005), kctd12.2 (previously known as right on; Gamse et al., 2005), solute carrier family 18 (vesicular acetylcholine), member 3b (slc18a3b; commonly known as vachtb; Hong et al., 2013), somatostatin1.1 (sst1.1; Thisse and Thisse, 2004) and tac1 (Hong et al., 2013).
Sectioning and microscopy
Larval and adult brains were embedded in 4% low melt agarose (Lonza) in PBS (100g/ml) and sectioned with a VT1000S vibratome (Leica Microsystems, Inc.) from 50 to 250 μm as indicated. For labeling cell nuclei, sections were stained in 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies) in PBS with 0.1% tween for 20 minutes and rinsed in PBS. Sections were mounted in either 50% glycerol (1:1 vol/vol with H2O), Aqua-Poly/Mount (Polysciences, Inc.) or Prolong Gold (Invitrogen) anti-fade mounting media under cover slips.
Bright-field images were captured by an Axiocam HRc digital camera mounted on an Axioskop (Carl Zeiss). Fluorescent images were collected using a Leica SP5 confocal microscope and processed either with ImageJ (National Institutes of Health) or Imaris (Bitplane).
RESULTS
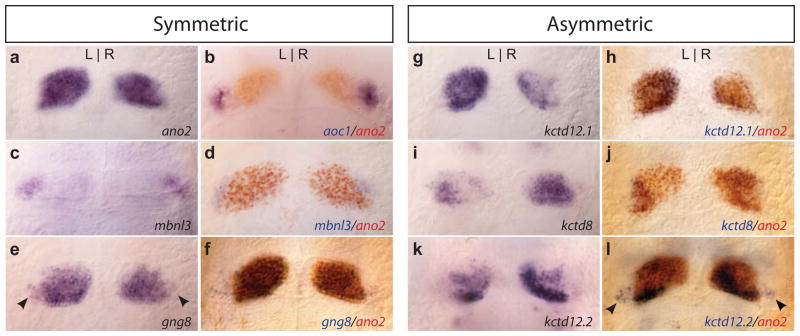
As summarized in Table 1, an increasing number of genes have been found to be expressed in the bilaterally paired dorsal and ventral habenulae of larval and adult zebrafish, and many of their homologues are transcribed in the corresponding medial and lateral habenular nuclei of rodents. For example, similar to expression of anoctamin 1 (ano1) in the medial habenula of the mouse (Quina et al., 2009), ano2 is transcribed in a bilaterally symmetric pattern throughout the dorsal nucleus of larval zebrafish (Fig. 1a), and is a useful marker for demarcating its boundaries. In zebrafish, the ventral nucleus was defined by its expression of the amine oxidase, copper containing 1 (aoc1) gene (Amo et al., 2010 and refer to Fig. 1b). We find that a zebrafish homologue of the muscleblind-like 3 (mbnl3) gene also exhibits bilaterally symmetric expression throughout the presumptive ventral nucleus (Fig. 1c, d), providing further evidence that the ventral habenulae have a distinct molecular identity. However, not all genes are transcribed exclusively in the dorsal or ventral nucleus. Transcripts for the guanine nucleotide binding protein (G protein), gamma 8 (gng8) gene, for instance, are distributed in a bilaterally symmetric pattern similar to ano2 (Fig. 1e, f) but are also found in a subset of neurons in both ventral nuclei (arrowheads in Fig. 1e). A transgenic line produced by integration of membrane-tagged green fluorescent protein gene into the gng8 locus (deCarvalho et al., 2013) labels the same pattern of dorsal and ventral habenular neurons (refer to Fig. 2a).
TABLE 1.
Gene expression in the zebrafish habenulae and homology with rodents
Listed are genes found to be transcribed in regions of the dorsal (D) and/or ventral (V) habenular nuclei for larval (La) or adult (A) zebrafish, as determined by RNA in situ hybridization. Questions marks indicate where published data are unclear. Expression of homologous genes in the medial (M) and/or lateral (L) habenular nuclei of rodents is also noted. References are provided for the zebrafish and rodent studies. AB atlas signifies the Allen Mouse Brain Atlas, available from: http://mouse.brain-map.org, ©2012 Allen Institute for Brain Science.
| Zebrafish Gene Name | Gene Symbol | Zebrafish Ha Domain | Stage | Zebrafish References | Rodent Ha Domain | Rodent References |
|---|---|---|---|---|---|---|
| adenylate cyclase activating polypeptide 1a | adcyap1a | D (left only) | La | Amo et al., 2010 | M, L | AB atlas |
| amine oxidase, copper containing 1 | aoc1 | V | La, A | Amo et al., 2010 | Neither | AB atlas |
| anoctamin 2 | ano2 | D | La | This study | M | AB atlas |
| Ca2+-dependent activator protein for secretion 2 | cadps2 | D | La | Gamse et al., 2003; Chen et al., 2009 | M, L | Sadakata et al., 2007 |
| calcium/calmodulin-dependent protein kinase IGb | camk1gb | D, V? | La | Tovin et al., 2012 | Neither | AB atlas |
| cAMP responsive element binding protein 1a | creb1a | D, V | A | Dworkin et al., 2007 | Neither | AB atlas |
| cell adhesion molecule 2a | cadm2a | D (L>R), V? | La, A | Pietri et al., 2007 | M | AB atlas |
| choline acetyltransferase | chatb | D (R>L), V | La, A | Hong et al., 2013 | M | Lauterborn et al., 1993 |
| chemokine (C-X-C motif), receptor 4b | cxcr4b | D | La | Carl et al., 2007; Roussinge et al., 2009 | Neither | AB atlas |
| collapsin response mediator protein | crmp1 | D | La | Jayasena et al., 2011 | M | AB atlas |
| contactin 2 | cntn2 | D (R>L), V? | La | Carl et al., 2007 | Neither | AB atlas |
| cytochrome P450, family 19, subfamily A, polypeptide 1 | cyp19a1b | D, V | A | Goto-Kazeto et al., 2004 | L | Shinoda et al., 1989 |
| developing brain homeobox 1b | dbx1b | D, V? | La | Wu et al., 2014 | Neither | AB atlas |
| ets variant gene 1 | etv1 | D (L>R) | La | Roussinge et al., 2006 | M | Quina et al., 2009 |
| extracellular leucine-rich repeat and fibronectin type III domain containing 1b | elfn1b | D (L>R) | La | Sollner and Wright, 2009 | M, L | Dolan and Mitchell, 2013 |
| F-box protein 7 | fbxo7 | D, V | A | Zhao et al., 2012 | M | AB atlas |
| follistatin-like 4 | fstl4 | D, V? | A | Tsuchimoto et al., 2005 | Neither | AB atlas |
| G protein-coupled receptor 161 | gpr161 | D, V? | La | Leung et al., 2008 | Neither | AB atlas |
| glutamate receptor, ionotropic, AMPA 1a,b | gria1a,b | D?, V | La | Hoppmann et al., 2008 | M, L | AB atlas |
| glutamate receptor, ionotropic, AMPA 4a,b | gria4a,b | D | La | Hoppmann et al., 2008 | M, L | AB atlas |
| guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O, a | gnao1a | D, V? | La | Huang et al., 2012 | M | AB atlas |
| guanine nucleotide binding protein, gamma 8 | gng8 | D, V | La | This study | M | Betty et al., 1998 |
| glutamate receptor, metabotropic 3 | grm3 | D (R>L) | La | Haug et al., 2013 | M, L | Ohishi et al., 1993 |
| glutamate receptor, metabotropic 4 | grm4 | D, V? | La | Haug et al., 2013 | Neither | AB atlas |
| glutamate receptor, metabotropic 6a | grm6a | D | La | Haug et al., 2013 | Neither | AB atlas |
| glutamate receptor, metabotropic 6b | grm6b | D (R>L) | La | Haug et al., 2013 | Neither | AB atlas |
| glutamate receptor, metabotropic 7 | grm7 | D | La | Haug et al., 2013 | L | Ohishi et al., 1995 |
| glutamate receptor, metabotropic 8b | grm8b | V | La | Haug et al., 2013 | Neither | AB atlas |
| 5-hydroxytryptamine (serotonin) receptor 1bd | htr1bd | D (L>R), V? | La, A | Norton, 2008 | Neither | AB atlas |
| hypocretin receptor | hcrtr | D | La, A | Appelbaum et al., 2009 | Neither | AB atlas |
| Kallmann syndrome 1a sequence | kal1a | D, V | A | Ayari et al., 2012 | NA | |
| Kallmann syndrome 1b sequence | kal1b | D?, V | A | Ayari et al., 2012 | NA | |
| kisspeptin1 | kiss1 | V | A | Kitashi et al., 2009; Ogawa 2012 | Neither | AB atlas |
| kisspeptin1 receptor | kiss1r | V | A | Servili et al., 2011 | M | AB atlas |
| Kruppel-like factor 4a | klf4a | V | La | Li et al., 2011 | Neither | AB atlas |
| leptin receptor | lepr | D, V | A | Liu et al., 2010 | Neither | AB atlas |
| platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit b | pafah1b1b | D | La | Drerup et al., 2010 | M, L | AB atlas |
| melanin-concentrating hormone receptor 1a | mchr1a | D, V | A | Berman et al., 2009 | Neither | AB atlas |
| monoamine oxidase | mao | D, V | A | Anichtchik et al., 2006 | M, L | AB atlas |
| muscleblind-like 3 (Drosophila) | mbnl3 | V | La | This study | Neither | AB atlas |
| nudE nuclear distribution gene E homolog like 1 (A. nidulans) A | ndel1a | D | La | Drerup et al., 2007 | M, L | AB atlas |
| neuropilin 1a | nrp1a | D (left only) | La | Kuan et al., 2007 | Neither | AB atlas |
| neuronal pentraxin 2a | nptx2a | D (L>R) | La, A | Agetsuma et al., 2010 | M | AB atlas |
| neuronal pentraxin 2b | nptx2b | D?, V | A | Appelbaum et al., 2010 | See nptx2a | |
| nuclear receptor subfamily, group A, member 2 | nr4a2 | D, V? | La | Blin, 2008 | M, L | Quina et al., 2009 |
| opsin 4xa | opn4xa | D | La | Matos-Cruz et al., 2011 | No homolog | |
| purkinje cell protein 4a | pcp4a | D, V | La, A | Mione et al., 2006 | M, L | AB atlas |
| PTEN induced putative kinase 1 | pink1 | D, V | A | Sallinen et al., 2010 | M, L | Taymans et al., 2006 |
| potassium channel tetramerization domain 12.1 | kctd12.1 | D (L>R) | La, A | Gamse et al., 2003 | M | Gamse et al., 2005 Metz et al., 2011 |
| potassium channel tetramerization domain 12.2 | kctd12.2 | D (R>L), V | La, A | Gamse et al., 2005 | See kctd12.1 | |
| potassium channel tetramerization domain 8 | ktcd8 | D (R>L) | La, A | Gamse et al., 2005 | M | Gamse et al., 2005 Metz et al., 2011 |
| POU domain, class 4, transcription factor 1 | pou4f1 | D (R>L) | La, A | Amo et al., 2010 | M, L | Quina et al., 2009 |
| prokineticin 2 | prok2 | D, V | A | Ayari et al., 2010 | M, L | Cheng et al., 2006 |
| protocadherin-10 | pcdh10a | V | A | Amo et al., 2010 | L | Aizawa et al., 2012 |
| purkinje cell peptide 4 | pcp4a | D, V | La, A | Mione et al., 2006 | M, L | AB atlas |
| reticulon 4 receptor | rtn4r | D, V? | La | Brösamle et al., 2009 | M | Josephson et al., 2002 |
| roundabout homolog 3 | robo3 | D, V? | La | Sollner and Wright, 2009 | M | Quina et al., 2009 |
| si:ch211-154o6.4 | D (R>L), V? | La | Amo et al., 2010 | No homolog | ||
| solute carrier family 5 (choline transporter) member 7 | slc5a7; hacta | D (R>L) | La, A | Hong et al., 2013 | M | AB atlas |
| solute carrier family 18 (vesicular acetylcholine), member 3b | slc18a3; vachtb | D (R>L), V | La, A | Hong et al. 2013 | M | Quina et al., 2009 |
| solute carrier family17 (sodium- dependent inorganic phosphate cotransporter), member 6a | slc17a6; vglut2b | D, V | La, A | Appelbaum 2009 | M, V | Aizawa et al., 2012 |
| solute carrier family17 (sodium- dependent inorganic phosphate cotransporter), member 6b | slc17a6; vglut2a | D, V | La, A | Bae et al., 2009; Appelbaum 2009 | M, V | Aizawa et al., 2012 |
| solute carrier family17 (sodium- dependent inorganic phosphate cotransporter), member 7 | slc17a7; vglut1 | D (L>R) | A | Bae et al., 2009 | M | Aizawa et al., 2012 |
| spondin 1b | spon1b | D, V | La, A | Gamse et al., 2003; Akle et al., 2012 | M | AB atlas |
| synuclein, gamma b | sncgb | D (L>R) | La, A | Milanese et al., 2012 Chen et al., 2009 | M | Quina et al., 2009 |
| tachykinin 1 | tac1 | D (R>L) | A | Hong et al., 2013 | M | Quina et al., 2009; Aizawa et al., 2012 |
| tachykinin 3a (formerly tac2a) | tac3 | D (R>L), V? | La, A | Biran et al., 2012; Ogawa et al., 2012 | No homolog | AB atlas |
| transient receptor potential cation channel, subfamily C, member 1 | trpc1 | D (L>R) | La | von Niederhäusern et al., 2013 | M, L | AB atlas |
| transient receptor potential cation channel, subfamily C, member 5a | trpc5a | D, V? | La | von Niederhäusern et al., 2013 | M, L? | Fowler et al., 2007 |
| transient receptor potential cation channel, subfamily M, member 3 | trpm3 | D, V | La | Kastenhuber et al., 2013 | M | Oberwinkler et al., 2005 |
| unc-51-like kinase 2 | ulk2 | D (R>L), V | La | Taylor et al., 2011 | Neither | AB atlas |
| uncoupling protein 2 | ucp2 | D, V | A | Tseng et al., 2011 | Neither | AB atlas |
| vertebrate ancient long opsin b | valopb | D, V? | La | Kojima et al., 2008 | Neither | AB atlas |
| wingless-type MMTV integration site family, member 4a | wnt4a | D | La | Hendricks et al., 2008 | Neither | AB atlas |
FIG. 1. Bilaterally symmetric and left-right asymmetric gene expression in the larval habenular nuclei.
(a) Symmetric expression of the ano2 gene delineates the boundaries of the dorsal habenular nuclei, whereas (b) aoc1 (Amo et al., 2010) and (c, d) mbnl3 transcripts localize to the ventral habenular nuclei. (e, f) Symmetric expression of gng8 encompasses the entire dorsal habenulae, and is found in a subset of cells in the ventral nucleus (arrowheads). (g) The kctd12.1 and (i,) kctd8 genes show predominant expression in the left or right side, respectively (Gamse et al., 2005), which is confined within the boundaries of the dorsal nuclei (h, j). (k) kctd12.2 transcripts are more abundant in the right dorsal nucleus (Gamse et al., 2005), and (l) also found bilaterally in the ventral nuclei (arrowheads). All larvae are 4 dpf.
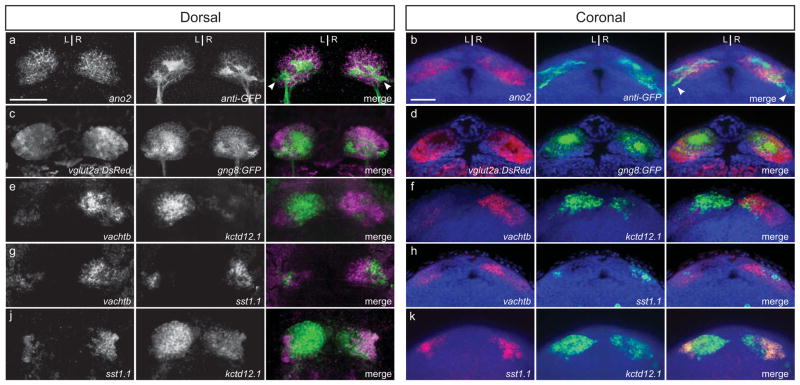
FIG. 2. L-R asymmetric cholinergic and peptidergic subdomains of the larval dorsal habenulae.
(a, b) gng8:GFP labeling detected by anti-GFP immunofluorescence is coextensive with ano2 in the dorsal habenular nuclei. The membrane-tagged GFP also labels a subpopulation of neurons in the ventral nucleus (arrowheads). (c, d) vglut2a:DsRed labels neurons throughout the dorsal and ventral habenulae. (e, f) vachtb and kctd21.1 are expressed in different neuronal populations, which are more intermingled in the right habenula than the left. (g, h) vachtb is transcribed in a non-overlapping pattern with sst1.1-expressing neurons located in a lateral subregion of the dorsal habenula. (i, j) The sst1.1-expressing cells are confined within the kctd12.1 domain and are more abundant in the right dorsal habenula than the left. All larvae are 4 dpf. Scale bars are 30 μm.
Additionally, a number of genes show L-R asymmetric expression in the dorsal habenular nuclei (e.g., Agetsuma et al., 2010; Aizawa et al., 2005; Amo et al., 2010; Gamse et al., 2005; Kuan et al., 2007; Taylor et al., 2011 and refer to Table 1). Members of the potassium channel tetramerization domain containing gene family were first discovered to exhibit L-R differences, with transcripts for kctd12.1 and kctd8 more extensive on the left and right sides, respectively (Gamse et al., 2005 and Fig. 1g, i). Double labeling confirms that expression of both genes is confined within the limits of the ano2-expressing dorsal habenular nuclei (Fig. 1h, j). kctd12.2 is not only expressed to a greater extent in the right dorsal nucleus (Fig. 1k), but also bilaterally in a subset of cells in the ventral habenular nuclei (Fig. 1l, arrowheads). Although gene expression patterns such as these appear to demarcate discrete subregions within the dorsal habenulae, how such asymmetric domains correlate with functional subnuclei is unclear.
Discrete asymmetric cholinergic and peptidergic subnuclei of the larval dorsal habenulae
In mammals, as described above, habenular subnuclei have been identified on the basis of neurotransmitter phenotype and neuronal connectivity. Recent work by Hong et al. (2013) demonstrated that the zebrafish larval habenula contains glutamatergic and cholinergic neurons as early as 4 days post-fertilization (dpf). We determined the position of these neuronal groups more systematically using the gng8:GFP transgenic background or with respect to the asymmetric expression of kctd12.1. In Tg(gng8:GFP) larvae, neurons labeled with membrane-tagged GFP are found throughout the dorsal habenulae (Fig. 2a, b) and more sparsely in the ventral nucleus (arrowheads). Co-labeling with vglut2a:DsRed,, indicates that these neurons are all glutamatergic (Fig. 2c, d). In the ventral nucleus, vglut2a:DsRed expression is more widespread than gng8:GFP.
Expression of the vesicular acetylcholine transporter b gene (vachtb) demarcates the L-R asymmetric cholinergic regions of the dorsal habenulae as well as the bilaterally symmetric ventral nuclei (Hong et al., 2013). Double fluorescence RNA in situ hybridization demonstrates that vachtb and kctd12.1 have complementary patterns of expression (Fig. 2e, f); thus, the kctd12.1-positive region corresponds to the non-cholinergic territory of the dorsal habenulae. In mammals, this non-cholinergic region contains substance P-expressing neurons. Transcripts for the zebrafish substance P precursor tachykinin (tac1) are not detected in the habenular region as early as 4 dpf, although they are present elsewhere in the brain (data not shown). However, we find that the neuropeptide encoding gene somatostatin1.1 (sst1.1) is expressed at this stage, in a lateral subdomain of the dorsal habenula (Fig. 2g, h). The sst1.1 neuronal population is located within the non-cholinergic kctd12.1-expressing territory and is larger on the right side of the brain than the left (Fig. 2i, j). Analyses of the combinatorial gene expression patterns in 4 dpf larvae, therefore, indicate that neurons of the dorsal habenulae are all glutamatergic, with a portion of them possessing either a cholinergic or sst1.1 identity. In the larval brain, the non-overlapping cholinergic and somatostatin neuronal populations are significantly larger in the right dorsal habenula compared to the left.
Asymmetric neurotransmitter-specific subpopulations are retained in the adult dorsal habenulae
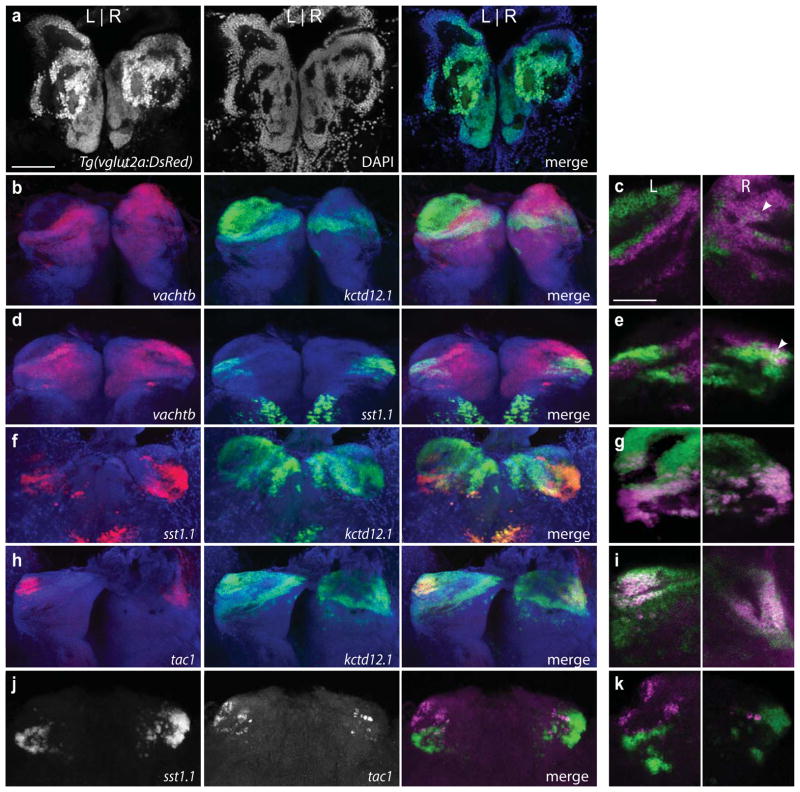
We next examined whether L-R asymmetric neuronal populations are maintained during the morphological changes that accompany formation of the adult zebrafish brain (refer to Amo et al., 2010). As in the larva, neurons throughout the dorsal and ventral habenular nuclei of adult zebrafish are glutamatergic as revealed by DsRed labeling from the vglut2a transgenic reporter (Fig. 3a). In the adult, the cholinergic territory is more extensive in the left habenula (Fig. 3b, c) compared to the larval stage (Fig. 2e, f). At the border, cholinergic and non-cholinergic cells are clearly distinct populations in the left dorsal nucleus, whereas some intermixing of vachtb and kctd12.1-expressing cells is observed in the right habenula (arrowhead, Fig. 3c). The sst1.1 and vachtb expression domains are also non-overlapping in the left habenula (Fig. 3d, e), although a subset of cells appears to express both genes in the right dorsal habenula (Fig. 3e, arrowhead). The neuropeptide encoding genes sst1.1 and tac1 are expressed by distinct neuronal populations within the kctd12.1-positive, non-cholinergic territory (Fig. 3f, g and 3h, i, respectively), (Fig. 3j, k). As in the larva, the sst1.1-expressing domain is larger in the right habenula (Fig. 3f, j). Overall, the cholinergic and peptidergic territories are preserved between the larval and adult stages; however, some neurons in the right habenula exhibit an overlap in sst1.1 and vachtb expression that was not detected at the larval stage.
FIG. 3. Neurotransmitter subpopulations of the adult dorsal habenulae.
(a) Tg(vglut2a:DsRed) is expressed throughout the dorsal and ventral habenulae. b, d, f, h, j are z-stack maximum projections of the dorsal habenula. c, e, g, i, k are magnified views of a single focal plane. (b) Complementary expression pattern of vachtb and kctd12.1 (z volume = 56 μm) with (c) no overlap on the left but some intermingling of cells in the right nucleus (arrowhead). (d) vachtb and sst1.1 are mainly expressed in different neuronal populations (z volume = 53 μm) except for (e) a small subset of co-expressing cells in the right habenula (arrowhead). (f, g) sst1.1 is mainly co-expressed within the kctd12.1 domain (z volume= 69 μm). (h, i) tac1 is also co-expressed with kctd12.1, (z volume= 64 μm) (j) sst1.1 and tac1 are transcribed in distinct neuronal populations (z volume= 14 μm). All images are of coronal sections and, except for Figs. j, k, were stained with DAPI, represented in the blue channel. Scale bar is 100 μm for a, b, d, f, h, j and 50 μm for c, e, g, i, k.
Partial correspondence of neurotransmitter-expressing subdomains with habenular transgenic reporters
Zebrafish transgenic lines that label habenular neurons with fluorescent proteins have been used to define dorsal habenular subregions, to trace axonal connections to the IPN, and in behavioral studies following neuronal ablation or inactivation (Agetsuma et al., 2010; Aizawa et al., 2005; Lee et al., 2010). Notably, Tg(brn3a:GFP) was designated as labeling the dHbM subnucleus and Tg(nptx2:Gal4) as driving expression in the dHbL (Agetsuma et al., 2010; Aizawa et al., 2005; Okamoto et al., 2012). After establishing the gene expression patterns and neurotransmitter identity of habenular subregions, we sought to determine how this regionalization corresponds with habenular labeling by these transgenes.
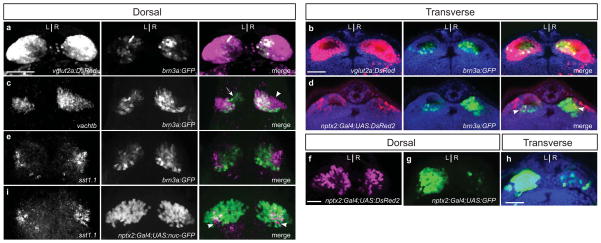
We mated Tg(brn3a:GFP) and Tg(vglut2:DsRed) carriers and observed that all GFP-positive cells in the habenulae of their larval progeny co-express DsRed (Fig. 4a, b), indicating that brn3a:GFP-positive cells are glutamatergic. In the dorsal habenulae of adults, the brn3a:GFP labeled region was found to overlap only partially with the cholinergic territory as defined by vachtb expression (Hong et al., 2013). We observed a similar pattern at the larval stage, with distinct neuronal populations that were either co-labeled, vachtb positive/brn3a:GFP negative (Fig. 4c, arrowhead) or vachtb negative/brn3a:GFP positive (Fig. 4c, arrow). Thus, the dorsal habenular subregion labeled by the brn3a:GFP transgene does not fully encompass a cholinergic subnucleus. Although the brn3a-GFP labeled neurons were designated as a distinct subnucleus in the adult habenula from that labeled by Tg(nptx2:Gal4); Tg(UAS:DsRed2) (Agetsuma et al., 2010), some neurons co-label with both fluorescent proteins in the larval dorsal habenula (Fig. 4d, arrowheads).
FIG. 4. Partial correspondence of neurotransmitter subdomains and habenular transgenic reporters.
(a, b) Coextensive labeling of habenulae by brn3a:GFP and vglut2:DsRed indicates that brn3a:GFP-positive cells are glutamatergic. (c) Detection of vachtb RNA by in situ hybridization and brn3a:GFP by anti-GFP immunolabeling reveals vachtb-positive/brn3a:GFP-negative (arrowhead), vachtb-negative/brn3a:GFP positive (arrow) as well as region of co-expression. (d) nptx2:Gal4;UAS:DsRed2-labeled neurons have areas of overlap at the boundaries of the brn3a:GFP-labeled population (arrowheads) (e) sst1.1-expressing neurons are distinct from the brn3a:GFP-labeled population, which were detected by anti-GFP immunolabeling. (f) The nptx2:Gal4;UAS:DsRed2 transgene labels neurons sparsely in the left and right habenulae. (g, h) 4xUAS:GFP activated by nptx2:Gal4 labels relatively more neurons in the left and less in the right dorsal habenular nucleus compared with UAS:DsRed2. (i) sst1.1 expression is confined to the lateral subregions of the nptx2:Gal4;14xUAS:GFP dorsal habenular domain (arrowheads) detected by anti-GFP immunolabeling. All larvae are 4 dpf. Scale bars are 30 μm.
Consistent with their location in the kctd21.1 positive non-cholinergic territory, sst1.1-expressing neurons were not co-labeled by the brn3a:GFP transgene (Fig. 4e). These cells were found in a similar region of the dorsal habenula as neurons labeled by the nptx2:Gal4 and UAS:DsRed2 transgenes. Upon determining whether the sst1.1 neurons co-express nptx2:Gal4, we unexpectedly found that the extent of labeling varied significantly depending on the UAS-regulated transgenic reporter that was activated by this Gal4 driver (compare Fig. 4f and g, h). However, despite the variability in labeling between lines, only a subset of sst1.1-expressing neurons were co-labeled by the nptx2:Gal4 driver for the three UAS-regulated reporters tested (Fig. 4i). Thus, expression driven by Tg(nptx2:Gal4) does not represent a discrete somatostatin neuronal population.
Habenular connectivity with the IPN
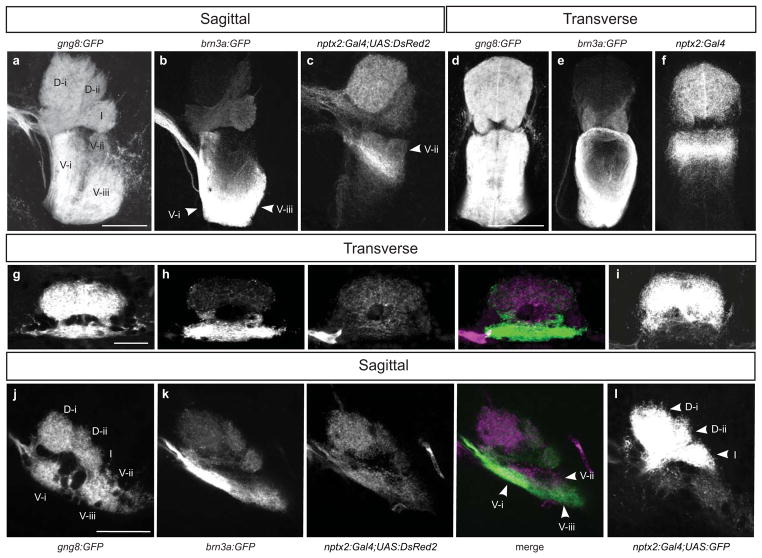
Although transgenic labeling in the dorsal habenulae does not appear to correspond to specific neuronal populations defined by neurotransmitter/neuropeptide expression, it is still useful for examining connectivity with the IPN. In zebrafish larvae and adults, the gng8:GFP transgene labels all dorsal habenular neurons (Fig. 2a) and their GFP-positive axons that terminate throughout the IPN (Fig. 5a, d, g, j). Because GFP is membrane-tagged in this line, discrete axonal densities can be recognized in a stereotypic pattern in sagittal sections of the adult IPN (Fig. 5a), with a minimum of 2 dorsal (D-i and D-ii) and 3 ventral clusters (V-i, V-ii and V-iii). An intermediate (I), horseshoe-shaped zone of innervating fibers separates the dorsal and ventral axonal bundles. The stereotypic morphology of axonal bundles at the IPN is already apparent at 4 dpf (Fig. 5j).
FIG. 5. Habenular connectivity with the IPN revealed by transgenic reporters.
gng8:GFP labeled habenular axons terminate in discrete, stereotypic bundles in the dorsal (D), intermediate (I) and ventral (V) IPN of (a, d) adult and (g, j) larval brains. (b, e) In the adult brain, brn3a:GFP labeled dorsal habenular neurons predominantly contribute to the D-ii, intermediate and V-i and V-iii bundles and (c, f) nptx2:Gal4;UAS:DsRed2 labeled axons terminate in the D-i, D-ii, intermediate and V-ii IPN bundles. (h, k) In the 4 dpf larval brain, IPN innervation patterns are similar to the adult when the UAS:DsRed2 reporter is used. (i, l) However, with Tg(4xUAS:GFP) regulated by the nptx2:Gal4 driver, labeled fibers preferentially innervate the dorsal and intermediate IPN. Scale bar is 100 μm for a–f and 30 μm for g–l.
Using Tg(brn3a:GFP) zebrafish, it had been previously shown that GFP labeled habenular axons predominantly terminate in the intermediate and ventral regions of the adult IPN (Aizawa et al., 2005) and axons labeled by Tg(nptx2:Gal4);Tg(UAS:DsRed2) innervate the dorsal, intermediate and ventral IPN (Agetsuma et al., 2010). We corroborated these results and found that, more specifically, in the ventral IPN of adult zebrafish, the brn3a:GFP terminals primarily contribute to the V-i and V-iii axon bundles (Figs. 5b, e) and those labeled by the nptx2:Gal4 driver and UAS:DsRed2 reporter innervate the V-ii bundle (Figs. 5c, f).
By mating Tg(brn3a:GFP) and Tg(nptx2:Gal4);Tg(UAS:DsRed2) carriers, the relative extent of GFP and DsRed2 axon terminals along the dorsoventral IPN was examined in transgenic larvae. At 4 dpf, a largely similar pattern of innervation was found in IPN subregions as in the adult (Fig. 5h, k). Because neurons in the left habenula were more extensively labeled when the nptx2:Gal4 transgenic driver was used to drive 4xUAS:GFP (Fig. 4g, h), we compared the profiles of innervating axons at the IPN using the two fluorescent reporter lines. Habenular efferents were weakly labeled by the UAS:DsRed2 transgene in subregions of the dorsal and ventral IPN (Fig. 5h, k). Significantly more GFP-labeled axon terminals were observed in the D-i, D-ii and, I regions with the 4xUAS:GFP line (Fig. 5i, l), consistent with this reporter being activated to a greater extent in the left habenula (Fig. 4g, h).
Because brn3a:GFP labels many cholinergic neurons but not sst1.1-expressing neurons, and the axons of brn3a:GFP positive neurons predominantly innervate the D-ii, I, V-i and V-iii regions of the IPN, we expect that these IPN regions receive cholinergic input. With only partial overlap between sst1.1- and nptx2:Gal4-expressing populations, it is difficult to infer the precise pattern of connectivity of this neuronal subgroup with certainty; however, efferents from the somatostatin neurons are expected to contribute to some of the same subregions of the IPN as the neuronal populations labeled by the nptx2 driver (Fig. 5k, l).
DISCUSSION
In the nervous system, it is generally assumed that discrete clusters of neurons exhibiting the same neurotransmitter phenotype fasciculate to innervate specific target regions and function together. In addition to their shared expression of particular neurotransmitters or neuropeptides, brain nuclei or their component subnuclei are defined by neuroanatomical features such as cell cluster size, density of cell packing, neuropil structure or stereotypic position. The significance of nuclear designations is especially relevant when modern genetic methods are employed to eliminate or modify brain regions for insights into their roles in physiology and behavior. These approaches have been applied to the study of the habenular nuclei in mammalian and non-mammalian systems (Agetsuma et al., 2010; Lee et al., 2010; Quina et al., 2009; Yamaguchi et al., 2013); however, detailed information about the precise subregions that are affected would allow more rigorous functional correlations.
The goal of this study was to generate a consolidated map of the habenular region in larval and adult zebrafish, by reexamining the expression of widely used genetic markers, describing the spatial expression patterns of newly identified genes, and determining how this information corresponds with neurotransmitter profiles and cell populations labeled by transgenic reporters. In addition, we further characterized the extent of L-R asymmetry in the dorsal habenular nuclei.
Designation of habenular subnuclei
The dorsal habenulae in the adult zebrafish brain have been subdivided into medial and lateral subnuclei on the basis of Tg(brn3a:GFP) labeling and by expression of kctd12.1 or directed by the Tg(nptx2:Gal4) driver (Agetsuma et al., 2010; Aizawa et al., 2005). The designation of the medial subdivision is further supported by experiments showing that brn3a:GFP positive and negative neurons have different birthdates, and that L-R differences in the timing of neurogenesis later contributes to the formation of asymmetric brn3a-labeled populations. Several lines of evidence, however, suggest that the regions defined by these transgenic tools may not represent discrete, functional subnuclei. Although initially defined as corresponding to distinct subnuclei, the kctd12.1 and brn3a domains do not appear to be completely segregated in the adult dorsal habenulae (Aizawa et al., 2005). In addition, the reported patterns of brn3a:GFP and nptx2:Gal4; UAS:DsRed2 labeled neurons fail to recapitulate endogenous gene expression, which appears more widespread in the habenular region for both genes (Agetsuma et al., 2010; Aizawa et al., 2005; Aizawa et al., 2007). Partial overlap in labeling from the brn3a:GFP and nptx2:Gal4 transgenes in the larval habenula further suggests that expression of these markers does not correspond to discrete subnuclei. While the brn3a:GFP population significantly overlaps with vachtb-expressing neurons in the larval dorsal habenula, a subset of cholinergic neurons are not labeled by this transgene. Similarly, the region of the dorsal habenulae labeled by the nptx2:Gal4 transgenic driver only partially overlaps with somatostatin1.1-expressing neurons. Together, these findings suggest that caution should be taken in interpreting brn3a:GFP and nptx2:Gal4; UAS:DsRed2 transgenic labeling as representative of functional habenular subnuclei at least for larval stages.
Distinct neuronal populations in the zebrafish dorsal habenula defined by neurotransmitter identity
The significance of subdomains of gene expression in the zebrafish habenulae is largely unknown. As a first step, we sought to determine how they might correlate with specific neuronal subgroups by examining the expression of zebrafish homologues for genes indicative of neurotransmitter identity. This approach avoids potential problems associated with using antibodies that are typically generated against mammalian antigens. We examined larval and adult stages, as some neurons are known to undergo changes in neurotransmitter phenotype over time (e.g., Landis and Keefe, 1983; Spitzer, 2012) and new neuronal subgroups may also arise.
The analysis of larval zebrafish revealed that cholinergic neurons are a distinct population from cells expressing kctd12.1, which encodes a protein shown to associate with gamma-aminobutyric B (GABAB) receptors to influence their pharmacology and signaling properties (Ivankova et al., 2013; Schwenk et al., 2010) and to promote formation of dense neuropil in the zebrafish dorsal habenula (Taylor et al., 2011). Instead, we found that the kctd12.1 domain contains somatostatin1.1-expressing peptidergic neurons.
All neurons of the zebrafish dorsal and ventral habenulae are glutamatergic. In the asymmetric dorsal habenulae of larval zebrafish, the left habenular nucleus is mainly composed of a glutamatergic-only subregion with additional small cholinergic and sst1.1 clusters. In contrast, the right nucleus largely consists of expanded cholinergic and sst1.1 subregions.
During maturation of the zebrafish brain to its adult form, the presumptive ventral nuclei become repositioned ventromedially (Amo et al., 2010), but the relative positions of subnuclear territories within the dorsal nuclei largely appear the same. Left-right differences in the size of neurotransmitter expressing populations, however, are less pronounced. Notably, at 4 dpf there is only a small cluster of cholinergic neurons in the left dorsal habenula, but by adulthood the proportion of neurons that express cholinergic genes is more similar between the left and right nucleus. In adults, both habenula also acquire a tac1-expressing subdomain (Hong et al., 2013), in addition to the cholinergic, sst1.1 and glutamatergic-only subregions.
Curiously, while the boundaries between these four neuronal populations are discrete in the left dorsal habenula, they are not as well demarcated on the right, due to intermixing of neurons expressing different neurotransmitters. It is the left habenula of zebrafish that receives preferential innervation from the parapineal organ, an accessory to the pineal organ, which in most individuals is asymmetrically positioned to the left of the pineal. The parapineal influences the gene expression and neuroanatomical properties of the left dorsal habenula that distinguish it from the right (Concha et al. 2003; Gamse et al., 2003). It is tempting to speculate that parapineal neurons may also serve to refine subnuclear organization, ensuring the formation of precise boundaries between peptidergic and cholinergic clusters in the left habenula. In certain frogs, the photoreceptive frontal organ sends asymmetric projections across the left habenular nucleus (Eldred et al., 1980; Guglielmotti and Cristino, 2006; Kemali and De Santis, 1983) and the left habenula contains distinct cholinergic and substance P subnuclei (Kemali and Guglielmotti, 1984; Marin et al., 1997). The right habenula also has cholinergic and substance P neurons (Kemali and Guglielmotti, 1984; Marin et al., 1997); however, as in zebrafish, the boundaries between these neuronal populations are not as clearly delineated as on the left. In mammals, where there is no structure analogous to the parapineal, deep pineal neurons project to both medial habenular nuclei (Korf et al., 1990), and both exhibit distinct cholinergic and substance P subnuclei (Cuello et al., 1978). Whether innervating parapineal or pineal neurons play a role in the refinement of the habenular map into discrete subnuclear compartments remains to be demonstrated experimentally.
From the analysis of neurotransmitter and neuropeptide producing neuronal populations, we present a schematic map of the dorsal habenulae of larval and adult zebrafish (Fig. 6a). It is important to stress that this is only a preliminary map, as it is highly likely that additional neuropeptides or transmitters are co-expressed with glutamatergic neurons and located in other subregions of the dorsal habenulae. For example, cells immunoreactive for thyrosine hydroxylase important in L-DOPA synthesis have been identified in the medial habenulae of alpaca (Marcos et al., 2013) and mRNA encoding orphanin FQ as well as this neuropeptide were localized to the rat medial habenulae (Neal et al., 1999). Indeed, as indicated in Table 1, genes encoding tachykinin 3a (Biran et al., 2012; Ogawa et al., 2012) and receptors for glutamate (Haug et al., 2013), serotonin (Norton et al., 2008) and hypocretin (Appelbaum et al., 2009) show enriched expression in subregions of the zebrafish dorsal habenulae, suggestive of other neuronal specializations.
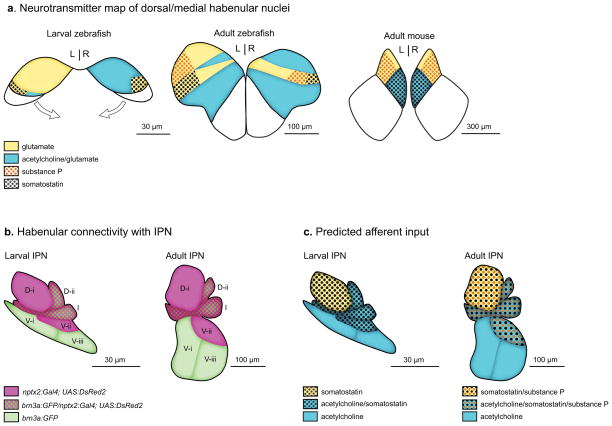
FIG. 6. Model of dorsal habenular neurotransmitter populations and their IPN connectivity.
(a) Transverse views of the dorsal/medial habenular nuclei. Prominent L- R differences in neurotransmitter distribution in the dorsal habenulae of larval zebrafish become less pronounced in the adult brain. In adult brains, the overall organization of glutamatergic, cholinergic and peptidergic populations is conserved between the dorsal habenula of zebrafish and medial habenula of the mouse (based on neuroanatomical data from Quina et al., 2009 and the Allen Mouse Brain Atlas). However, somatostatin and cholinergic neurons are in distinct populations in the zebrafish habenulae, which has not been reported for rodents. (b, c) Sagittal views of stereotypic axon terminal bundles at the interpeduncular nucleus of larval and adult zebrafish. In (b), habenular axon terminals labeled by either the brn3a:GFP or nptx2:Gal4 transgenes are mostly enriched in different subregions of the IPN, (c) Predicted afferent input to the IPN. The projections of cholinergic neurons are limited to the ventral and intermediate IPN; whereas those of peptidergic (sst1.1 and tac1) neurons are expected to be found at the dorsal IPN, intermediate IPN and/or V-ii region of the ventral IPN.
Designating subnuclei in the zebrafish habenulae on the basis of neurotransmitter identity enables a more direct comparison with mammalian brains, in efforts to uncover the habenular subregions and circuitry modulating a wide variety of behaviors. In rodents, the medial habenulae are bilaterally symmetric and contain glutamatergic neurons (Barroso-Chinea et al., 2007; Qin and Luo, 2009), which have been subdivided into dorsal and ventral subnuclei depending on whether they utilize substance P or acetylcholine (Aizawa et al., 2012; Contestabile et al., 1987; Cuello et al., 1978; Lecourtier and Kelly, 2007; Quina et al., 2009). Recent work reveals an additional glutamatergic-only subnucleus, which is found medially to the substance P subnucleus (Aizawa et al., 2012). The location and organization of subnuclei designated by neurotransmitter phenotype in the dorsal habenulae of adult zebrafish is strikingly similar to that of the rodent medial habenulae. An exception is the presence of somatostatin neurons, which has not yet been described for the rodent medial habenula. Somatostatin-immunopositive fibers are found only in the lateral habenulae of the postnatal rat (Shiosaka et al., 1981). However, images from the Allen Brain Atlas indicate that the sst gene is transcribed in neurons in the medial habenula of the mouse, in the same region where cholinergic neurons are located. Further studies are needed to ascertain the location of these neurons relative to the cholinergic population, in order to determine whether there is a distinct somatostatin-producing subnucleus as in the zebrafish dorsal habenula.
Connectivity of dorsal habenular subnuclei with the IPN
Neurons of the medial habenula of mammals project their axons through the fasciculus retroflex to terminate at the interpeduncular nucleus (refer to Kappers et al., 1936; Sutherland, 1982)). From the accumulations of terminating fibers, the structure of the IPN has been well defined in rodents as consisting of 3 unpaired (rostral, apical, central) and 4 paired (intermediate, lateral, rostral lateral, dorsal lateral) subdivisions in a consensus nomenclature put forward by Lenn and Hamill (1984) Lesioning studies, histochemistry and immunoreactivity have revealed the presence of a wide variety of neurotransmitters and neuropeptides in axons terminating within particular subdivisions of the IPN (reviewed by Morley, 1986).
Visualization of a transgenic line with expression of membrane-tagged GFP under the control of gng8 regulatory sequences (deCarvalho et al., 2013) provides a complete picture of innervation of the zebrafish IPN by dorsal habenular neurons. As with previous dye labeling and immunolabeling experiments (Aizawa et al., 2005; Gamse et al., 2005; Tomizawa et al., 2001), the axon terminals of habenular neurons can be readily distinguished as contributing to bundles in dorsal, intermediate and ventral regions of the larval and adult IPN. However, because our understanding of the zebrafish is far from complete, it is premature to assign a nomenclature analogous to that proposed for mammals (Lenn and Hamill, 1984). We, therefore, designated IPN afferent input as consisting of two dorsal (D-i and D-ii) and three ventral (V-i, V-ii, and V-iii) axonal densities and an intermediate fiber bundle (I) that separates them on the basis of the stereotypic morphology of GFP labeled afferents (and refer to Fig. 11 in Agetsuma et al., 2010). Moreover, we find that the pattern of innervating fibers is already in place in the IPN of 4 dpf larvae. This simplified naming strategy must be modified as more information is obtained about the precise homology with mammalian IPN subdivisons and the distribution of synaptic terminals expressing different neurotransmitters.
Previous work had demonstrated that that brn3a:GFP and nptx2:Gal4 transgenic lines primarily label different neuronal populations in the dorsal habenulae of adult zebrafish, which innervate largely distinct dorsoventral regions of the IPN (Agetsuma et al., 2010). Overall, our findings support this connectivity map for the adult, with the potential for overlapping input in the D-ii and I IPN subregions (shown in Fig. 6b). In 4 dpf larvae, these transgenes label innervating fibers throughout the IPN, although they are more concentrated in certain subregions at this early stage. For example, brn3a:GFP labeled axons predominate in the V-i density and to a lesser extent in V-iii. Because the pattern of neuronal labeling in the larval habenula depends on the UAS reporter under nptx2:Gal4 control, the location of labeled axons also varied between lines. With the UAS:DsRed2 reporter that labeled neurons sparsely in both habenulae, axonal endings were enriched in the D-i and V-ii bundles, relative to the brn3a:GFP labeled terminals. However, when a significantly larger group of neurons in the left habenula was labeled by 4xUAS:GFP, innervating axons were primarily detected in the D-i, D-ii and I regions of the IPN.
Differences in the pattern of IPN innervation between larval and adult stages, such as the strongly labeled V-ii terminal bundle present in nptx2:Gal4 transgenic adults or the increase in brn3a:GFP labeled terminals in the V-iii region, are likely the outcome of differential neurogenesis in subregions of the developing dorsal habenular nuclei and continued axonal outgrowth to target regions. Understanding how and when new neuronal populations arise, such as the expanded cholinergic cluster on the left or the bilateral tac1-expressing groups, will help clarify how the habenulo-interpeduncular connectivity map evolves over time.
We can predict the most likely sites of cholinergic and peptidergic afferent input to the IPN from the partial overlap between neurotransmitter expression and transgene labeling in dorsal habenular neurons. As depicted schematically in Fig. 6c, cholinergic neurons are expected to terminate in the D-ii, I and V IPN regions, owing to the majority of them being labeled by brn3a:GFP. Based on partial overlap with the nptx2:Gal4 driver and lack of co-expression with brn3a:GFP, the contribution of sst1.1-expressing neurons would be limited to the dorsal IPN, intermediate IPN, and V-ii region of the ventral IPN. A similar profile is expected for substance P neurons in the adult brain based on the location of tac1-expressing neurons within the brn3a:GFP negative region (Hong et al., 2013). While the proposed connectivity map provides a useful guide, it is an oversimplification that rests on indirect inferences and will need to be validated by the production and analysis of transgenic lines that selectively label each neurotransmitter-expressing population of the dorsal habenulae.
L-R asymmetry in IPN connections
The value of having a framework to classify afferent input is that it helps resolve a discrepancy concerning how left and right habenular neurons connect with the IPN in the zebrafish brain. One model suggests that the left habenula primarily projects to the dorsal IPN and the right habenula innervates the ventral IPN in a laterotopic manner (Aizawa et al., 2005; Bianco et al., 2008). Results from dye labeling, immunolabeling and transgenic labeling of habenular axons, however, indicate a more complicated scenario with differing proportions of neurons from the left and right sides of the brain contributing to both the dorsal and ventral IPN (Agetsuma et al., 2010; Beretta et al., 2012; Gamse et al., 2005; Kuan et al., 2007; Okamoto et al., 2012). For the right habenula of the larval brain, we hypothesize that the significantly larger cholinergic subnucleus innervates the ventral IPN. In the left habenula, the smaller cholinergic neuronal cluster would also project to the ventral IPN while the larger population of non-cholinergic neurons would innervate the dorsal IPN.
This revised model can explain two previous puzzling observations. First, when the parapineal is ablated and the left dorsal habenula adopts the molecular profile and neuroanatomical properties of the right dorsal habenula, the vast majority of habenular efferents project to the ventral IPN (Gamse et al., 2005; Kuan et al., 2007). These projections are presumed to emanate from the large cholinergic subnucleus that, following loss of the parapineal, is present in the left as well as the right dorsal habenula (K. Santhakumar, T. deCarvalho and M.E. Halpern, unpublished observations). However, a small axonal bundle remains at the dorsal IPN (Gamse et al., 2005) that may represent the efferents of peptidergic neurons present in both habenulae. Second, a study analyzing the projections of individual neurons described distinct axonal morphologies for cells labeled in the left dorsal habenula versus the right (Bianco et al., 2008). Axons with the left morphology (L-typical) exhibit a greater degree of arborization and innervate an extensive region of the IPN and those with the right morphology (R-typical) show less axonal branching and innervate a more dorsoventrally restricted region. Even though neurons with a L-typical arbor predominate in the left habenula and those with a R-typical arbor are the majority in the right habenula, neurons having the opposite axonal morphology were also observed (Bianco et al., 2008). We propose that the subset of neurons in the right habenula with L-typical arbors contain the sst-1 subpopulation that innervates the dorsal IPN; whereas neurons in the left habenula with R-typical arbors correspond, in part, to the small cholinergic group that projects ventrally. In this model, neurons from both sides of the brain that utilize the same neurotransmitter innervate the same regions of the IPN.
The neurotransmitter profile of the zebrafish dorsal habenular nuclei serves as a useful guide to understand habenular function, but also points to many outstanding questions. Knowledge of how subsets of habenular axons form connections at the appropriate subregion of the IPN is lacking. Selective expression of Neuropilin1a in only the left dorsal habenula allows neurons to respond to Semaphorin3b signaling and directs them to innervate the dorsal IPN (Kuan et al., 2007). However, more cues must be involved in steering habenular axons at the IPN and determining their precise connectivity along its dorsoventral axis. Once at the target, the interplay between habenular efferents and IPN neuronal populations is largely unknown. In larval zebrafish, IPN neurons can differ greatly in their morphology (Bianco et al., 2008) and cells with elaborate processes may receive diverse input from innervating habenular fibers. Although recent studies have focused on the modulatory role of dorsal habenular neurons in fear responses (Agetsuma et al., 2010; Lee et al., 2010), the exact neuronal subtypes involved remain unclear. Specific functions and microcircuits involving the distinct cholinergic and somatostatin-expressing subsets of neurons must also be determined. A greater understanding of the neurotransmitter identity, subnuclear organization, and connectivity of the zebrafish dorsal habenulae will ultimately reveal why the left and right nuclei differ and how this specialization might influence behavior.
Acknowledgments
Contract Grant Sponsor: NIH, Contract grant numbers: F32 MH09198, R01 HD042215; Contract Grant Sponsor: University of Virginia
We thank Harold Burgess and Hitoshi Okamoto for generously sharing transgenic lines, Lea Fortuno, Michelle Macurak and Estela Monge for technical assistance, and members of the Halpern laboratory for helpful input.
References
- Agetsuma M, Aizawa H, Aoki T, Nakayama R, Takahoko M, Goto M, Sassa T, Amo R, Shiraki T, Kawakami K, Hosoya T, Higashijima S, Okamoto H. The habenula is crucial for experience-dependent modification of fear responses in zebrafish. Nat Neurosci. 2010;13:1354–1356. doi: 10.1038/nn.2654. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Bianco IH, Hamaoka T, Miyashita T, Uemura O, Concha ML, Russell C, Wilson SW, Okamoto H. Laterotopic representation of left-right information onto the dorso-ventral axis of a zebrafish midbrain target nucleus. Curr Biol. 2005;15:238–243. doi: 10.1016/j.cub.2005.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Cui W, Tanaka K, Okamoto H. Hyperactivation of the habenula as a link between depression and sleep disturbance. Front Hum Neurosci. 2013;7:826. doi: 10.3389/fnhum.2013.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizawa H, Goto M, Sato T, Okamoto H. Temporally regulated asymmetric neurogenesis causes left-right difference in the zebrafish habenular structures. Dev Cell. 2007;12:87–98. doi: 10.1016/j.devcel.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Aizawa H, Kobayashi M, Tanaka S, Fukai T, Okamoto H. Molecular characterization of the subnuclei in rat habenula. J Comp Neurol. 2012;520:4051–4066. doi: 10.1002/cne.23167. [DOI] [PubMed] [Google Scholar]
- Akle V, Guelin E, Yu LL, Brassard-Giordano H, Slack BE, Zhdanova IV. F-Spondin/spon1b expression patterns in developing and adult zebrafish. PLoS One. 2012:7. doi: 10.1371/journal.pone.0037593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amo R, Aizawa H, Takahoko M, Kobayashi M, Takahashi R, Aoki T, Okamoto H. Identification of the zebrafish ventral habenula as a homolog of the mammalian lateral habenula. J Neurosci. 2010;30:1566–1574. doi: 10.1523/JNEUROSCI.3690-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres KH, Von During M, Veh RW. Subnuclear organization of the rat habenular complexes. J Comp Neurol. 1999;407:130–150. doi: 10.1002/(sici)1096-9861(19990428)407:1<130::aid-cne10>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Anichtchik O, Sallinen V, Peitsaro N, Panula P. Distinct structure and activity of monoamine oxidase in the brain of zebrafish (Danio rerio) J Comp Neurol. 2006;498:593–610. doi: 10.1002/cne.21057. [DOI] [PubMed] [Google Scholar]
- Appelbaum L, Wang G, Yokogawa T, Skariah GM, Smith SJ, Mourrain P, Mignot E. Circadian and homeostatic regulation of structural synaptic plasticity in hypocretin neurons. Neuron. 2010;68:87–98. doi: 10.1016/j.neuron.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelbaum L, Wang GX, Maro GS, Mori R, Tovin A, Marin W, Yokogawa T, Kawakami K, Smith SJ, Gothilf Y, Mignot E, Mourrain P. Sleep-wake regulation and hypocretin-melatonin interaction in zebrafish. Proc Natl Acad Sci U S A. 2009;106:21942–21947. doi: 10.1073/pnas.906637106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayari B, El Hachimi KH, Yanicostas C, Landoulsi A, Soussi-Yanicostas N. Prokineticin 2 expression is associated with neural repair of injured adult zebrafish telencephalon. J Neurotrauma. 2010;27:959–972. doi: 10.1089/neu.2009.0972. [DOI] [PubMed] [Google Scholar]
- Ayari B, Landoulsi A, Soussi-Yanicostas N. Localization and characterization of kal 1.a and kal 1.b in the brain of adult zebrafish (Danio rerio) Brain Res Bull. 2012;88:345–353. doi: 10.1016/j.brainresbull.2012.03.006. [DOI] [PubMed] [Google Scholar]
- Bae YK, Kani S, Shimizu T, Tanabe K, Nojima H, Kimura Y, Higashijima S, Hibi M. Anatomy of zebrafish cerebellum and screen for mutations affecting its development. Dev Biol. 2009;330:406–426. doi: 10.1016/j.ydbio.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Barroso-Chinea P, Castle M, Aymerich MS, Perez-Manso M, Erro E, Tunon T, Lanciego JL. Expression of the mRNAs encoding for the vesicular glutamate transporters 1 and 2 in the rat thalamus. J Comp Neurol. 2007;501:703–715. doi: 10.1002/cne.21265. [DOI] [PubMed] [Google Scholar]
- Beretta CA, Dross N, Guiterrez-Triana JA, Ryu S, Carl M. Habenula circuit development: past, present, and future. Front Neurosci. 2012;6:51. doi: 10.3389/fnins.2012.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman JR, Skariah G, Maro GS, Mignot E, Mourrain P. Characterization of two melanin-concentrating hormone genes in zebrafish reveals evolutionary and physiological links with the mammalian MCH system. J Comp Neurol. 2009;517:695–710. doi: 10.1002/cne.22171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betty M, Harnish SW, Rhodes KJ, Cockett MI. Distribution of heterotrimeric G-protein beta and gamma subunits in the rat brain. Neuroscience. 1998;85:475–486. doi: 10.1016/s0306-4522(97)00623-4. [DOI] [PubMed] [Google Scholar]
- Bianco IH, Carl M, Russell C, Clarke JDW, Wilson SW. Brain asymmetry is encoded at the level of axon terminal morphology. Neural Dev. 2008:3. doi: 10.1186/1749-8104-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco IH, Wilson SW. The habenular nuclei: a conserved asymmetric relay station in the vertebrate brain. Philos Trans R Soc Lond B Biol Sci. 2009;364:1005–1020. doi: 10.1098/rstb.2008.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biran J, Palevitch O, Ben-Dor S, Levavi-Sivan B. Neurokinin Bs and neurokinin B receptors in zebrafish-potential role in controlling fish reproduction. Proc Natl Acad Sci U S A. 2012;109:10269–10274. doi: 10.1073/pnas.1119165109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin M, Norton W, Bally-Cuif L, Vernier P. NR4A2 controls the differentiation of selective dopaminergic nuclei in the zebrafish brain. Mol Cell Neurosci. 2008;39:592–604. doi: 10.1016/j.mcn.2008.08.006. [DOI] [PubMed] [Google Scholar]
- Braitenberg V, Kemali M. Exceptions to bilateral symmetry in the epithalamus of lower vertebrates. J Comp Neurol. 1970;138:137–146. doi: 10.1002/cne.901380203. [DOI] [PubMed] [Google Scholar]
- Brosamle C, Halpern ME. Nogo-Nogo receptor signalling in PNS axon outgrowth and pathfinding. Mol Cell Neurosci. 2009;40:401–409. doi: 10.1016/j.mcn.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Carl M, Bianco IH, Bajoghli B, Aghaallaei N, Czerny T, Wilson SW. Wnt/Axin1/beta-catenin signaling regulates asymmetric nodal activation, elaboration, and concordance of CNS asymmetries. Neuron. 2007;55:393–405. doi: 10.1016/j.neuron.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Cheng CH, Chen GD, Hung CC, Yang CH, Hwang SPL, Kawakami K, Wu BK, Huang CJ. Recapitulation of zebrafish sncga expression pattern and labeling the habenular complex in transgenic zebrafish using green fluorescent protein reporter gene. Dev Dyn. 2009;238:746–754. doi: 10.1002/dvdy.21877. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Leslie FM, Zhou QY. Expression of prokineticins and their receptors in the adult mouse brain. J Comp Neurol. 2006;498:796–809. doi: 10.1002/cne.21087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Wilson SW. Asymmetry in the epithalamus of vertebrates. J Anat. 2001;199:63–84. doi: 10.1046/j.1469-7580.2001.19910063.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha ML, Russell C, Regan JC, Tawk M, Sidi S, Gilmour DT, Kapsimali M, Sumoy L, Goldstone K, Amaya E, Kimelman D, Nicolson T, Grunder S, Gomperts M, Clarke JD, Wilson SW. Local tissue interactions across the dorsal midline of the forebrain establish CNS laterality. Neuron. 2003;39:423–438. doi: 10.1016/s0896-6273(03)00437-9. [DOI] [PubMed] [Google Scholar]
- Contestabile A, Villani L, Fasolo A, Franzoni MF, Gribaudo L, Oktedalen O, Fonnum F. Topography of cholinergic and substance-P pathways in the habenulo-interpeduncular system of the rat. An immunocytological and microchemical approach. Neuroscience. 1987;21:253–270. doi: 10.1016/0306-4522(87)90337-x. [DOI] [PubMed] [Google Scholar]
- Cuello AC, Emson PC, Paxinos G, Jessell T. Substance-P containing and cholinergic projections from habenula. Brain Res. 1978;149:413–429. doi: 10.1016/0006-8993(78)90484-5. [DOI] [PubMed] [Google Scholar]
- deCarvalho TN, Akitake CM, Thisse C, Thisse B, Halpern ME. Aversive cues fail to activate fos expression in the asymmetric olfactory-habenula pathway of zebrafish. Front Neural Circuits. 2013;7:98. doi: 10.3389/fncir.2013.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan J, Mitchell KJ. Mutation of Elfn1 in mice causes seizures and hyperactivity. PLoS One. 2013;8:e80491. doi: 10.1371/journal.pone.0080491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll CA, Burkart JT, Hope KD, Halpern ME, Gamse JT. Subnuclear development of the zebrafish habenular nuclei requires ER translocon function. Dev Biol. 2011;360:44–57. doi: 10.1016/j.ydbio.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drerup CM, Wiora HM, Morris JA. Characterization of the overlapping expression patterns of the zebrafish LIS1 orthologs. Gene Expr Patterns. 2010;10:75–85. doi: 10.1016/j.gep.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Dworkin S, Heath JK, DeJong-Curtain TA, Hogan BM, Lieschke GJ, Malaterre J, Ramsay RG, Mantamadiotis T. CREB activity modulates neural cell proliferation, midbrain-hindbrain organization and patterning in zebrafish. Dev Biol. 2007;307:127–141. doi: 10.1016/j.ydbio.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Eckenrode TC, Barr GA, Battisti WP, Murray M. Acetylcholine in the interpeduncular nucleus of the rat: normal distribution and effects of deafferentation. Brain Res. 1987;418:273–286. doi: 10.1016/0006-8993(87)90095-3. [DOI] [PubMed] [Google Scholar]
- Eldred WD, Finger TE, Nolte J. Central projections of the frontal organ of Rana pipiens, as demonstrated by the anterograde transport of horseradish peroxidase. Cell Tissue Res. 1980;211:215–222. doi: 10.1007/BF00236444. [DOI] [PubMed] [Google Scholar]
- Fowler MA, Sidiropoulou K, Ozkan ED, Phillips CW, Cooper DC. Corticolimbic expression of TRPC4 and TRPC5 channels in the rodent brain. PLoS One. 2007;2:e573. doi: 10.1371/journal.pone.0000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamse JT, Kuan YS, Macurak M, Brosamle C, Thisse B, Thisse C, Halpern ME. Directional asymmetry of the zebrafish epithalamus guides dorsoventral innervation of the midbrain target. Development. 2005;132:4869–4881. doi: 10.1242/dev.02046. [DOI] [PubMed] [Google Scholar]
- Gamse JT, Thisse C, Thisse B, Halpern ME. The parapineal mediates left-right asymmetry in the zebrafish diencephalon. Development. 2003;130:1059–1068. doi: 10.1242/dev.00270. [DOI] [PubMed] [Google Scholar]
- Geisler S, Andres KH, Veh RW. Morphologic and cytochemical criteria for the identification and delineation of individual subnuclei within the lateral habenular complex of the rat. J Comp Neurol. 2003;458:78–97. doi: 10.1002/cne.10566. [DOI] [PubMed] [Google Scholar]
- Gorelick DA, Watson W, Halpern ME. Androgen receptor gene expression in the developing and adult zebrafish brain. Dev Dyn. 2008;237:2987–2995. doi: 10.1002/dvdy.21700. [DOI] [PubMed] [Google Scholar]
- Goto-Kazeto R, Kight KE, Zohar Y, Place AR, Trant JM. Localization and expression of aromatase rnRNA in adult zebrafish. Gen Comp Endocrinol. 2004;139:72–84. doi: 10.1016/j.ygcen.2004.07.003. [DOI] [PubMed] [Google Scholar]
- Guglielmotti V, Cristino L. The interplay between the pineal complex and the habenular nuclei in lower vertebrates in the context of the evolution of cerebral asymmetry. Brain Res Bull. 2006;69:475–488. doi: 10.1016/j.brainresbull.2006.03.010. [DOI] [PubMed] [Google Scholar]
- Gurdjian E. Olfactory connections in the albino rat, with special reference to the stria medullaris and the anterior commissure. J Comp Neurol. 1925;38:127–163. [Google Scholar]
- Haug MF, Gesemann M, Mueller T, Neuhauss SC. Phylogeny and expression divergence of metabotropic glutamate receptor genes in the brain of zebrafish (Danio rerio) J Comp Neurol. 2013;521:1533–1560. doi: 10.1002/cne.23240. [DOI] [PubMed] [Google Scholar]
- Hendricks M, Mathuru AS, Wang H, Silander O, Kee MZL, Jesuthasan S. Disruption of Esrom and Ryk identifies the roof plate boundary as an intermediate target for commissure formation. Mol Cell Neurosci. 2008;37:271–283. doi: 10.1016/j.mcn.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Nauta WJH. Afferent connections of habenular nuclei in rat. A horseradish peroxidase study, with a note on fiber-of-passage problem. J Comp Neurol. 1977;173:123–145. doi: 10.1002/cne.901730107. [DOI] [PubMed] [Google Scholar]
- Herkenham M, Nauta WJH. Efferent connections of the habenular nuclei in the rat. J Comp Neurol. 1979;187:19–47. doi: 10.1002/cne.901870103. [DOI] [PubMed] [Google Scholar]
- Herrick JC. Ambystoma tigrinum. Chicago, IL: University of Chicago Press; 1948. The brain of the tiger salamander; p. 409. [Google Scholar]
- Hikosaka O. The habenula: from stress evasion to value-based decision-making. Nat Rev Neurosci. 2010;11:503–513. doi: 10.1038/nrn2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikosaka O, Sesack SR, Lecourtier L, Shepard PD. Habenula: crossroad between the basal ganglia and the limbic system. J Neurosci. 2008;28:11825–11829. doi: 10.1523/JNEUROSCI.3463-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E, Santhakumar K, Akitake CA, Ahn SJ, Thisse C, Thisse B, Wyart C, Mangin JM, Halpern ME. Cholinergic left-right asymmetry in the habenulo-interpeduncular pathway. Proc Natl Acad Sci U S A. 2013;110:21171–21176. doi: 10.1073/pnas.1319566110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppmann V, Wu JJ, Soviknes AM, Helvik JV, Becker TS. Expression of the eight AMPA receptor subunit genes in the developing central nervous system and sensory organs of zebrafish. Dev Dyn. 2008;237:788–799. doi: 10.1002/dvdy.21447. [DOI] [PubMed] [Google Scholar]
- Huang YY, Haug MF, Gesemann M, Neuhauss SC. Novel expression patterns of metabotropic glutamate receptor 6 in the zebrafish nervous system. PloS One. 2012;7:e35256. doi: 10.1371/journal.pone.0035256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivankova K, Turecek R, Fritzius T, Seddik R, Prezeau L, Comps-Agrar L, Pin JP, Fakler B, Besseyrias V, Gassmann M, Bettler B. Up-regulation of GABA(B) receptor signaling by constitutive assembly with the K+ channel tetramerization domain-containing protein 12 (KCTD12) J Biol Chem. 2013;288:24848–24856. doi: 10.1074/jbc.M113.476770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwahori N. Golgi study on habenular nucleus of cat. J Comp Neurol. 1977;171:319–344. doi: 10.1002/cne.901710303. [DOI] [PubMed] [Google Scholar]
- Jayasena CS, Trinhle A, Bronner M. Live imaging of endogenous Collapsin response mediator protein-1 expression at subcellular resolution during zebrafish nervous system development. Gene Expr Patterns. 2011;11:395–400. doi: 10.1016/j.gep.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Josephson A, Trifunovski A, Widmer HR, Widenfalk J, Olson L, Spenger C. Nogo-receptor gene activity: cellular localization and developmental regulation of mRNA in mice and humans. J Comp Neurol. 2002;453:292–304. doi: 10.1002/cne.10408. [DOI] [PubMed] [Google Scholar]
- Kappers CUA, Huber GC, Crosby E. The Comparative Anatomy of the Nervous System of Vertebrates Including Man. New York: The Macmillan Company; 1936. [Google Scholar]
- Kastenhuber E, Gesemann M, Mickoleit M, Neuhauss SC. Phylogenetic analysis and expression of zebrafish transient receptor potential melastatin family genes. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:1236–1249. doi: 10.1002/dvdy.24020. [DOI] [PubMed] [Google Scholar]
- Kemali M, De Santis A. The extracranial portion of the pineal complex of the frog (frontal organ) is connected to the pineal, the hypothalamus, the brain stem and the retina. Exp Brain Res. 1983;53:193–196. doi: 10.1007/BF00239412. [DOI] [PubMed] [Google Scholar]
- Kemali M, Guglielmotti V. The distribution of substance P in the habenulo-interpeduncular system of the frog shown by an immunohistochemical method. Arch Ital Biol. 1984;122:269–280. [PubMed] [Google Scholar]
- Kitahashi T, Ogawa S, Parhar IS. Cloning and expression of kiss2 in the zebrafish and medaka. Endocrinology. 2009;150:821–831. doi: 10.1210/en.2008-0940. [DOI] [PubMed] [Google Scholar]
- Klemm WR. Habenular and interpeduncularis nuclei: shared components in multiple-function networks. Med Sci Monit. 2004;10:RA261–RA273. [PubMed] [Google Scholar]
- Kojima D, Torii M, Fukada Y, Dowling JE. Differential expression of duplicated VAL-opsin genes in the developing zebrafish. J Neurochem. 2008;104:1364–1371. doi: 10.1111/j.1471-4159.2007.05093.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf HW, Sato T, Oksche A. Complex relationships between the pineal organ and the medial habenular nucleus-pretectal region of the mouse as revealed by S-antigen immunocytochemistry. Cell Tissue Res. 1990;261:493–500. doi: 10.1007/BF00313528. [DOI] [PubMed] [Google Scholar]
- Kuan YS, Yu HH, Moens CB, Halpern ME. Neuropilin asymmetry mediates a left-right difference in habenular connectivity. Development. 2007;134:857–865. doi: 10.1242/dev.02791. [DOI] [PubMed] [Google Scholar]
- Landis SC, Keefe D. Evidence for neurotransmitter plasticity in vivo: developmental changes in properties of cholinergic sympathetic neurons. Dev Biol. 1983;98:349–372. doi: 10.1016/0012-1606(83)90365-2. [DOI] [PubMed] [Google Scholar]
- Lauterborn JC, Isackson PJ, Montalvo R, Gall CM. In situ hybridziation localization of choline-acetyltransferase mRNA in the adult rat brain and spinal cord. Mol Brain Res. 1993;17:59–69. doi: 10.1016/0169-328x(93)90073-x. [DOI] [PubMed] [Google Scholar]
- Lecourtier L, Kelly PH. A conductor hidden in the orchestra? Role of the habenular complex in monoamine transmission and cognition. Neuroscience and Biobehav Reviews. 2007;31:658–672. doi: 10.1016/j.neubiorev.2007.01.004. [DOI] [PubMed] [Google Scholar]
- Lee A, Mathuru AS, Teh C, Kibat C, Korzh V, Penney TB, Jesuthasan S. The habenula prevents helpless behavior in larval zebrafish. Curr Biol. 2010;20:2211–2216. doi: 10.1016/j.cub.2010.11.025. [DOI] [PubMed] [Google Scholar]
- Lenn NJ, Hamill GS. Subdivisions of the interpeduncular nucleus: a proposed nomenclature. Brain Res Bull. 1984;13:203–204. doi: 10.1016/0361-9230(84)90023-6. [DOI] [PubMed] [Google Scholar]
- Leung T, Humbert JE, Stauffer AM, Giger KE, Chen H, Tsai HJ, Wang C, Mirshahi T, Robishaw JD. The orphan G protein-coupled receptor 161 is required for left-right patterning. Dev Biol. 2008;323:31–40. doi: 10.1016/j.ydbio.2008.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li IC, Chan CT, Lu YF, Wu YT, Chen YC, Li GB, Lin CY, Hwang SP. Zebrafish kruppel-like factor 4a represses intestinal cell proliferation and promotes differentiation of intestinal cell lineages. PloS One. 2011;6:e20974. doi: 10.1371/journal.pone.0020974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Chen Y, Copeland D, Ball H, Duff RJ, Rockich B, Londraville RL. Expression of leptin receptor gene in developing and adult zebrafish. Gen Comp Endocrinol. 2010;166:346–355. doi: 10.1016/j.ygcen.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahler J, Driever W. Expression of the zebrafish intermediate neurofilament Nestin in the developing nervous system and in neural proliferation zones at postembryonic stages. BMC Dev Biol. 2007;7:89. doi: 10.1186/1471-213X-7-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marburg O. The structure and fiber connections of the human habenula. J Comp Neurol. 1944;80:211–233. [Google Scholar]
- Marcos P, Arroyo-Jimenez MM, Lozano G, Gonzalez-Fuentes J, Lagartos-Donate MJ, Aguilar LA, Covenas R. Mapping of tyrosine hydroxylase in the diencephalon of alpaca (Lama pacos) and co-distribution with somatostatin-28 (1–12) J Chem Neuroanat. 2013;50–51:66–74. doi: 10.1016/j.jchemneu.2013.02.006. [DOI] [PubMed] [Google Scholar]
- Marin O, Smeets W, Gonzalez A. Distribution of choline acetyltransferase immunoreactivity in the brain of anuran (Rana perezi, Xenopus laevis) and urodele (Pleurodeles waltl) amphibians. J Comp Neurol. 1997;382:499–534. doi: 10.1002/(sici)1096-9861(19970616)382:4<499::aid-cne6>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Matos-Cruz V, Blasic J, Nickle B, Robinson PR, Hattar S, Halpern ME. Unexpected diversity and photoperiod dependence of the zebrafish melanopsin system. PloS One. 2011;6:e25111. doi: 10.1371/journal.pone.0025111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz M, Gassmann M, Fakler B, Schaeren-Wiemers N, Bettler B. Distribution of the auxiliary GABAB receptor subunits KCTD8, 12, 12b, and 16 in the mouse brain. J Comp Neurol. 2011;519:1435–1454. doi: 10.1002/cne.22610. [DOI] [PubMed] [Google Scholar]
- Milanese C, Sager JJ, Bai Q, Farrell TC, Cannon JR, Greenamyre JT, Burton EA. Hypokinesia and reduced dopamine levels in zebrafish lacking beta- and gamma1-synucleins. J Biol Chem. 2012;287:2971–2983. doi: 10.1074/jbc.M111.308312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mione M, Lele Z, Kwong CT, Concha ML, Clarke JD. Expression of pcp4a in subpopulations of CNS neurons in zebrafish. J Comp Neurol. 2006;495:769–787. doi: 10.1002/cne.20907. [DOI] [PubMed] [Google Scholar]
- Miyasaka N, Morimoto K, Tsubokawa T, Higashijima S, Okamoto H, Yoshihara Y. From the olfactory bulb to higher brain centers: genetic visualization of secondary olfactory pathways in zebrafish. J Neurosci. 2009;29:4756–4767. doi: 10.1523/JNEUROSCI.0118-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley BJ. The Interpeduncular Nucleus. Int Rev Neurobiol. 1986;28:157–182. doi: 10.1016/s0074-7742(08)60108-7. [DOI] [PubMed] [Google Scholar]
- Neal CR, Jr, Mansour A, Reinscheid R, Nothacker HP, Civelli O, Watson SJ., Jr Localization of orphanin FQ (nociceptin) peptide and messenger RNA in the central nervous system of the rat. J Comp Neurol. 1999;406:503–547. [PubMed] [Google Scholar]
- Norton WH, Folchert A, Bally-Cuif L. Comparative analysis of serotonin receptor (HTR1A/HTR1B families) and transporter (slc6a4a/b) gene expression in the zebrafish brain. J Comp Neurol. 2008;511:521–542. doi: 10.1002/cne.21831. [DOI] [PubMed] [Google Scholar]
- Oberwinkler J, Lis A, Giehl KM, Flockerzi V, Philipp SE. Alternative splicing switches the divalent cation selectivity of TRPM3 channels. J Biol Chem. 2005;280:22540–22548. doi: 10.1074/jbc.M503092200. [DOI] [PubMed] [Google Scholar]
- Ogawa S, Ng KW, Ramadasan PN, Nathan FM, Parhar IS. Habenular Kiss1 neurons modulate the serotonergic system in the brain of zebrafish. Endocrinology. 2012;153:2398–2407. doi: 10.1210/en.2012-1062. [DOI] [PubMed] [Google Scholar]
- Ohishi H, Akazawa C, Shigemoto R, Nakanishi S, Mizuno N. Distributions of the mRNAs for L-2-amino-4-phosphonobutyrate-sensitive metabotropic glutamate receptors, mGluR4 and mGluR7, in the rat brain. J Comp Neurol. 1995;360:555–570. doi: 10.1002/cne.903600402. [DOI] [PubMed] [Google Scholar]
- Ohishi H, Shigemoto R, Nakanishi S, Mizuno N. Distribution of the mRNA for a metabotropic glutamate receptor (mGluR3) in the rat brain: an in situ hybridization study. J Comp Neurol. 1993;335:252–266. doi: 10.1002/cne.903350209. [DOI] [PubMed] [Google Scholar]
- Okamoto H, Agetsuma M, Aizawa H. Genetic dissection of the zebrafish habenula, a possible switching board for selection of behavioral strategy to cope with fear and anxiety. Dev Neurobiol. 2012;72:386–394. doi: 10.1002/dneu.20913. [DOI] [PubMed] [Google Scholar]
- Pietri T, Easley-Neal C, Wilson C, Washbourne P. Six cadm/SynCAM genes are expressed in the nervous system of developing zebrafish. Dev Dyn. 2008;237:233–246. doi: 10.1002/dvdy.21397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C, Luo M. Neurochemical phenotypes of the afferent and efferent projections of the mouse medial habenula. Neuroscience. 2009;161:827–837. doi: 10.1016/j.neuroscience.2009.03.085. [DOI] [PubMed] [Google Scholar]
- Quina LA, Wang S, Ng L, Turner EE. Brn3a and Nurr1 mediate a gene regulatory pathway for habenula development. J Neurosci. 2009;29:14309–14322. doi: 10.1523/JNEUROSCI.2430-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigne M, Bianco IH, Wilson SW, Blader P. Nodal signalling imposes left-right asymmetry upon neurogenesis in the habenular nuclei. Development. 2009;136:1549–1557. doi: 10.1242/dev.034793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussigne M, Blader P. Divergence in regulation of the PEA3 family of ETS transcription factors. Gene Expr Patterns. 2006;6:777–782. doi: 10.1016/j.modgep.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Sadakata T, Washida M, Morita N, Furuichi T. Tissue distribution of Ca2+-dependent activator protein for secretion family members CAPS1 and CAPS2 in mice. J Histochem Cytochem. 2007;55:301–311. doi: 10.1369/jhc.6A7033.2006. [DOI] [PubMed] [Google Scholar]
- Sallinen V, Kolehmainen J, Priyadarshini M, Toleikyte G, Chen YC, Panula P. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol Dis. 2010;40:93–101. doi: 10.1016/j.nbd.2010.06.001. [DOI] [PubMed] [Google Scholar]
- Schwenk J, Metz M, Zolles G, Turecek R, Fritzius T, Bildl W, Tarusawa E, Kulik A, Unger A, Ivankova K, Seddik R, Tiao JY, Rajalu M, Trojanova J, Rohde V, Gassmann M, Schulte U, Fakler B, Bettler B. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465:231–235. doi: 10.1038/nature08964. [DOI] [PubMed] [Google Scholar]
- Servili A, Le Page Y, Leprince J, Caraty A, Escobar S, Parhar IS, Seong JY, Vaudry H, Kah O. Organization of two independent kisspeptin systems derived from evolutionary-ancient kiss genes in the brain of zebrafish. Endocrinology. 2011;152:1527–1540. doi: 10.1210/en.2010-0948. [DOI] [PubMed] [Google Scholar]
- Shinoda K, Yagi H, Fujita H, Osawa Y, Shiotani Y. Screening of aromatase-containing neurons in rat forebrain: an immunohistochemical study with antibody against human placental antigen X-P2 (hPAX-P2) J Comp Neurol. 1989;290:502–515. doi: 10.1002/cne.902900405. [DOI] [PubMed] [Google Scholar]
- Shiosaka S, Takatsuki K, Sakanaka M, Inagaki S, Takagi H, Senba E, Kawai Y, Tohyama M. Ontogeny of somatostatin-containing neuron system of the rat: immunohistochemical observations. I. Lower brainstem. J Comp Neurol. 1981;203:173–188. doi: 10.1002/cne.902030203. [DOI] [PubMed] [Google Scholar]
- Sollner C, Wright GJ. A cell surface interaction network of neural leucine-rich repeat receptors. Genome biology. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer NC. Activity-dependent neurotransmitter respecification. Nat Rev Neurosci. 2012;13:94–106. doi: 10.1038/nrn3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland RJ. The dorsal diencephalic conduction system - a review of the anatomy and functions of the habenular complex. Neurosci Biobehav Rev. 1982;6:1–13. doi: 10.1016/0149-7634(82)90003-3. [DOI] [PubMed] [Google Scholar]
- Taylor RW, Qi JY, Talaga AK, Ma TP, Pan L, Bartholomew CR, Klionsky DJ, Moens CB, Gamse JT. Asymmetric inhibition of Ulk2 causes left-right differences in habenular neuropil formation. J Neurosci. 2011;31:9869–9878. doi: 10.1523/JNEUROSCI.0435-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taymans JM, Van den Haute C, Baekelandt V. Distribution of PINK1 and LRRK2 in rat and mouse brain. J Neurochem. 2006;98:951–961. doi: 10.1111/j.1471-4159.2006.03919.x. [DOI] [PubMed] [Google Scholar]
- Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission. 2004 ( http://zfin.org)
- Tomizawa K, Katayama H, Nakayasu H. A novel monoclonal antibody recognizes a previously unknown subdivision of the habenulo-interpeduncular system in zebrafish. Brain Res. 2001;901:117–127. doi: 10.1016/s0006-8993(01)02313-7. [DOI] [PubMed] [Google Scholar]
- Tovin A, Alon S, Ben-Moshe Z, Mracek P, Vatine G, Foulkes NS, Jacob-Hirsch J, Rechavi G, Toyama R, Coon SL, Klein DC, Eisenberg E, Gothilf Y. Systematic identification of rhythmic genes reveals camk1gb as a new element in the circadian clockwork. PLoS Genetics. 2012;8:e1003116. doi: 10.1371/journal.pgen.1003116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YC, Chen RD, Lucassen M, Schmidt MM, Dringen R, Abele D, Hwang PP. Exploring uncoupling proteins and antioxidant mechanisms under acute cold exposure in brains of fish. PloS One. 2011;6:e18180. doi: 10.1371/journal.pone.0018180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchimoto M, Yasuo S, Funada M, Aoki M, Sasagawa H, Yoshimura T, Tadauchi O, Cameron SA, Kitagawa Y, Kadowaki T. Conservation of novel Mahya genes shows the existence of neural functions common between Hymenoptera and Deuterostome. Dev Genes Evol. 2005;215:564–574. doi: 10.1007/s00427-005-0021-z. [DOI] [PubMed] [Google Scholar]
- Viswanath H, Carter AQ, Baldwin PR, Molfese DL, Salas R. The medial habenula: still neglected. Front Hum Neurosci. 2013;7:931. doi: 10.3389/fnhum.2013.00931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Niederhausern V, Kastenhuber E, Stauble A, Gesemann M, Neuhauss SC. Phylogeny and expression of canonical transient receptor potential (TRPC) genes in developing zebrafish. Developmental dynamics : an official publication of the American Association of Anatomists. 2013;242:1427–1441. doi: 10.1002/dvdy.24041. [DOI] [PubMed] [Google Scholar]
- Wagner F, Stroh T, Veh RW. Correlating habenular subnuclei in rat and mouse using topographical, morphological and cytochemical criteria. J Comp Neurol. 2014 doi: 10.1002/cne.23554. [DOI] [PubMed] [Google Scholar]
- Walker C. Haploid Screens and Gamma-Ray Mutagenesis. In: Detrich HW, Westerfield M, Zon LI, editors. The Zebrafish, Genetics and Genomics. San Diego: Academic Press; 1999. pp. 44–68. [Google Scholar]
- Wu SY, de Borsetti NH, Bain EJ, Bulow CR, Gamse JT. Mediator subunit 12 coordinates intrinsic and extrinsic control of epithalamic development. Dev Biol. 2014;385:13–22. doi: 10.1016/j.ydbio.2013.10.023. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Danjo T, Pastan I, Hikida T, Nakanishi S. Distinct roles of segregated transmission of the septo-habenular pathway in anxiety and fear. Neuron. 2013;78:537–544. doi: 10.1016/j.neuron.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Zondervan-van der Linde H, Severijnen LA, Oostra BA, Willemsen R, Bonifati V. Dopaminergic neuronal loss and dopamine-dependent locomotor defects in Fbxo7-deficient zebrafish. PLoS One. 2012;7:e48911. doi: 10.1371/journal.pone.0048911. [DOI] [PMC free article] [PubMed] [Google Scholar]