Abstract
Background
The two North Atlantic eel species, the European and the American eel, represent an ideal system in which to study parallel selection patterns due to their sister species status and the presence of ongoing gene flow. A panel of 80 coding-gene SNPs previously analyzed in American eel was used to genotype European eel individuals (glass eels) from 8 sampling locations across the species distribution. We tested for single-generation signatures of spatially varying selection in European eel by searching for elevated genetic differentiation using FST-based outlier tests and by testing for significant associations between allele frequencies and environmental variables.
Results
We found signatures of possible selection at a total of 11 coding-gene SNPs. Candidate genes for local selection constituted mainly genes with a major role in metabolism as well as defense genes. Contrary to what has been found for American eel, only 2 SNPs in our study correlated with differences in temperature, which suggests that other explanatory variables may play a role. None of the genes found to be associated with explanatory variables in European eel showed any correlations with environmental factors in the previous study in American eel.
Conclusions
The different signatures of selection between species could be due to distinct selective pressures associated with the much longer larval migration for European eel relative to American eel. The lack of parallel selection in North Atlantic eels could also be due to most phenotypic traits being polygenic, thus reducing the likelihood of selection acting on the same genes in both species.
Keywords: Adaptation, European eel, Genetic-by-environment associations, Parallel selection, Single nucleotide polymorphisms
Background
Parallel adaptive changes under replicated environmental conditions have been particularly valuable for understanding evolutionary processes in natural populations. One of the classical questions in evolutionary biology concerns whether different species and populations within species will adapt to the same agent of selection in the same way or whether the response will involve different traits and genes [1,2]. Parallel genotypic adaptation appears to be frequent and occurs at all taxonomic levels from microbes and plants to humans [3,4] and is likely to result in changes at a relatively small number of genes [5]. For instance, the study of Colosimo et al. [6] demonstrated that selection on a single gene, ectodysplasin (Eda), is responsible for the parallel reduction of armor plates in freshwater populations of threespine stickleback Gasterosteus aculeatus. However, more complex physiological processes relevant in the context of parallel freshwater adaptation of threespine sticklebacks are influenced by several genes, each of small effect [7-10]. Using a survey of the published literature on parallel adaptation of independent lineages of natural populations, Conte et al. [5] concluded that divergence at loci under selection is most likely to be based on standing genetic variation derived from a common ancestor rather than mutations occurring de novo after divergence. Hence, probability of gene reuse is plausibly higher in closely related species, which are likely to show similar divergence at loci subjected to similar selection pressures [11].
An excellent opportunity to test for genetic parallelism exists in the two North Atlantic eel species, the European eel (Anguilla anguilla) and the American eel (A. rostrata). Both species are morphologically almost indistinguishable, with the number of vertebrae being regarded as the best diagnostic character between species [12]. Divergence time between the two species remains largely unresolved, encompassing between 1.5 and 5.8 million years [13-15]. Remarkably, although mitochondrial DNA lineages of the two species are reciprocally monophyletic [13], differentiation at nuclear loci is surprisingly low (FST = 0.055 [16]; FST = 0.018 [17]; FST = 0.06 [18]; FST = 0.09 [19]), suggestive of ongoing gene flow. In this sense, it is well established that the spawning grounds of the two species overlap in the Sargasso Sea and there is also overlap in spawning time [20]. European and American eels are known to hybridize, with hybrids observed almost exclusively in Iceland [21-23]. Hence, the sister species status of European and American eel and the low but biologically significant gene flow makes them an adequate system in which to test the occurrence of selection at homologous loci within each species.
North Atlantic eels have a catadromous life cycle and after spawning in the Sargasso Sea, larvae are transported by the Gulf Stream and other currents to the shores of North America and Europe/North Africa, respectively. Upon reaching the continental shelf, larvae metamorphose into glass eels, which complete the migration into riverine, estuarine and coastal feeding habitats and grow up as yellow eels. After a highly variable feeding stage, yellow eels metamorphose into partially mature silver eels that migrate back to the Sargasso undertaking a journey of about 2,000 km for the American eel and 5,000-6,000 km for the European eel. Upon arriving in the Sargasso Sea, eels reproduce and die [24]. During the continental phase, eels occupy a broad range of habitats from the Caribbean to Greenland in the western Atlantic (American eel) and from Morocco to Iceland in the eastern Atlantic (European eel). The presence of eels in extremely heterogenous environments in terms of temperature (i.e. from subtropical to subarctic), salinity (i.e. from freshwater to marine), substrate, depth or productivity along their geographic distribution makes them ideal species in which to study the consequences of spatially varying selective pressures that often result in local adaptation of ecologically important traits [1,25,26]. Beginning with Levene [27], who introduced the first theoretical model for examining the impact of diversifying selection in space, a number of studies have shown that balancing selection due to spatial heterogeneity is an important mechanism responsible for the maintenance of genetic polymorphism (reviewed in [28]). Genetic variation in a spatially heterogenous environment may be maintained even when dispersal results in complete mixing of the gene pool [1]. However, under such a panmixia scenario, in which offspring are distributed to environments at random independently of the environment experienced by the parents, local selection cannot to lead to local adaptation [29]. In the case of eels, owing to panmixia in both European [19,30] and American eel [31] and random larval dispersal across habitats, heritable trans-generational local adaptation is not possible although single-generation footprints of selection can still be detected. In this sense, significant geographic clines at allozyme loci have been detected in both European [32] and American eel [33]. In the most comprehensive study to date, Gagnaire et al. [26] found evidence for spatially varying selection at 13 coding genes in American eel showing significant correlations between allele frequencies and environmental variables (latitude, longitude and temperature) across the entire species range.
In this study, we tested for single-generation signatures of spatial varying selection in European eel and compared the results to those obtained by Gagnaire et al. [26]. We genotyped glass eels from 8 sampling locations across the geographic distribution of the species, using the same set of SNPs analyzed by Gagnaire et al. [26] in American eel. We used two main analytical approaches, one that identifies outliers as those markers with greater differentiation among all SNPs and a second based on determining positive associations between allele frequencies and environmental factors. Following the positive associations observed by Gagnaire et al. [26] in American eel, variables used in our study were degrees North latitude, degrees East/West longitude and sea-surface temperature at river mouth. We specifically wanted to test whether the same genes were under spatially varying selection in both European and American eel, hence providing evidence for parallel patterns of local selection, or whether the response involved a different set of genes. Considering their sister species status and the existence of gene flow between species, together with the similar environmental conditions they encounter [34], we hypothesize that the two North Atlantic eel species show parallel patterns of selection at the same loci.
Results
Genetic diversity values for all genotyped individuals at 80 SNPs are summarized in Table 1. 17 out of 80 loci were monomorphic and 63 were polymorphic in European eel, although frequency of the most common allele was >0.95 at 27 loci. Diversity indices were higher in American eel (Ho = 0.302; He = 0.306; P95 = 0.896; P99 = 0.922) than in European eel (Ho = 0.149; He = 0.157; P95 = 0.429; P99 = 0.610), suggestive of a strong ascertainment bias effect due to the fact that SNPs were identified in American eel.
Table 1.
Details of all genes and loci studied, including observed (H o ) and expected (H e ) heterozygosities at all loci in American (AR) and European eel (AA)
|
Locus |
Gene |
AR |
|
AA |
|
|---|---|---|---|---|---|
| H o | H e | H o | H e | ||
| 40S_S18_1401 |
40s ribosomal protein s18 |
0.389 |
0.375 |
0.344 |
0.312 |
| 60S_L10A_21874 |
60s ribosomal protein L10a |
0.250 |
0.219 |
0.282 |
0.306 |
| ACT_A3B_8646 |
Actinin alpha 3b |
0.300 |
0.255 |
0.006 |
0.006 |
| ACTB_21752 |
Beta-actin |
0.474 |
0.411 |
0.213 |
0.244 |
| ACYL_13914 |
Acyl carrier protein |
0.421 |
0.388 |
0.320 |
0.310 |
| ADH_3 |
Alcohol dehydrogenase class-3 |
0.263 |
0.361 |
0.229 |
0.351 |
| ADSS_L1_15447 |
Adenylosuccinate synthetase isozyme 1 |
0.158 |
0.229 |
0.016 |
0.016 |
| ALD_R |
Aldose reductase |
1.000 |
0.500 |
0.395 |
0.345 |
| ALDH_2_16634 |
Aldehyde dehydrogenase 2 |
0.947 |
0.499 |
0.468 |
0.428 |
| ANK_R_13478 |
Ankyrin repeat domain-comtaining protein 1 |
0.250 |
0.289 |
0.000 |
0.000 |
| ANN_A11_16176 |
Annexin A11 |
0.200 |
0.180 |
0.171 |
0.172 |
| ANX_2_249 |
Annexin A2-A |
0.100 |
0.375 |
0.016 |
0.034 |
| ARF_4_18099 |
ADP-ribsylation factor 4 |
0.444 |
0.346 |
0.019 |
0.019 |
| ATP_BC_259 |
ATP-bindincasette sub-family A member 1 |
0.450 |
0.439 |
0.022 |
0.022 |
| BPNT_1_18778 |
3′(5′),5′-biphosphate nucleotidase 1 |
0.263 |
0.411 |
0.025 |
0.025 |
| CLIC_5_10148 |
Chloride intracellular channel 5 |
0.250 |
0.219 |
0.521 |
0.492 |
| COI_17591 |
Cytochrome oxidase subunit I |
0.000 |
0.000 |
0.010 |
0.009 |
| COP_9_18132 |
266S protease regulatory subunit 7 |
0.300 |
0.255 |
0.035 |
0.034 |
| CSDE_1_11069 |
Cold shock domain-containing protein E1 |
0.316 |
0.266 |
0.019 |
0.019 |
| CSDE_1_19713 |
Cold shock domain-containing protein E1 |
0.474 |
0.411 |
0.066 |
0.064 |
| CST_21113 |
Cystatin precursor |
0.421 |
0.499 |
0.379 |
0.393 |
| CYT_BC1_9061 |
Cytochrome b-c1 complex subunit 2 |
0.200 |
0.180 |
0.000 |
0.000 |
| EF_1G_4796 |
Translation elongation factor 1 gamma |
0.400 |
0.320 |
0.000 |
0.000 |
| EF2_10494 |
Translation elongation factor 2 |
0.200 |
0.180 |
0.000 |
0.000 |
| EIF_3F_341 |
Translation elongation factor 3 subunit F |
0.211 |
0.332 |
0.113 |
0.146 |
| EIF_3J_11587 |
Translation elongation factor 3 subunit J |
0.300 |
0.375 |
0.079 |
0.082 |
| FER_H_20955 |
Ferritin heavy subunit |
0.421 |
0.432 |
0.009 |
0.009 |
| FGB_47 |
Fibrinogen Beta Chain |
0.300 |
0.255 |
0.000 |
0.000 |
| GAPDH_20355 |
Glyceraldehyde-3-phoshpate dehydrogenase |
0.200 |
0.180 |
0.222 |
0.197 |
| GDE1_2508 |
Glycerophosphochlorine phosphodiesterase |
0.389 |
0.313 |
0.000 |
0.000 |
| GOG_B1_15792 |
Golgin sub-family B member 1 |
0.150 |
0.289 |
0.050 |
0.049 |
| GPX_4_19607 |
Glutathione peroxidase 4 |
0.100 |
0.095 |
0.000 |
0.000 |
| HMG_T_9973 |
High mobility group-T protein |
0.050 |
0.049 |
0.025 |
0.025 |
| HSP_90A_15666 |
Heat shock protein 90 alpha |
0.158 |
0.229 |
0.063 |
0.061 |
| HSP_90B_21100 |
Heat shock protein 90 beta |
0.150 |
0.139 |
0.107 |
0.107 |
| HSPE_1_17854 |
10 kDa heat shock protein |
0.368 |
0.362 |
0.009 |
0.009 |
| IF_RF2_19747 |
Interferon regulatory factor 2 |
0.150 |
0.139 |
0.000 |
0.000 |
| JAM_3_13916 |
Junctional adhesion molecule 3b |
0.053 |
0.051 |
0.131 |
0.165 |
| KRT_13_20618 |
Keratin |
0.350 |
0.499 |
0.325 |
0.317 |
| KRT_A_15738 |
Keratin alpha-like |
0.000 |
0.000 |
0.022 |
0.022 |
| LBL_L2_20921 |
No hit |
0.000 |
0.000 |
0.000 |
0.000 |
| LDH_B_9441 |
Lactase dehydrogenase B |
0.600 |
0.455 |
0.025 |
0.025 |
| MDH_1393 |
Malate dehydrogenase |
0.263 |
0.450 |
0.298 |
0.493 |
| MYH_14857 |
Superfast myosin heavy chain |
0.300 |
0.255 |
0.563 |
0.496 |
| NADH_4_21742 |
NADH dehydrogenase subunit 4 |
0.000 |
0.0180 |
0.000 |
0.000 |
| NADH_5_17101 |
NADH dehydrogenase subunit 5 |
0.000 |
0.255 |
0.000 |
0.019 |
| NADH1_10_21119 |
NADH dehydrogenase 1 alpha subunit 10 |
0.300 |
0.455 |
0.000 |
0.000 |
| NCP_2_15547 |
Nucleolar complex protein 2 |
0.350 |
0.289 |
0.085 |
0.087 |
| NEX_19953 |
Nexilin |
0.450 |
0.489 |
0.476 |
0.496 |
| NGD_21138 |
Neuroguidin |
0.450 |
0.469 |
0.238 |
0.273 |
| NRAP_1541 |
Nebulin-related anchoring protein |
0.500 |
0.455 |
0.031 |
0.031 |
| PA2G4_2600 |
Proliferation associated protein 2G4 |
0.444 |
0.401 |
0.442 |
0.481 |
| PFN_15113 |
Profilin-2 |
0.400 |
0.375 |
0.000 |
0.000 |
| PGD_18096 |
6-phosphogluconate dehydrogenase |
0.000 |
0.000 |
0.042 |
0.053 |
| PGI_1 |
Phosphoglucose isomerase-1 |
0.211 |
0.188 |
0.338 |
0.348 |
| PGI_2 |
Phosphoglucose isomerase-2 |
0.368 |
0.478 |
0.456 |
0.498 |
| PGK_1_11454 |
Phosphoglycerate kinase 1 |
0.474 |
0.362 |
0.090 |
0.092 |
| PRP_40_16504 |
Pre-mRNA-processing factor 40 homolog A |
0.150 |
0.219 |
0.205 |
0.214 |
| PSA_4_21534 |
Proteasome subunit alpha type-4 |
0.350 |
0.439 |
0.484 |
0.498 |
| PSME_1_21196 |
Proteasome activator |
0.235 |
0.458 |
0.441 |
0.457 |
| RFC_3_18186 |
Replication factor C subunit 3 |
0.350 |
0.289 |
0.416 |
0.368 |
| RTF_1_21288 |
RNA polymerase-associated protein RTF1 homolog |
0.632 |
0.432 |
0.339 |
0.374 |
| SDH_O |
Sorbitol dehydrogenase |
0.000 |
0.000 |
0.000 |
0.000 |
| SLC_25A5_19808 |
ADP/ATP translocase 2 |
0.450 |
0.499 |
0.006 |
0.006 |
| SM_22_6449 |
Transgelin |
0.550 |
0.499 |
0.074 |
0.083 |
| SN4_TDR_374 |
Taphylococcal nuclease domain-containing protein 1 |
0.526 |
0.465 |
0.182 |
0.191 |
| TENT_02_11046 |
No hit |
0.111 |
0.105 |
0.318 |
0.362 |
| TENT_03_12589 |
Collagen type XXVIII alpha 1 a |
0.150 |
0.219 |
0.041 |
0.040 |
| TENT_05_19704 |
No hit |
0.500 |
0.461 |
0.013 |
0.013 |
| TENT_06_16512 |
Protein Phosphatase regulatory subunit |
0.105 |
0.266 |
0.510 |
0.484 |
| TENT_07_21161 |
No hit |
0.700 |
0.451 |
0.744 |
0.477 |
| TNNT_2E_20968 |
Troponin T2e |
0.000 |
0.000 |
0.168 |
0.196 |
| TRIM_35_8416 |
Tripartite motif-contaning protein 35 |
0.368 |
0.450 |
0.361 |
0.426 |
| TTN_B_20952 |
Titin b |
0.421 |
0.332 |
0.003 |
0.003 |
| TUB_A_19211 |
Tubulin alpha 2 |
0.550 |
0.489 |
0.010 |
0.009 |
| UBI_A52_5049 |
Ubiquitin A-52 residue ribosomal protein fusion product 1 |
0.474 |
0.362 |
0.058 |
0.056 |
| UGP_2_2128 |
UDP-glucose pyrophosphorylase 2 |
0.316 |
0.808 |
0.236 |
0.771 |
| UGP_A_2307 |
Glycerol-3-phosphate transporter subunit |
0.600 |
0.334 |
0.733 |
0.466 |
| UNA_SINE2_16912 |
Eel Short interspersed elements |
0.000 |
0.000 |
0.000 |
0.000 |
| ZETA_15177 | Tyr 3-monooxygenase/Trp 5-monooxygenase activation protein | 0.421 | 0.388 | 0.246 | 0.245 |
Three loci deviated significantly from Hardy-Weinberg expectations after Bonferroni correction: locus UGP_2_2128, showing a deficit of heterozygotes, and loci TENT_0721161 and UGP_A_2307, showing an excess of heterozygotes. However, those loci were not excluded from the analysis as they could reflect selection.
Two loci (ALD_R and PSME_1_21196) were genotyped at all locations except Tiber, so all tests for selection were conducted considering all 8 locations and 78 loci (excluding ALD_R and PSME_1_21196) and on a restricted data set with 7 locations (excluding Tiber) and 80 loci.
Overall genetic differentiation was low (FST = 0.0079). Using STRUCTURE, a scenario with two clusters (K = 2) corresponding to the two species was the most likely, with no substructuring within species (Figure 1). In the same analysis, a total of 5 individuals from Iceland were identified as admixed individuals with 90% probability intervals that did not overlap with zero. While the SNPs used in this study were not species-diagnostic, results were concordant with the study of Pujolar et al. [23], in which the same individuals were identified as admixed on the basis of 86 species-diagnostic SNPs, encompassing four F1 hybrids and one second generation backcross. Hybrids were only observed in Iceland and were absent in the remaining European locations. All hybrid individuals were removed from further analyses.
Figure 1.
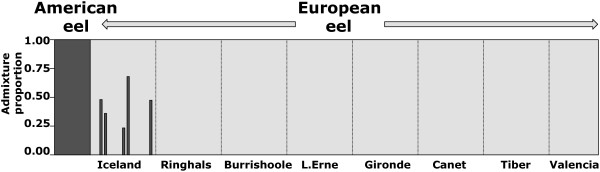
Admixture analysis using STRUCTURE. Individuals were assigned assuming the presence of two groups (K = 2). Each vertical line represents one individual, partitioned into segments according to the proportion of European eel (light) and American eel (dark).
The selection detection workbench LOSITAN identified three outlier loci possibly under diversifying selection using the full data set, GAPDH_20355, MYH_14857 and ALDH_2_16634, with p < 0.05 (Table 2). When using the restricted data set with 7 locations, a further outlier was also identified, ALD_R (p = 0.000). Using the complete data set with 8 locations BAYESCAN identified a single outlier, GAPDH_20355, showing a high FST value of 0.123 relative to the background FST (Table 2). At this locus, the alpha coefficient was positive, suggestive of diversifying selection. When using the restricted data set with 7 locations, ALD_R was also identified as outlier in addition to GAPDH_20355, showing a high FST value of 0.091 and a positive alpha coefficient.
Table 2.
Detection of outlier loci using the F ST -outlier approach implemented in LOSITAN and BAYESCAN based on the full data set with 8 locations (78 loci) and a reduced data set with 7 locations (80 loci)
|
LOSITAN |
|
|
|||
|---|---|---|---|---|---|
| Locus | Gene | Het | F ST | p value | |
|
8 populations |
|
|
|
|
|
| MYH_14857 |
Superfast myosin heavy chain |
0.490 |
0.042 |
0.018 |
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
0.213 |
0.246 |
0.000 |
|
| ALDH_2_16634 |
Aldehyde dehydrogenase 2 |
0.428 |
0.032 |
0.047 |
|
|
7 populations |
|
|
|
|
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
0.231 |
0.256 |
0.000 |
|
| ALDH_2_16634 |
Aldehyde dehydrogenase 2 |
0.435 |
0.036 |
0.012 |
|
| ALD_R |
Aldose reductase |
0.346 |
0.156 |
0.000 |
|
|
BAYESCAN |
|
|
|
|
|
|
Locus |
Gene |
BPP |
q value |
alpha |
F
ST
|
|
8 populations |
|
|
|
|
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
1.000 |
0.000 |
3.376 |
0.123 |
|
7 populations |
|
|
|
|
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
1.000 |
0.000 |
2.424 |
0.116 |
| ALD_R | Aldose reductase | 1.000 | 0.000 | 2.150 | 0.091 |
Heterozygosity and FST values are detailed for all outlier loci detected in LOSITAN. Bayesian posterior probabilities (BPP), q values, alpha coefficients and FST values are detailed for all outlier loci detected in BAYESCAN.
A generalized linear model between allelic frequencies and explanatory variables using the full data set revealed significant associations with temperature (2 loci), latitude (3 loci) and longitude (5 loci) (Table 3). One locus (GAPDH_20355) showed a significant association with both latitude and longitude. No interactions with explanatory variables were found at any locus. Locus ALD_R showed a positive association with longitude when using the restricted data set with 7 locations (p = 0.016).
Table 3.
Statistical associations between allele frequencies and a set of three explanatory variables (TEMP, temperature; LAT, latitude; LON, longitude) assessed using generalized linear models (GLM) and BAYENV based on the full data set with 8 locations (78 loci) and a reduced data set with 7 locations (80 loci)
|
GLM |
|
|
|---|---|---|
| Locus | Gene | p value |
|
8 populations |
|
|
| TRIM_35_8416 |
Tripartite motif-containing protein 35 |
TEMP (r = 0.78; p = 0.023) |
| NEX_19953 |
Nexilin |
LAT (r = 0.80; p = 0.018) |
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
LAT (r = 0.72; p = 0.045) + LON (r = 0.74; p = 0.042) |
| KRT_13_20618 |
Keratin |
TEMP (r = 0.81; p = 0.015) |
| UBI_A52_5049 |
Ubiquitin A-52 |
LON (r = 0.72; p = 0.044) |
| PGK_1_11454 |
Phosphoglycerate kinase |
LON (r = 0.76; p = 0.031) |
| PSA_4_21534 |
Proteasome subunit alpha type-4 |
LAT (r = 0.80; p = 0.018) |
| ALDH_2_16634 |
Aldehyde dehydrogenase 2 |
LON (r = 0.72; p = 0.044) |
| CST_21113 |
Cystatin precursor |
LON (r = 0.72 p = 0.043) |
|
7 populations |
|
|
| TRIM_35_8416 |
Tripartite motif-containing protein 35 |
TEMP (r = 0.77; p = 0.042) |
| NEX_19953 |
Nexilin |
LAT (r = 0.76; p = 0.045) |
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
LON (r = 0.73; p = 0.046) |
| KRT_13_20618 |
Keratin |
TEMP (r = 0.82; p = 0.023) |
| PSA_4_21534 |
Proteasome subunit alpha type-4 |
LAT (r = 0.76; p = 0.047) |
| ALD_R |
Aldose reductase |
LON (r = 0.82; p = 0.016) |
|
BAYENV |
|
|
|
Locus |
Gene |
BF |
|
8 populations |
|
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
LAT (BF = 7.450) |
|
7 populations |
|
|
| GAPDH_20355 |
Glyceraldehyde-3-phosphate dehydrogenase |
LAT (BF = 4.501) |
| ALD_R | Aldose reductase | LON (BF = 3.295) |
Correlation coefficients and p-values are detailed for all loci showing significant associations using GLM. Bayes Factors (BF) are presented for all positive associations in BAYENV.
On the other hand, one positive association was found using BAYENV, locus GAPDH_20355 and latitude, with a Bayes Factor of 7.450 (Table 3), while no associations were found for the rest of loci (Bayes Factor <3). In addition, a positive association was found between locus ALD_R and longitude, with a Bayes Factor of 3.295, when considering the reduced data set with 7 locations.
Overall, we identified a total of 11 candidate loci, 4 from outlier tests plus 7 additional loci from the analysis of association of allelic frequencies with explanatory variables.
Discussion
Signatures of local selection in European eel
The observation of a small set of SNPs showing significantly high genetic differentiation in comparison with the background FST and significant associations between allele frequencies and environmental variables is consistent with the action of spatially varying selection associated with the highly heterogenous habitats that glass eels colonize throughout their geographic range. Our findings fit the general prediction that selective pressures frequently vary in space, often resulting in local selection of ecologically adaptive traits [1].
Overall, we found signatures of selection at a total of 11 loci. We found discordances between the two approaches used (FST outlier tests vs. environmental-association methods), with few SNPs identified as targets of selection by both methods and with a higher number of candidates identified using a generalized linear model. This is in agreement with recent studies showing that SNPs positively correlated with environmental variables were not FST outliers [35-37]. It has been suggested that SNP-environment associations are more sensitive to detect subtle clines in allele frequencies than FST outlier tests and that both approaches might be complementary but not concordant when testing for selection [38].
Nevertheless, three loci in our study (GAPDH, ALDH2 and ALD_R) showed higher genetic differentiation than the background FST together with significant associations between allele frequencies and environmental variables. All three are genes with major metabolic functions: GAPDH (Glyceraldehide 3-phosphate dehydrogenase) is part of the glycolysis pathway and catalyzes the conversion of glyceraldehyde 3-phosphate to D-glycerate 1,3-bisphosphate; ALDH2 (Aldehyde dehydrogenase 2) belongs to the aldehyde dehydrogenase family of enzymes that catalyze acetaldehyde to acetic acid and is the second enzyme of the major oxidative pathway of alcohol metabolism; ALD_R (Aldose reductase) catalyzes the reduction of glucose to sorbitol, the first step in the polyol pathway of glucose metabolism. Besides these genes, a positive environment correlation was observed for PGK (Phosphoglycerate kinase), which is a transferase enzyme in glycolysis acting in the first ATP-generating step of the glycolytic pathway. Surprisingly, none of the above genes linked to metabolism showed positive associations with temperature, arguably an environmental variable of key importance influencing enzymatic activities and metabolic pathways [39,40]. In eels, decreased metabolic activities have been observed below certain threshold temperatures in both European [41] and American eel [42], and distinct behaviour patterns such as upstream migration of glass eels have been shown to be temperature-related [43]. Since it could be argued that temperature at other time intervals might be more relevant than the 30 day-interval used in our study, we re-conducted a generalized linear model between allelic frequencies at GAPDH, ALDH2 and ALD_R with temperature using other time intervals (10 days, 3 months, 6 months, 12 months). No significant associations were found at any of the locus, which suggests that other agents of selection than temperature could underlie the significant associations found. In the case of aldose reductase, this is an enzyme induced by hyperosmolarity stress [44]. Spatially varying selection in European eel regarding osmoregulation seems plausible, since eels occupy highly variable habitats across Europe in terms of salinity, including both fresh and salt-water (i.e. marine and brackish) habitats [45].
Besides genes involved in metabolic functions, several genes involved in defense response showed a positive environment correlation, including TRIM35 (Tripartite motif-containing protein 35), CST (Cystatin precursor), PSA4 (Proteasome subunit alpha-4) and UBIA52 (Ubiquitin A52), all involved in catalytic activity. Interestingly, TRIM35 is a gene implicated in processes associated with innate immunity [46]. Together with other TRIM family genes, TRIM35 is located on a region of significantly elevated genetic diversity (LG XIII) in the threespine stickleback, which suggests that the polymorphism increase on LG XIII has been likely driven by selection on innate immunity genes [7]. While allele frequencies at TRIM35 were positively correlated with temperature, allele frequencies at CST, UBIA52 and PSA4 were associated with geographic coordinates. As in the case of metabolism, it is possible that other explanatory variables (e.g. productivity, oxygen level, salinity, pollution and parasite load) may play a role in defense response rather than temperature or geographic coordinates.
No apparent parallel patterns of selection in North Atlantic eels
The contrasting pattern of spatially varying selection observed in European eel (this study) and American eel [26] using the same panel of candidate SNPs suggests no common genetic-by-environment associations between North Atlantic eels. Using generalized linear models, Gagnaire et al. [26] found significant associations with environmental variables at 8 loci within glass eels (ACP, ANX2, GPX4, HSP90A, MDH, NRAP, PRP40 and UGP2), none of which are common with the 10 loci that showed significant associations in our study using the same statistical approach (ALDH, ALD_R, CST, GAPDH, KRT, NEX, PGK, PSA4, TRIM35, UBIA52) and also conducted on glass eels.
However, two loci (TRIM35 and CST) showed some evidence of selection in both species. In American eel, TRIM35 showed the highest FST value detected between localities (FST = 0.174) although no correlation with environmental variables was detected at this locus. CST showed signatures of selection within cohorts of juveniles but not within glass eels [26].
While most loci under selection in Gagnaire et al. [26] represented metabolic genes associated with temperature, those genes with a major role in metabolism in our study (GAPDH, ALD_R, ALDH, PGK) did not show a positive association with temperature despite the similar temperature ranges encountered by both species (European eel: 4.2-15.1°C, American eel: 3.4-19.8°C). However, the different signatures of selection between species could be due to distinct selective pressures associated with the much longer larval migration for European eel than for American eel, with estimates ranging from 7 months to 2 years for European eel depending on the assumptions and methods used, whereas estimates for American eel range between 6 and 12 months [47]. The one extra year that possibly European eel larvae spend in the open sea could impose a different set of selective agents relative to American eel.
In contrast with our findings, the recent survey of Conte et al. [5] on published literature of repeated phenotypic evolution in natural populations concluded that the probability of gene reuse was high (on average 55%). However, the survey was based on candidate gene studies, which might have biased upward the reuse estimates. The lack of parallel selection patterns in North Atlantic eels is unanticipated owing to the sister species status of European and American eel and the permeable barrier to gene flow between species. A recent study using a RAD-sequencing approach to identify diagnostic markers between the two species found a small proportion of fixed SNPs (<0.5%), while most of the SNPs showed low non-significant differentiation that suggest that most of the genome is homogenized by gene flow [23]. One possible explanation to the lack of parallel selection is that complex phenotypic traits affected by local selection might have a highly polygenic basis, hence influenced by several genes, each with a small contribution to the ultimate function [48,49]. Parallel selection is more likely to occur when the adaptive response is controlled by a single gene, i.e. the Eda gene and armor plate reduction [6,50] and the Kit ligand Kitlg gene and pigmentation [51] in threespine sticklebacks or the melonacortin-1 receptor Mc1r and colour pattern in beach mice [52]. More complex traits are likely to involve a higher number of genes, thus reducing the likelihood of selection acting on the same genes in multiple species or locations, as it has been argued in the case of osmoregulation in threespine sticklebacks [7,8]. Similarly, partial parallel patterns of genetic differentiation have been observed between two whitefish sympatric species pairs (a normal benthic and a dwarf limnetic) across lakes, suggestive of polygenic adaptation [53-55].
Conclusions
The distinct signatures of selection between North American eels could be attributable to differences in larval migration between species. Alternatively, the fact that many genes of small effect likely shape adaptive pathways (i.e. metabolism, growth, osmoregulation, pathogen resistance) could explain the private signatures of spatially varying selection with no shared genetic-by-environment associations between European and American eel. As an alternative to candidate loci approaches, high-density genome-wide scans using next-generation sequencing and genotyping-by-sequencing approaches [7,56] might be more adequate. A recent study using RAD (Restriction site Associated DNA) sequencing generated a SNP resource for European eel consisting of 82,425 loci and over 375,000 SNPs [57] that provides a valuable tool for future studies on parallel selection in both North Atlantic eels on a genome-wide scale.
Methods
Ethical statement
No experiments were conducted on the animals and animal manipulation was limited to sacrificing fish, using the least painful method to obtain tissue samples for DNA extraction. In all cases, in order to minimize the suffering of the animals used in the study, fish were deeply anaesthetized with MS-222 (3-amonobenzoic acid ethyl ester) or 2-phenoxyethanol 1% and then painlessly sacrificed. All procedures were conducted by technical staff, who had all the necessary fishing and animal ethics permits (please see in the Additional file 1: Appendix 1).
Sampling
A total of 321 European eel (Anguilla anguilla) individuals were collected at 8 locations across the geographical distribution of the species, from Iceland to the Mediterranean Sea (Table 4; Figure 2). All individuals were glass eels caught by electrofishing (Iceland) and fyke nets (remaining localities). Individuals from Iceland were collected at four separate sampling sites in southwestern Iceland, but pooled to increase sample size. Additionally, 20 American eel (Anguilla rostrata) individuals collected at Rivière Blanche (Québec, Canada), Mira River (Nova Scotia, Canada), Wye River (MD, US), Medomak River (ME, US) and Boston Harbor (MA, US) were used for comparison. Genomic DNA was extracted using standard phenol-chloroform extraction.
Table 4.
Sampling details including number of individuals per sampling location (N), latitude, longitude and sea-surface temperature at river mouth averaged across the 30 days preceding the sampling date
| Country | Location | N | Latitude | Longitude | Date | Temp (°C) |
|---|---|---|---|---|---|---|
| Spain |
Valencia |
44 |
39°46′ N |
0°24′ W |
15 January 2010 |
15.05 |
| Italy |
Tiber |
39 |
41°73′ N |
12°23′ E |
30 December 2007 |
14.17 |
| South France |
Canet |
40 |
42°70′ N |
3°15′ E |
23 January 2008 |
13.24 |
| West France |
Gironde |
40 |
44°86′ N |
0°42′ W |
16 April 2008 |
11.26 |
| Ireland |
Burrishoole |
39 |
53°90′ N |
9°58′ W |
14 March 2005 |
9.57 |
| Northern Ireland |
Lough Erne |
39 |
54°46′ N |
7°77′ W |
1 July 2008 |
13.85 |
| Sweden |
Ringhals |
40 |
57°21′ N |
12°27′ E |
15 March 2008 |
4.19 |
| Iceland |
|
40 |
|
|
May – June 2001 |
8.45 |
| |
Stokkseyri |
10 |
63°81′ N |
21°04′ W |
|
|
| |
Vifilsstadvatn |
10 |
64°07′ N |
21°87′ W |
|
|
| |
Seljar |
10 |
64°56′ N |
22°31′ W |
|
|
| Vogslækur | 10 | 64°69′ N | 22°33′ W |
Figure 2.
Sampling locations of European eel (circles) and American eel (stars) individuals used for comparison.
SNP genotyping
We examined the panel of 100 coding-gene SNPs developed by Gagnaire et al. [26] in American eel. 20 out of the 100 primer sets did not give good amplification products in European eel and were excluded. All individuals were genotyped at 80 coding-gene SNPs: 47 SNPs that were detected as outliers between samples from Florida and Québec using RNA-sequencing data (including 4 SNPs identified within allozyme-coding genes showing clinal variation in Williams et al. [33]) and 33 SNPs that were not outliers (Table 2). SNP genotyping was conducted using the Kbioscience Competitive Allele-Specific PCR genotyping system (KASPar) (Kbioscience, Hoddeston, UK).
Data analysis
Allele frequencies, measures of genetic diversity including polymorphism at the 95% (P95) and 99% level (P99), observed (Ho) and expected (He) heterozygosities and deviations from Hardy-Weinberg equilibrium were calculated using GENEPOP [58]. In all cases, significance levels were corrected for multiple comparisons using the sequential Bonferroni technique [59].
Overall genetic differentiation (FST) was calculated in GENEPOP. Population structure was further investigated using STRUCTURE v.2.3.4 [60], which also allowed us to test the presence of hybrids in the data set. We assumed an admixture model, uncorrelated allele frequencies and we did not use population priors. Given that two panmictic species were analyzed, we assumed k = 2 and conducted 10 replicates to check the consistency of results. A burn-in length of 100,000 steps followed by one million additional iterations was performed.
We used two different approaches to test for evidence of local selection. First, we searched for elevated population differentiation using FST-based outlier analyses. We used the selection detection workbench LOSITAN [61], which uses a coalescent-based simulation approach to identify outliers based on the distributions of heterozygosity and FST[62]. First, LOSITAN was run using all SNPs to estimate the mean neutral FST as recommended by Antao et al. [61]. After the first run, the mean neutral FST was re-computed by removing those SNPs outside the confidence interval in order to obtain a better approximation of the mean neutral FST. This mean was then used to conduct a second and final run of LOSITAN using all SNPs. The analysis was performed on the whole data set divived according to sampling location. An estimate of p value was obtained for each SNP. We used a threshold of 0.95 and a false discovery rate of 0.1 to minimize the number of false positives.
Outlier SNPs were also detected using BAYESCAN [63], a Bayesian method based on a logistic regression model that separates locus-specific effects of selection from population-specific effects of demography. Outlier analysis was conducted on the whole data set divided according to sampling location. BAYESCAN runs were implemented using default values for all parameters, including a total of 100,000 iterations after an initial burn-in of 50,000 steps. Posterior probabilities, q values and alpha coefficients (positive values indicate diversifying selection, negative values are indicative of balancing selection) were calculated. A q-value of 10% was used for significance.
As an alternative to FST-outlier tests, our second approach for identifying targets of local selection was to test for significant statistical associations between allelic frequencies and environmental variables following Gagnaire et al. [26], using a generalized linear model in r. Environmental variables used included degrees North latitude, degrees East/West longitude, and sea-surface temperature at river mouth averaged across the 30 days preceding the sampling date. Sea-surface temperature data were retrieved from the IRI (International Research Institute for Climate and Society) Climate Data Library (http://iridl.ldeo.columbia.edu/) database “NOAA NCDC OISST version2 AVHRR SST: Daily Sea Surface Temperature”. We also searched for SNP-environment associations using BAYENV [38], which tests for covariance between candidate SNP allele frequencies and environment variables that exceed the expected covariances under genetic drift. First, SNP frequencies at all loci were used to describe how allele frequencies covary across populations, hence avoiding population-specific effects of demography (even though a panmixia scenario is most likely to apply for European eel). After the covariance matrix was estimated, the program determined the Bayes factors for the environmental variables of interest. Bayes Factors >3 were considered indicative of an allele frequency correlation with an environmental variable.
Availability of supporting data
The data set supporting the results of this article is available from Dryad: http://datadryad.org/resource/doi:10.5061/dryad.jn800/1.
Competing interests
The authors declare no competing interest.
Authors’ contributions
MMH and LB conceived and designed the project. MGU and JMP conducted population genetics analyses with help from MMH, TDA, ALF and MWJ. MGU and JMP wrote the manuscript with contributions from MMH, LB, PAP, ALF, MWJ, TDA, JF, PKB and BJ. All authors read and approved the final version of the manuscript.
Supplementary Material
Supplementary ethical statement.
Contributor Information
Malene G Ulrik, Email: maleneu@hotmail.com.
José Martín Pujolar, Email: jmartin@biology.au.dk.
Anne-Laure Ferchaud, Email: annelaure.ferchaud@biology.au.dk.
Magnus W Jacobsen, Email: magnus.jacobsen@biology.au.dk.
Thomas D Als, Email: tda@hum-gen.au.dk.
Pierre Alexandre Gagnaire, Email: papagnai@univ-montp2.fr.
Jane Frydenberg, Email: jane.frydenberg@biology.au.dk.
Peder K Bøcher, Email: peder.bocher@biology.au.dk.
Bjarni Jónsson, Email: bjarni@holar.is.
Louis Bernatchez, Email: Louis.Bernatchez@bio.ulaval.ca.
Michael M Hansen, Email: michael.m.hansen@biology.au.dk.
Acknowledgements
We thank Eleonora Ciccotti, Russell Poole, Javier Lobón-Cervia, Eric Feunteun, Francoise Daverat and Håkan Wickström for providing samples, Annie Brandstrup for technical assistance, Virginia Settepani for help in preparing the figures and Mads F. Schou for help in statistical analysis. We acknowledge funding from the Danish Council for Independent Reasearch, Natural Sciences (grant 09–072120 to MMH).
References
- Kawecki TJ, Ebert D. Conceptual issues in local adaptation. Ecol Lett. 2004;7:1225–1241. doi: 10.1111/j.1461-0248.2004.00684.x. [DOI] [Google Scholar]
- Stapley J, Reger J, Feulner PGD, Smadja C, Galindo J, Ekblom R, Bennison C, Ball AD, Beckerman AP, Slate J. Adaptation genomics: the next generation. Trends Ecol Evol. 2010;12:705–712. doi: 10.1016/j.tree.2010.09.002. [DOI] [PubMed] [Google Scholar]
- Wood TE, Burke JM, Reiseberg LH. Parallel genotypic adaptation: when evolution repeats itself. Genetica. 2005;123:157–170. doi: 10.1007/s10709-003-2738-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, Meyer A. Adaptation in the age of ecological genomics: insights from parallelism and convergence. Trends Ecol Evol. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- Conte GL, Arnegard ME, Peichel CL, Schluter D. The probability of genetic parallelism and convergence in natural populations. Proc R Soc Lond B. 2012;279:5039–5047. doi: 10.1098/rspb.2012.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colosimo PF, Hosemann KE, Balabhadra W, Villarreal G, Dickson M, Grimwood J, Schmutz J, Myers RM, Schluter D, Kingsley DM. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science. 2005;307:1928–1933. doi: 10.1126/science.1107239. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Etter PD, Stiffler N, Johnson EA, Cresko WA. Population genomics of parallel adaptation in threespine stickleback using sequenced RAD tags. PLoS Genet. 2010;6:e1000862. doi: 10.1371/journal.pgen.1000862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFaveri J, Shikano T, Shimada Y, Goto A, Merilä J. Global analysis of genes involved in freshwater adaptation in threespine sticklebacks (Gasterosteus aculeatus) Evolution. 2011;65:1800–1807. doi: 10.1111/j.1558-5646.2011.01247.x. [DOI] [PubMed] [Google Scholar]
- Jones FC, Chan YF, Schmutz J, Grimwood J, Brady SD, Southwick AM, Absher DM, Myers RM, Reimchen T, Deagle BE, Schluter D, Kingslet DM. A genome-wide SNP genotyping array reveals patterns of global and repeated species-pair divergence in sticklebacks. Curr Biol. 2012;22:83–90. doi: 10.1016/j.cub.2011.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, Birney E, Searle S, Schmutz J, Grimwood J, Dickson MC, Myers RM, Miller CT, Summers BR, Knecht AK, Brady SD, Zhang H, Pollen AA, Howes T, Amemiya C. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault S, Owens GL, Rieseberg LH. Shared selective pressure and local genomic landscape lead to repeatable patterns of genomic divergence in sunflowers. Mol Ecol. 2014;23:311–324. doi: 10.1111/mec.12600. [DOI] [PubMed] [Google Scholar]
- Boëtius J. Atlantic Anguilla: a presentation of old and new data of total number of vertebrae with special reference to the occurrence of Anguilla rostrata in Europe. Dana. 1980;1:93–112. [Google Scholar]
- Avise JC, Helfman GS, Saunders NC, Hales LS. Mitochondrial DNA differentiation in North Atlantic eels: population genetic consequences of an unusual life history pattern. Proc Natl Acad Sci U S A. 1986;83:4350–4354. doi: 10.1073/pnas.83.12.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mineguishi Y, Aoyama J, Inoue JG, Miya M, Nishida M, Tsukamoto K. Molecular phylogeny and evolution of the freshwater eels genus Anguilla based on the whole mitochondrial genome sequences. Mol Phylogenet Evol. 2005;34:134–146. doi: 10.1016/j.ympev.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Jacobsen MW, Pujolar JM, Gilbert TP, Mayar JVM, Bernatchez L, Als TD, Hansen MM. Speciation and demographic history of Atlantic eels (Anguilla anguilla and A. rostrata) revealed by mitogenome sequencing. Heredity. 2014. in press. [DOI] [PMC free article] [PubMed]
- Mank JE, Avise JC. Microsatellite variation and differentiation in North Atlantic eels. J Hered. 2003;94:30–34. doi: 10.1093/jhered/esg062. [DOI] [PubMed] [Google Scholar]
- Wirth T, Bernatchez L. Decline of Atlantic eels: a fatal synergy? Proc R Soc Lond B. 2003;270:681–688. doi: 10.1098/rspb.2002.2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Albert V, Jónsson B, Bernatchez L. Natural selection influences AFLP intraspecific variability and introgression patterns in Atlantic eels. Mol Ecol. 2009;18:1678–1691. doi: 10.1111/j.1365-294X.2009.04142.x. [DOI] [PubMed] [Google Scholar]
- Als TD, Hansen MM, Maes GE, Castonguay M, Riemann L, Aerestrup K, Munk P, Sparholt T, Hanel R, Bernatchez L. All roads lead to home: panmixia of European eel in the Sargasso Sea. Mol Ecol. 2011;20:1333–1346. doi: 10.1111/j.1365-294X.2011.05011.x. [DOI] [PubMed] [Google Scholar]
- McCleave JD, Kleckner RC, Castonguay M. Reproductive sympatry of American and European eels and implications for migration and taxonomy. Am Fish Soc Symp. 1987;1:286–297. [Google Scholar]
- Avise JC, Nelson WS, Arnold J, Koehn RK, Williams GC, Thorsteinsson V. The evolutionary genetic status of Icelandic eels. Evolution. 1990;44:1254–1262. doi: 10.2307/2409286. [DOI] [PubMed] [Google Scholar]
- Albert V, Jónsson B, Bernatchez L. Natural hybrids in Atlantic eels (Anguilla anguilla, A. rostrata): evidence for successful reproduction and fluctuating abundance in space and time. Mol Ecol. 2006;15:1903–1916. doi: 10.1111/j.1365-294X.2006.02917.x. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, Jacobsen MW, Als TD, Frydenberg J, Magnussen E, Jónsson B, Jiang X, Cheng L, Bekkevold D, Maes GE, Bernatchez L, Hansen MM. Assessing patterns of hybridization between North Atlantic eels using diagnostic single-nucleotide polymorphisms. Heredity. 2014. in press. [DOI] [PMC free article] [PubMed]
- Van den Thillart G, Rankin JC, Dufour S. Spawning migration of the European eel: reproduction index, a useful tool for conservation management. Dordecht, The Netherlands: Springer; 2009. [Google Scholar]
- Fraser DJ, Weir LK, Bernatchez L, Hansen MM, Taylor EB. Extend and scale of local adaptation in salmonid fishes: review and meta-analysis. Heredity. 2011;106:404–420. doi: 10.1038/hdy.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Normandeau E, Côté C, Hansen MM, Bernatchez L. The genetic consequences of spatially varying selection in the panmictic American eel (Anguilla rostrata) Genetics. 2012;190:725–736. doi: 10.1534/genetics.111.134825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levene H. Genetic equilibrium when more than one ecological niche is available. Am Nat. 1953;87:331–333. doi: 10.1086/281792. [DOI] [Google Scholar]
- Hedrick PW. Genetic polymorphism in heterogeneous environments: the age of genomics. Annu Rev Ecol Evol Syst. 2006;37:67–93. doi: 10.1146/annurev.ecolsys.37.091305.110132. [DOI] [Google Scholar]
- Yeaman S, Otto SP. Establishment and maintenance of adaptive genetic divergence under migration, selection and drift. Evolution. 2011;65:2123–2129. doi: 10.1111/j.1558-5646.2011.01277.x. [DOI] [PubMed] [Google Scholar]
- Dannewitz J, Maes GE, Johansson L, Wickström H, Volckaert FAM, Jarvi T. Panmixia in the European eel: a matter of time. Proc R Soc Lond B. 2005;272:1129–1137. doi: 10.1098/rspb.2005.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Côté C, Gagnaire PA, Bourret V, Verrault G, Castonguay M, Bernatchez L. Population genetics of the American eel (Anguilla rostrata): FST = 0 and North Atlantic Oscillation effects on demographic fluctuations of a panmictic species. Mol Ecol. 2013;22:1763–1776. doi: 10.1111/mec.12142. [DOI] [PubMed] [Google Scholar]
- Maes GE, Volckaert FAM. Clinal genetic variation and isolation by distance in the European eel Anguilla anguilla. Biol J Linn Soc. 2002;77:509–522. doi: 10.1046/j.1095-8312.2002.00124.x. [DOI] [Google Scholar]
- Williams GC, Koehn RK, Mitton JB. Genetic differentiation without isolation in the American eel, Anguilla rostrata. Evolution. 1973;27:192–204. doi: 10.2307/2406960. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Serrao EA, Pearson GA, Riginos C, Rawson PD, Hilbish TJ, Brawley SH, Trussell GC, Carrington E, Wethey DS, Grahame JW, Bonhomme F, Rand DM. Ecological genetics in the North Atlantic: environmental gradients and adaptation at specific loci. Ecology. 2008;89:S91–S107. doi: 10.1890/07-1162.1. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, Alkorta-Aranburu G, Beall C, Gebremedhin A, Sukernik R, Utermann G, Pritchard J, Coop G, Di Rienzo A. Colloquium paper: human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A. 2010;107:8924–8930. doi: 10.1073/pnas.0914625107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma XF, Hall D, Onge KR, Jansson S, Ingvarsson PK. Genetic differentiation, clinal variation and phenotypic associations with growth cessation across the Populus tremula photoperiodic pathway. Genetics. 2010;186:1033–1044. doi: 10.1534/genetics.110.120873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller SR, Levsen N, Olson MS, Tiffin P. Local adaptation in the flowering-time gene network of balsam poplar, Populus balsamifera. Mol Biol Evol. 2012;29:3143–3152. doi: 10.1093/molbev/mss121. [DOI] [PubMed] [Google Scholar]
- Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somero GN. Adaptation of enzymes to temperature: searching for basic “strategies”. Comp Biochem Physiol B Biochem Mol Biol. 2004;139:321–333. doi: 10.1016/j.cbpc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Lim ST, Kay RM, Bailey GS. Lactate dehydrogenase isozymes in salmonid fish: evidence for unique and rapid functional divergence of duplicated H-4 lactate dehydrogenases. J Biol Chem. 1975;10:1790–1800. [PubMed] [Google Scholar]
- Nyman L. Some effects of temperature on eel (Anguilla) behaviour. Rep Inst Freshw Res Drottingholm. 1972;52:90–102. [Google Scholar]
- Walsh PJ, Foster GD, Moon TW. The effects of temperature and metabolism of the American eel Anguilla rostrata: compensation in the summer and torpor in the winter. Physiol Zool. 1983;56:532–540. [Google Scholar]
- Linton ED, Jónsson B, Noakes DLG. Effects of water temperature on the swimming and climbing behavior of glass eels Anguilla spp. Environ Biol Fishes. 2007;78:189–192. doi: 10.1007/s10641-005-1367-9. [DOI] [Google Scholar]
- Henry DN, Delmonte M, Greene DA, Killen PD. Altered aldose reductase regulation in human retinal-pigment epithelial-cells. J Clin Invest. 1993;92:617–623. doi: 10.1172/JCI116629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daverat F, Limburg K, Thibaut I, Shiao JC, Dodson JJ, Caron F, Tzeng WN, Iizuka Y, Wickström H. Phenotypic plasticity of habitat use by three temperate eel species Anguilla anguilla, A. japonica and A. rostrata. Mar Ecol Prog Ser. 2006;308:231–241. [Google Scholar]
- Carthagena L, Bergamaschi A, Luna JM, David A, Uchil PD, Margottin-Goguet F, Mothes W, Hazan U, Transy C, Pancino G, Nisole S. Human TRIM gene expression in response to interferons. PLoS One. 2009;4:e4894. doi: 10.1371/journal.pone.0004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonhommeau S, Blanke B, Tréguier AM, Grima N, Rivot E, Vermard Y, Greiner E, Le Pape O. How fast can the European eel (Anguilla anguilla) larvae cross the Atlantic Ocean? Fish Oceanogr. 2009;18:371–385. doi: 10.1111/j.1365-2419.2009.00517.x. [DOI] [Google Scholar]
- Orr HA. The genetic theory of adaptation: a brief history. Nature Rev Genet. 2005;6:119–127. doi: 10.1038/nrg1523. [DOI] [PubMed] [Google Scholar]
- Wainwright PC, Alfaro ME, Bolnick DI, Hulsey CD. Many-to-one mapping of form to function: a general principle in organismal design? Integr Comp Biol. 2005;45:256–262. doi: 10.1093/icb/45.2.256. [DOI] [PubMed] [Google Scholar]
- Raeymaekers JAM, Konijnendijk N, Larmuseau MHD, Hellemans B, De Meester L, Volckaert FAM. A gene with major phenotypic effects as a target for selection versus homogenizing gene flow. Mol Ecol. 2014;23:162–181. doi: 10.1111/mec.12582. [DOI] [PubMed] [Google Scholar]
- Miller CT, Beleza S, Pollen AA, Schluter D, Kittles RA, Shriver MD, Kingsley DM. Cis-regulatory changes in Kit ligand expression and parallel evolution of pigmentation in sticklebacks and humans. Cell. 2007;131:1179–1189. doi: 10.1016/j.cell.2007.10.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoekstra HE, Hirschmann RJ, Bundey RA, Insel PA, Crossland JP. A single amino acid mutation contributes to adaptive beach mouse color pattern. Science. 2006;313:101–104. doi: 10.1126/science.1126121. [DOI] [PubMed] [Google Scholar]
- Roger SM, Bernatchez L. The genetic architecture of ecological speciation and the association with signatures of selection in natural lake whitefish (Coregonus sp.) species pairs. Mol Biol Evol. 2007;24:1423–1438. doi: 10.1093/molbev/msm066. [DOI] [PubMed] [Google Scholar]
- Bernatchez L, Renault S, Whiteley AR, Derome N, Jeukens J, Landry L, Lu G, Nolte AW, Østbye K, Rogers SM, St-Cyr J. On the origin of species: insights from the ecological genomics of whitefish. Phil Trans R Soc B. 2010;367:354–363. doi: 10.1098/rstb.2009.0274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagnaire PA, Pavey SA, Normandeau E, Bernatchez L. The genetic architecture of reproductive isolation during speciation-with-gene-flow in lake whitefish species pairs assesed by RAD sequencing. Evolution. 2013;67:2483–2497. doi: 10.1111/evo.12075. [DOI] [PubMed] [Google Scholar]
- Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML. Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nature Rev Genet. 2011;12:499–510. doi: 10.1038/nrg3012. [DOI] [PubMed] [Google Scholar]
- Pujolar JM, Jacobsen MW, Frydenberg J, Als TD, Larsen PF, Maes GE, Zane L, Jian JB, Cheng L, Hansen MM. A resource of genome-wide single-nucleotide polymorphisms generated by RAD tag sequencing in the critically endangered European eel. Mol Ecol Resour. 2013;13:706–714. doi: 10.1111/1755-0998.12117. [DOI] [PubMed] [Google Scholar]
- Raymond M, Rousset F. GENEPOP (version 1.2): a population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–249. [Google Scholar]
- Rice WR. Analyzing tables and statistical tests. Evolution. 1989;43:223–225. doi: 10.2307/2409177. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antao T, Lopes A, Lopes RJ, Beja-Pereira A, Luikart G. LOSITAN- a workbench to detect molecular adaptation based on a FST-outlier method. BMC Bioinformatics. 2008;9:323. doi: 10.1186/1471-2105-9-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaumont MA, Nichols RA. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond B. 1996;263:1619–1626. doi: 10.1098/rspb.1996.0237. [DOI] [Google Scholar]
- Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary ethical statement.


