Abstract
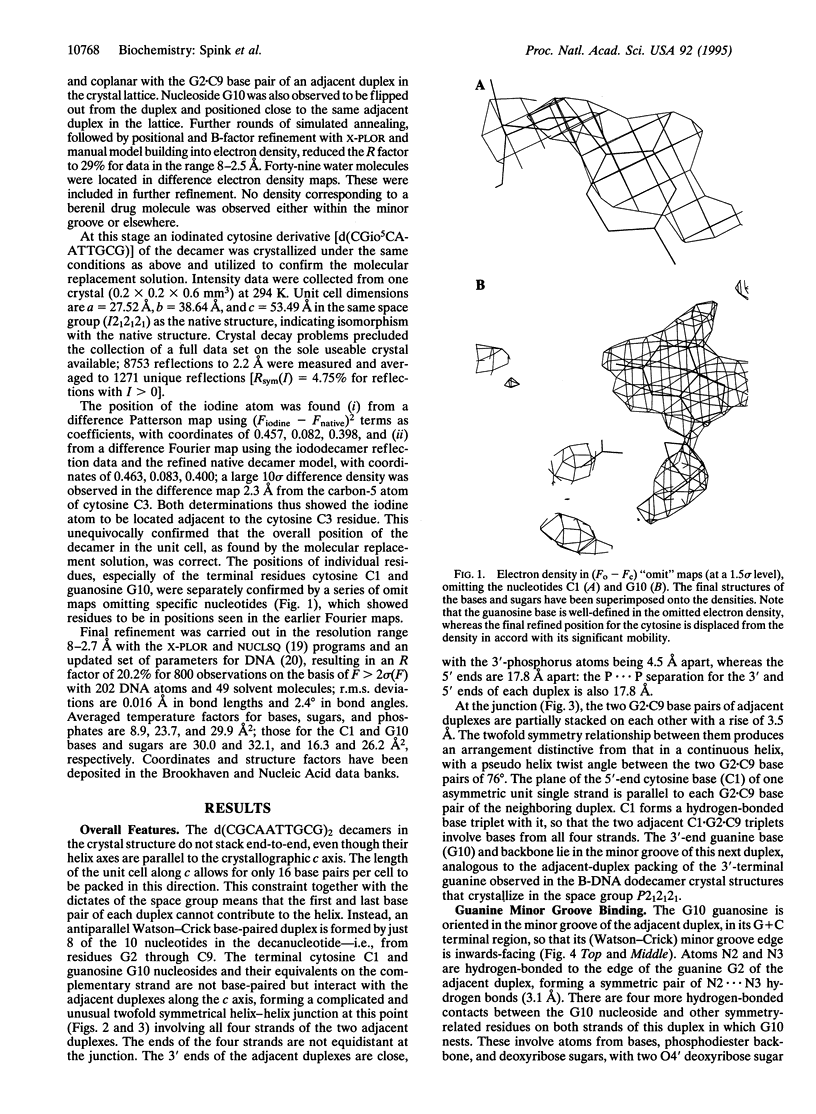
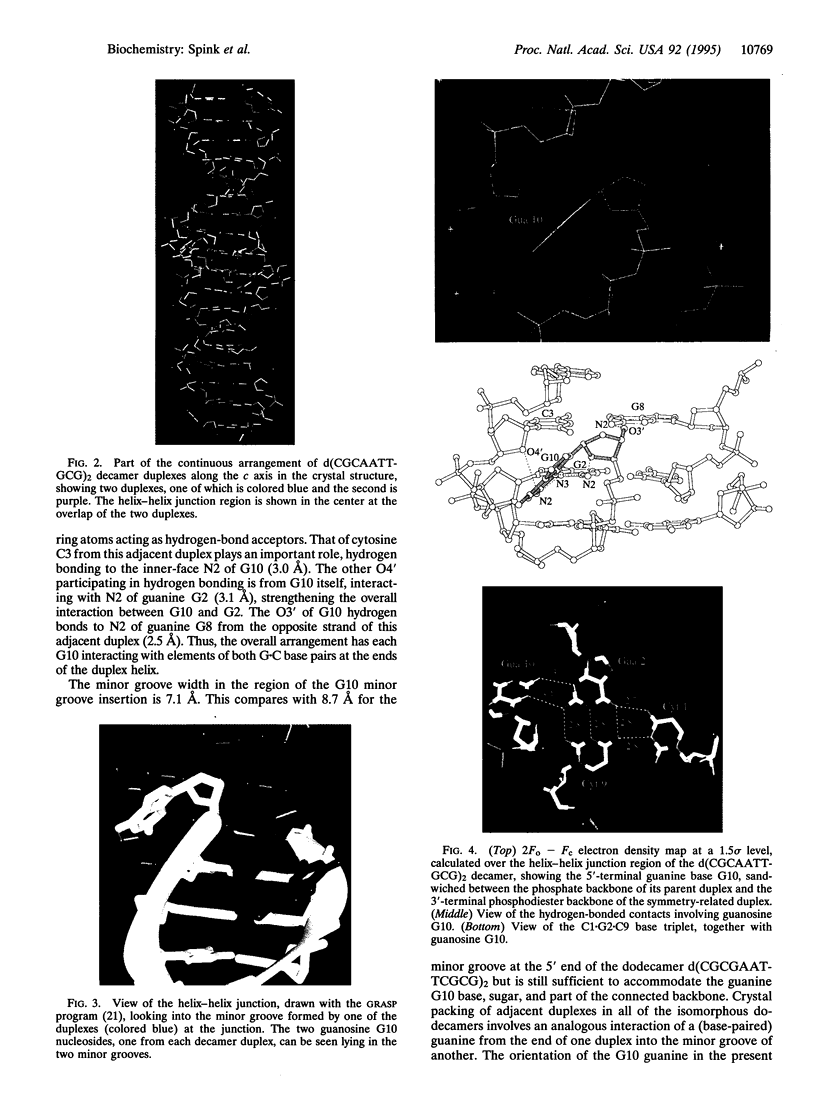
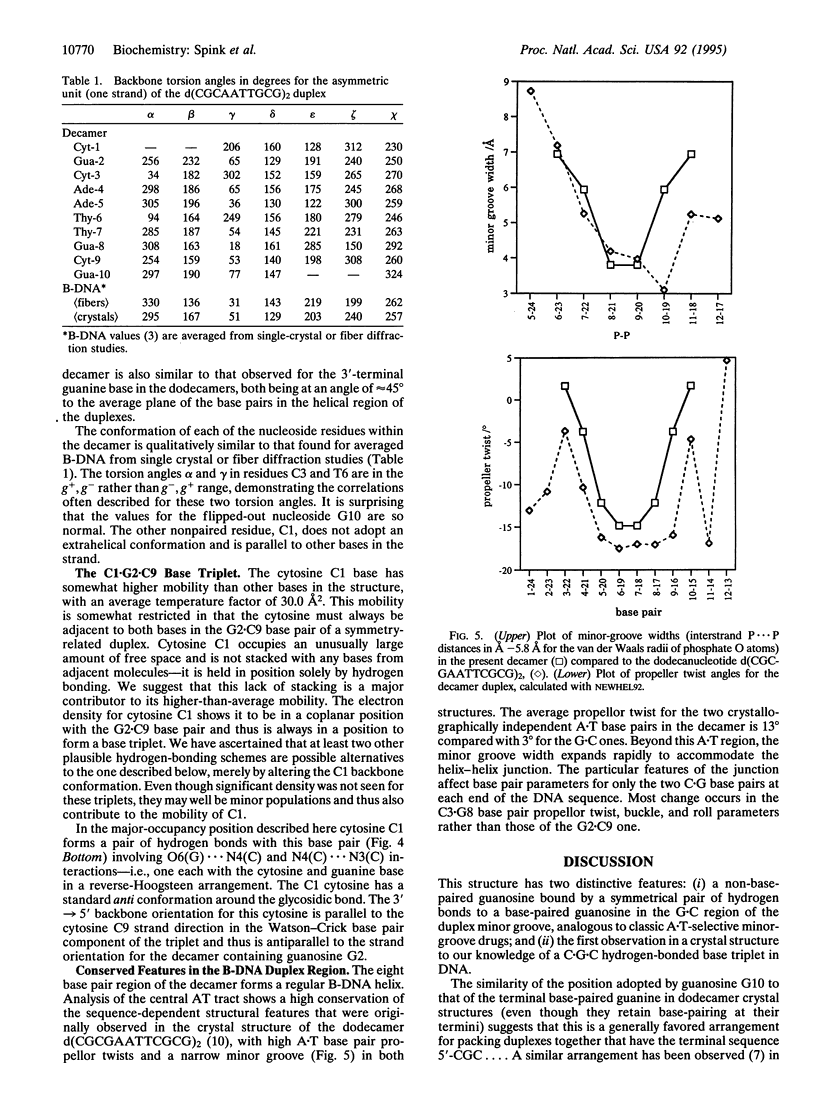
The crystal structure of the decanucleotide d(CGCAATTGCG)2 has been solved by a combination of molecular replacement and heavy-atom procedures and has been refined to an R factor of 20.2% at 2.7 A. It is not a fully base-paired duplex but has a central core of eight Watson-Crick base pairs flanked by unpaired terminal guanosines and cytosines. These participate in hydrogen-bonding arrangements with adjacent decamer duplexes in the crystal lattice. The unpaired guanosines are bound in the G+C regions of duplex minor grooves. The cytosines have relatively high mobility, even though they are constrained to be in one region where they are involved in base-paired triplets with G.C base pairs. The 5'-AATT sequence in the duplex region has a narrow minor groove, providing further confirmation of the sequence-dependent nature of groove width.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brady S. T. Motor neurons and neurofilaments in sickness and in health. Cell. 1993 Apr 9;73(1):1–3. doi: 10.1016/0092-8674(93)90151-f. [DOI] [PubMed] [Google Scholar]
- Brünger A. T., Kuriyan J., Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987 Jan 23;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- Chattopadhyaya R., Grzeskowiak K., Dickerson R. E. Structure of a T4 hairpin loop on a Z-DNA stem and comparison with A-RNA and B-DNA loops. J Mol Biol. 1990 Jan 5;211(1):189–210. doi: 10.1016/0022-2836(90)90020-M. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E. DNA structure from A to Z. Methods Enzymol. 1992;211:67–111. doi: 10.1016/0076-6879(92)11007-6. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Dickerson R. E., Goodsell D. S., Neidle S. "...the tyranny of the lattice...". Proc Natl Acad Sci U S A. 1994 Apr 26;91(9):3579–3583. doi: 10.1073/pnas.91.9.3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. J., Jenkins T. C., Neidle S. Crystal structure of a pentamidine-oligonucleotide complex: implications for DNA-binding properties. Biochemistry. 1992 Aug 11;31(31):7104–7109. doi: 10.1021/bi00146a011. [DOI] [PubMed] [Google Scholar]
- Geierstanger B. H., Mrksich M., Dervan P. B., Wemmer D. E. Design of a G.C-specific DNA minor groove-binding peptide. Science. 1994 Oct 28;266(5185):646–650. doi: 10.1126/science.7939719. [DOI] [PubMed] [Google Scholar]
- Goodsell D. S., Kaczor-Grzeskowiak M., Dickerson R. E. The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T-A steps. J Mol Biol. 1994 May 27;239(1):79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L., Frolow F., Appella E., Hope H., Rabinovich D., Sussman J. L. Three-dimensional structures of bulge-containing DNA fragments. J Mol Biol. 1992 May 20;225(2):397–431. doi: 10.1016/0022-2836(92)90929-e. [DOI] [PubMed] [Google Scholar]
- Kennard O., Salisbury S. A. Oligonucleotide X-ray structures in the study of conformation and interactions of nucleic acids. J Biol Chem. 1993 May 25;268(15):10701–10704. [PubMed] [Google Scholar]
- Klimasauskas S., Kumar S., Roberts R. J., Cheng X. HhaI methyltransferase flips its target base out of the DNA helix. Cell. 1994 Jan 28;76(2):357–369. doi: 10.1016/0092-8674(94)90342-5. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Yoon C., Goodsell D., Pjura P., Dickerson R. E. Binding of an antitumor drug to DNA, Netropsin and C-G-C-G-A-A-T-T-BrC-G-C-G. J Mol Biol. 1985 Jun 25;183(4):553–563. doi: 10.1016/0022-2836(85)90171-8. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski S. C., Eggleston A. K. Homologous pairing and DNA strand-exchange proteins. Annu Rev Biochem. 1994;63:991–1043. doi: 10.1146/annurev.bi.63.070194.005015. [DOI] [PubMed] [Google Scholar]
- Larsen T. A., Kopka M. L., Dickerson R. E. Crystal structure analysis of the B-DNA dodecamer CGTGAATTCACG. Biochemistry. 1991 May 7;30(18):4443–4449. doi: 10.1021/bi00232a010. [DOI] [PubMed] [Google Scholar]
- Malinina L., Urpí L., Salas X., Huynh-Dinh T., Subirana J. A. Recombination-like structure of d(CCGCGG). J Mol Biol. 1994 Oct 28;243(3):484–493. doi: 10.1006/jmbi.1994.1674. [DOI] [PubMed] [Google Scholar]
- Narayana N., Ginell S. L., Russu I. M., Berman H. M. Crystal and molecular structure of a DNA fragment: d(CGTGAATTCACG). Biochemistry. 1991 May 7;30(18):4449–4455. doi: 10.1021/bi00232a011. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp K. A., Honig B. Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins. 1991;11(4):281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- Privé G. G., Yanagi K., Dickerson R. E. Structure of the B-DNA decamer C-C-A-A-C-G-T-T-G-G and comparison with isomorphous decamers C-C-A-A-G-A-T-T-G-G and C-C-A-G-G-C-C-T-G-G. J Mol Biol. 1991 Jan 5;217(1):177–199. doi: 10.1016/0022-2836(91)90619-h. [DOI] [PubMed] [Google Scholar]
- Quintana J. R., Grzeskowiak K., Yanagi K., Dickerson R. E. Structure of a B-DNA decamer with a central T-A step: C-G-A-T-T-A-A-T-C-G. J Mol Biol. 1992 May 20;225(2):379–395. doi: 10.1016/0022-2836(92)90928-d. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan I., Patel D. J. DNA triplexes: solution structures, hydration sites, energetics, interactions, and function. Biochemistry. 1994 Sep 27;33(38):11405–11416. doi: 10.1021/bi00204a001. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B., Sundaralingam M. High resolution crystal structure of the A-DNA decamer d(CCCGGCCGGG). Novel intermolecular base-paired G*(G.C) triplets. J Mol Biol. 1993 May 20;231(2):431–444. doi: 10.1006/jmbi.1993.1292. [DOI] [PubMed] [Google Scholar]
- Spink N., Brown D. G., Skelly J. V., Neidle S. Sequence-dependent effects in drug-DNA interaction: the crystal structure of Hoechst 33258 bound to the d(CGCAAATTTGCG)2 duplex. Nucleic Acids Res. 1994 May 11;22(9):1607–1612. doi: 10.1093/nar/22.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabernero L., Verdaguer N., Coll M., Fita I., van der Marel G. A., van Boom J. H., Rich A., Aymamí J. Molecular structure of the A-tract DNA dodecamer d(CGCAAATTTGCG) complexed with the minor groove binding drug netropsin. Biochemistry. 1993 Aug 24;32(33):8403–8410. doi: 10.1021/bi00084a004. [DOI] [PubMed] [Google Scholar]
- Westhof E., Dumas P., Moras D. Crystallographic refinement of yeast aspartic acid transfer RNA. J Mol Biol. 1985 Jul 5;184(1):119–145. doi: 10.1016/0022-2836(85)90048-8. [DOI] [PubMed] [Google Scholar]
- Yuan H., Quintana J., Dickerson R. E. Alternative structures for alternating poly(dA-dT) tracts: the structure of the B-DNA decamer C-G-A-T-A-T-A-T-C-G. Biochemistry. 1992 Sep 1;31(34):8009–8021. [PubMed] [Google Scholar]
- Zhurkin V. B., Raghunathan G., Ulyanov N. B., Camerini-Otero R. D., Jernigan R. L. A parallel DNA triplex as a model for the intermediate in homologous recombination. J Mol Biol. 1994 Jun 3;239(2):181–200. doi: 10.1006/jmbi.1994.1362. [DOI] [PubMed] [Google Scholar]