Abstract
Epigenetic alterations have been identified as a major characteristic in human cancers. Advances in the field of epigenetics have contributed significantly in refining our knowledge of molecular mechanisms underlying malignant transformation. DNA methylation and microRNA expression are epigenetic mechanisms that are widely altered in human cancers including hepatocellular carcinoma (HCC), the third leading cause of cancer related mortality worldwide. Both DNA methylation and microRNA expression patterns are regulated in developmental stage specific-, cell type specific- and tissue-specific manner. The aberrations are inferred in the maintenance of cancer stem cells and in clonal cell evolution during carcinogenesis. The availability of genome-wide technologies for DNA methylation and microRNA profiling has revolutionized the field of epigenetics and led to the discovery of a number of epigenetically silenced microRNAs in cancerous cells and primary tissues. Dysregulation of these microRNAs affects several key signalling pathways in hepatocarcinogenesis suggesting that modulation of DNA methylation and/or microRNA expression can serve as new therapeutic targets for HCC. Accumulative evidence shows that aberrant DNA methylation of certain microRNA genes is an event specifically found in HCC which correlates with unfavorable outcomes. Therefore, it can potentially serve as a biomarker for detection as well as for prognosis, monitoring and predicting therapeutic responses in HCC.
Keywords: DNA methylation, MicroRNA, Epigenetics, Hepatocellular carcinoma, Biomarker
Core tip: A comprehensive review of the literature revealed that epigenetic inactivation of microRNA genes is a frequent event in hepatocellular carcinoma (HCC). Hypermethylation of microRNA genes can discriminate HCC from benign liver tumors and correlates with poor prognosis, representing a promising new diagnostic and prognostic marker in HCC. Aberrant DNA methylation of microRNA genes affects several key signaling pathways important in hepatocarcinogenesis and for maintenance of cancer stem cell phenotype.
INTRODUCTION
Human cancers develop through gradual accumulation and mutual interaction of genetic and epigenetic alterations[1]. Genetic alterations, both germline and somatic mutations, have been recognized as an important aspect in carcinogenesis[2]. Epigenetics, on the other hand, refers to inherited modifications that influence gene expression and phenotype manifestation without any changes in the DNA sequence. Epigenetic mechanisms consist of several processes i.e., DNA methylation, histone modification, and expression of non-coding RNA. Recent findings suggest that epigenetic alterations occur at much higher rates and more diverse in cancer cells compared to DNA mutations[3]. Among other epigenetic factors, aberrant DNA methylation is the longest known and best studied. DNA methylation cooperates with other epigenetic marks such as histone modifications and non-coding RNAs in the complex regulation of gene expression[4].
MicroRNAs, a major class of small non-coding RNAs, are well-conserved very small RNA molecules (20-22 nucleotides) that can negatively modulate gene expression post-transcriptionally. Accumulating evidence over the past decades highlights the importance of microRNAs as key regulators of many important physiological processes such as cell proliferation, differentiation, apoptosis, and embryonic development[5]. Dysregulation of microRNA expression has been inferred in numerous diseases as well as in cancer. It has later been found that the expression of certain microRNAs is regulated by DNA methylation. This points out that several layers of epigenetic mechanisms are involved in the regulation of gene expression[6,7]. Aberrations of DNA methylation and microRNA expression have been inferred to play an important role in the initiation and progression of human hepatocellular carcinoma (HCC)[8,9].
HCC is the most common type of primary liver cancer that ranks as the fifth most frequent cancer and the third leading cause of cancer mortality worldwide[10]. Epidemiological studies have revealed several key risk factors for the development of HCC such as hepatitis B virus infection, chronic hepatitis and cirrhosis, chronic alcoholic consumption, exposure of dietary aflatoxin, and cigarette smoking[11]. HCC is frequently diagnosed at a late stage in individuals with severe liver dysfunction. Therefore, options for chemotherapeutic and adjuvant therapies are often limited. In addition, lack of early detection markers and drug-resistance may contribute to the high mortality rate in HCC[12]. Although surgical resection and liver transplantation provide improvement of the 5-year survival up to 65%, most HCC cases are diagnosed at an intermediate or advance stage when surgical procedure is not an option anymore[11,12]. The understanding of cellular and molecular mechanisms leading to overt malignant liver tumors is very important in order to develop early detection markers as well as to improve clinical outcome and develop new therapeutic targets for patients with HCC. Over the last decade, the involvement of DNA methylation and microRNAs in liver carcinogenesis has been shown in many studies. DNA methylation at certain genomic loci has been established as a potential marker for sub-classification, diagnosis, prognosis, and therapeutic targets in HCC (as previously reviewed[9,13,14]) and so does microRNA expression patterns (as reviewed in[8,15,16]). In this paper, we focus on the interplay between DNA methylation and microRNA dysregulation, the involved pathways during liver carcinogenesis, and their potential benefits for certain clinical applications in HCC.
DNA METHYLATION IN LIVER CARCINOGENESIS
In the mammalian genome, covalent addition of a methyl group to nucleotides takes place at cytosine located next to guanine (CpG dinucleotide)[17]. CpG dinucleotides are frequently enriched in certain genomic regions[18] that span the range of 0.5-5 kb known as “CpG islands”. Nearly 70% of annotated gene promoters in the human genome are characterized by a high CpG content[19]. DNA methylation at CpG islands located upstream of gene promoter is associated with differential expression of the gene. DNA methylation can mediate gene silencing through direct inhibition of the binding of methylation-dependent transcriptional activators or indirectly by altering the affinity of proteins involved in the chromatin remodeling[4,20]. During embryogenesis, DNA methylation plays a role in the regulation of expression of some genes involved in the differentiation of pluripotent cells[21]. However, recent evidence shows that non-CpG methylation is prevalently observed in embryonic stem cells and during neuronal development[22,23]. Nearly 25% of DNA methylation in embryonic stem cells is in non-CpG nulceotides that disappears upon differentiation[22].
Aberrant DNA methylation is frequently found in cancer cells in comparison to healthy cells[24]. Studies in primary tumor specimens showed that differential DNA methylation can be found in almost every type of cancer[25]. Methylation changes are manifested as hypomethylation and/or hypermethylation. Loss of methylation primarily affects repetitive genomic elements and gene bodies while hypermethylation mostly occurs at the promoters of tumor suppressor genes. Both loss and gain of DNA methylation are often found concurrently in cancer and are likely to be driven by different mechanisms involving chromatin reorganization and DNA replication timing[26].
Almost half of the human genome consists of repetitive elements, in which long interspersed nucleotide element-1 (LINE-1) and ALU already contribute to nearly 17% and 11%, respectively[27]. CpG dinucleotides located within repetitive transposable elements are typically methylated in healthy tissues. DNA methylation at the repetitive sequence is a natural protective mechanism to suppress their activation. Global loss of methylation contributes to the transformation from dysplastic to malignant nodules and is reported to be gradually altered during colorectal cancer progression[28,29]. Demethylation can cause reactivation of transposable elements and insertion to a new location leading to genetic translocations[30], insertions, exon deletions, and chromosomal loss[31]. Alterations of DNA repair pathways and error-prone DNA replication have also been described as a result of LINE-1 demethylation[32,33].
In liver carcinogenesis, hypomethylation of LINE-1, ALU, and SAT2 seems to play a significant role. Loss of methylation at SAT2 precedes LINE-1 and ALU demethylation and occurs at an early stage of HCC[34]. DNA methylation levels of LINE-1 are lower in hepatitis virus and aflatoxin associated HCC[35,36]. Genome-wide loss of methylation correlates with chromosomal instability and poorer prognosis in HCC[37]. In addition, hypomethylation of LINE-1 elements in circulating DNA of HCC patients correlate with the advanced disease and worse survival[38]. A report involving 305 HCC cases and 1254 healthy individuals revealed that hypomethylation of SAT2 detected in white blood cells was associated with increased susceptibility for HCC[39]. Recently, Shukla et al[40] demonstrated that endogenous LINE-1 retrotransposons can propagate oncogenic activation in HCC. L1 insertion caused MCC (mutated in colon cancer) ablation leading to activation of β-catening/Wnt signaling and interrupted inhibition of oncogene ST18.
DNA methylation is mediated by a family of DNA methyltransferase enzymes (DNMTs). Being widely expressed in various tissues, DNMT1 has an ability to induce both de novo and maintenance of methylation. DNMT3A and DNMT3B are mainly involved in the de novo methyltransferase[41]. Mechanisms underlying establishment of DNA methylation have been widely known. However, how methylation is removed from DNA remains to be clarified. Multiple pathways including both active and passive mechanisms seem to be involved in DNA demethylation[42,43]. Improper functions of DNMTs and other components of the methylation machinery can lead to aberrant DNA methylation. Compelling evidences have shown that dysregulation of establishment and removal of DNA methylation is involved in hepatocarcinogenesis[34,44].
In contrast to hypomethylation, promoter hypermethylation is associated with inhibition of gene expression. CpG islands located at the gene promoters are commonly unmethylated. Increased DNA methylation at the CpG island-associated gene promoters are common features in cancer cells[45]. Hypermethylation is related to transcriptional inhibition and loss of gene function. In HCC, hypermethylation mainly affects tumor suppressor genes, particularly those that are involved in cell proliferation, cell differentiation, DNA repair, cellular metabolism, cell adhesion and metastasis. Table 1 summarizes genes that are frequently hypermethylated in primary HCC specimens[46-109]. Although alterations in gene body DNA methylation have been overlooked in the past, it seems that it is not associated with gene repression. Gene body methylation is suggested as a mechanism for silencing repetitive DNA elements and regulating exon splicing[46].
Table 1.
List of genes commonly silenced by DNA hypermethylation in primary hepatocellular carcinoma tumor samples
| Gene | Cellular function | Freq | Ref. |
| 14-3-3 ε | Cell cycle, mitogenic signaling | 89% | [47] |
| APC | Cell proliferation, migration, apoptosis | 44%-71% | [48-52] |
| ARHI | Cell proliferation and invasion | 47% | [53] |
| ASS | Cell cycle and cell invasion | - | [54] |
| BASP1 | Apoptosis | 50% | [55] |
| BLU | Apoptosis | 81% | [56] |
| CADM1 | Cell adhesion and cell differentiation | 41% | [57] |
| CASP8 | Apoptosis | 34% | [58] |
| CAV1 | Cell cycle and proliferation | 56% | [59] |
| CCND2 | Cell cycle | 24%-68% | [48,50] |
| CDH1 | Cell adhesion and metastasis | 34% | [52] |
| CDKN2A | Cell cycle | 30%-70% | [49,50,60,61] |
| CFTR | Intercellular transport | 77%-98% | [50,62] |
| CHFR | Cell cycle, protein degradation | 35% | [63] |
| COX-2 | Immune response, cell migration | 35% | [64] |
| CSRP1 | Cell proliferation and differentiation | 56% | [59] |
| DKK1 | Cell proliferation | 36% | [52] |
| DKK3 | Cell differentiation, embryonic development | - | [65] |
| DLC-1 | Cell proliferation | 24%-35% | [52,66] |
| DLEC1 | Cell proliferation | 71% | [67] |
| E2F1 | Cell proliferation, migration, and differentiation | 71% | [68] |
| FAM43B | Cell proliferation | 60% | [69] |
| FBLN1 | Cell adhesion and migration | 50% | [70] |
| FHIT | Cell proliferation and apoptosis | 65% | [56] |
| GSTP1 | Cell metabolism and detoxification | 70%-76% | [50,60,61,71] |
| HAI-2/PB | Cell proliferation and invasion | 80% | [72] |
| HDPR1 | Cell proliferation, differentiation, cellular signaling | 51% | [73] |
| HINT1 | Apoptosis | 55% | [74] |
| IGFBP-3 | Cell proliferation and growth | 33% | [75] |
| KL | Cell proliferation and growth | 81% | [76] |
| KLK10 | Cell proliferation, survival, and cellular signaling | 55% | [77] |
| LIFR | Signal transduction | 48% | [78] |
| MAGE-A1 | Cell differentiation, embryonic development | 53% | [79] |
| MAGE-A3 | Cell differentiation, embryonic development | 74% | [79] |
| MAT1A | Cell metabolism | 85% | [80] |
| MT1G | Cell proliferation and apoptosis | 60% | [81] |
| MTM1 | Cell differentiation | 100% | [82] |
| MTSS1 | Cell migration and metastasis | 80% | [83] |
| MUC2 | Immune system | 62% | [84] |
| NORE1B | Cell proliferation and growth | 62% | [85] |
| NQO1 | Cell metabolism | 50% | [86] |
| OXGR1 | Cell growth and metabolism | 78% | [77] |
| p14ARF | Cell proliferation and apoptosis | 40%-42% | [68,71] |
| p15INK4B | Cell cycle | 22%-61% | [71,87] |
| P16 | Cell cycle | 37%-83% | [48,71,87] |
| P21 | Cell cycle | 63% | [68] |
| P27 | Cell cycle | 48% | [68] |
| P300 | Cell proliferation and differentiation | 68% | [68] |
| P53 | Cell proliferation, apoptosis, and DNA repair | 14% | [68] |
| P73 | DNA repair and apoptosis | 35% | [71] |
| PCDH10 | Cell adhesion and migration | 76% | [88] |
| PER3 | DNA repair | 60% | [89] |
| PRDM2 | Cell metabolism | - | [90] |
| PTEN | Cell proliferation and migration | 16%-22% | [87,91] |
| RASSF1A | Cell proliferation, migration, and apoptosis | 64%-88% | [48,51] |
| RB | Cell cycle, apoptosis | 24%-32% | [68,87] |
| RECK | Cell invasion and metastasis | 55% | [92] |
| RELN | Cell adhesion | 37% | [93] |
| RIZ1 | Cell proliferation and apoptosis | 29%-67% | [48,94,95] |
| RUNX-3 | Apoptosis | 38%-48% | [50-52] |
| SFRP1 | Cell proliferation and differentiation | 37%-45% | [51,52,96] |
| SFRP2 | Cell proliferation and differentiation | 48%-54% | [97] |
| SFRP5 | Cell proliferation and differentiation | 39% | [96] |
| SLIT2 | Cell migration | 83% | [98] |
| SOCS1 | Cell growth and survival | 39%-72% | [9,48,90,99] |
| SOX1 | Cell proliferation | 57% | [100] |
| SOX17 | Cell differentiation and embryonic development | 82% | [101] |
| SPARC | Cell growth and invasion | 75% | [102] |
| SPINT2 | Cell proliferation and growth | 60% | [50] |
| SRD5A2 | Cell proliferation and androgenic physiology | 50% | [55] |
| Survivin | Apoptosis and cell proliferation | 33% | [58] |
| TFPI2 | Matrix remodelling | 47% | [103] |
| TIP30 | Apoptosis and metastasis | 47% | [104] |
| UCHL1 | Cell metabolism and protein degradation | 44% | [105] |
| UNC5C | Cell migration | 26% | [106] |
| Vimentin | Cell migration and signaling | 56% | [107] |
| WIF-1 | Cell proliferation | 49%-61% | [52,65,108] |
| WT1 | Cell proliferation and survival | 54% | [68] |
| ZHX2 | Cell proliferation, differentiation, and development | 47% | [109] |
Freq: Frequency of hypermethylation.
Epigenetic aberrations have been inferred as key factors during a multi-step process of HCC development. Hypermethylation of APC, RASSF1A, and SOCS1 genes has already been detected in chronic hepatitis and cirrhosis. Both level and the frequency of methylation continuously increase in dysplastic liver nodules and HCC[9,64]. Also GSTP1, CDKN2A, COX2, HIC1, and RUNX3 are frequently methylated in dysplastic liver nodules[9,64,90,110]. Gain of methylation at CDH1, CASP8, MINT, SFRP2, and TIMP3 genes is observed in early and late stage of HCC[64,90]. These data support the notion that DNA methylation aberrations emerge at the early stage of hepatocarcinogenesis and gradually increase in combination with accumulation of genetic events such as P53 mutations and copy number alterations during progression to the advanced stage of HCC.
Concurrent hypermethylation at several genes in HCC also leads to the emerging concept of CpG island methylator phenotype (“CIMP”). This concept was originally described in colorectal and gastric cancer illustrating cancer development through simultaneous inactivation of tumor suppressor and DNA repair genes by DNA methylation[111]. It was shown that CIMP can be used as an independent prognostic factor[111]. Although the concept is still under discussion in HCC given that gene panel for the classification and definition for the phenotype are not yet universally accepted, CIMP positive HCCs have been generally associated with poor clinical outcome[68,71,112,113]. Some reports have also indicated that DNA methylation profiles can be used for molecular sub-classification of HCC to improve the prognosis and the prediction of therapeutic outcomes. The future challenges for routine application in patient-based service will be not only the definition of a consensus gene panel but also the standardization of the methodology for methylation analysis. As shown in Table 1, there is high variation in the frequency of gene hypermethylation reported among different cohorts because of various techniques used for methylation analysis. In addition, many studies do not define “hypermethylation” or use dissimilar definition for hypermethylation.
MICRORNAS IN LIVER CARCINOGENESIS
The past decade has witnessed the important discovery of small non-coding single strand RNAs known as microRNAs that have revolutionized our view of regulatory networks within eukaryotic cells[114]. MicroRNAs negatively modulate gene expression through binding to target messenger RNAs, typically in the 3’ untranslated region. Partial complementary binding of a mature microRNA affects the stability of the target mRNA leading to transcriptional inhibition. In contrast, complete complementary binding can lead to direct endonucleolytic mRNA cleavage. Currently, around 1600 human microRNAs are identified and registered in miRBase (www.mirbase.org) and are predicted to target almost 30% of the total human genes[115]. MicroRNAs regulate important physiological processes such as embryonic development, cell cycle checkpoint, cell proliferation, migration, differentiation, and apoptosis[116,117]. Dysregulation of microRNA expression is involved in a number of diseases including developmental disorders, neurological diseases, cardiovascular disorders, as well as cancer.
The contribution of microRNAs in the process of malignant transformation has been well characterized. They act as oncogenes or tumor suppressors depending on the target genes and their cellular functions. In HCC, a substantial number of reports have shown frequent and extensive dysregulation of microRNA expression in different stages of liver cancer progression[118]. With the availability of tools for genome-wide expression analysis such as microarray and deep sequencing, profiling of microRNA expression has also revealed some unique signatures that are clinically valuable for diagnosis, prognosis, staging, and prediction of therapeutic responses in the majority of human cancers including HCC[16,119]. In addition, aberrant microRNA expression has also been associated with proliferative and self-renewal potential in liver cancer stem cells[120-122].
Differentially microRNA expression in primary HCC specimens has been comprehensively reported and reviewed[16,123,124]. Upregulation of miR-17-92 cluster, miR-21, miR-221, miR-222, and miR-224 is consistently reported in HCC by many studies[16,125,126]. Meanwhile, let-7 family, miR-29, miR-122, miR-124, miR-199a/b, miR-200 family are frequently downregulated in HCC[16,123]. Some important molecular networks such as Wnt/β-catenin, Ras, transforming growth factor-β (TGF-β), and JAK/STAT signaling pathways are being activated due to the changes of microRNA expression in HCC[16,127]. Recent studies using massive parallel sequencing in HCC cell lines[128] and primary specimens[129] showed basal microRNA expression in hepatocytes and healthy liver as well as the deregulation in chronic hepatitis and HCC samples. MiR-122 was most abundantly expressed in liver (approximately 50% of total microRNAs) and frequently down-regulated in HCC[128,129]. MiR-199a/b was down-regulated in all HCC patients under study (n = 40) and significantly correlated with shorter survival[129].
In addition, expression patterns of 3-6 microRNAs have been suggested to be able to discriminate HCC from the adjacent liver tissue, chronic cirrhosis, and benign liver lesions[126,130]. Furthermore, the expression profile of 20 microRNAs can be used as a metastatic predictor and correlates with survival as well as relapse rates in HCC[131]. Recent reports showed that different panels of microRNAs were differentially regulated in metastatic HCCs[118,132]. Accumulating evidence also shows that differential microRNA expression is of great use for predicting disease survival and recurrence in HCC[133-135]. Patterns of microRNA expression have also been suggested to have clinical value to predict therapeutic response to interferon[133,136,137], doxorubicin[138,139], adriamycin and vincristine[140], 5-fluorouracil[136,141], and sorafenib[142-144]. In addition, association between microRNA expression and multidrug resistance in HCC has also been reported[16,145].
CROSS-TALK OF DNA METHYLATION AND MICRORNA EXPRESSION IN HCC
Differential expression of microRNAs in cancer cells can be caused by several mechanisms including genetic instability (amplification, deletion, or translocation). Approximately 50%-70% of microRNA genes are located at fragile genomic sites that are frequently affected by copy number alterations[146,147]. Dysregulation of microRNA expression driven by some oncogenes such as c-Myc is also evident[148,149]. Myc overexpression modulates the expression of let-7a, miR-100, miR-371, and miR-373 and the expression patterns of these four microRNAs can identify a subclass of HCC with aggressive metastatic behavior[150]. In addition, a number of transcription factors regulate microRNA transcription and their dysregulation in cancer cells affect in turn the expression of microRNA. P53, for example, has been demonstrated to mediate repression of miR-125a/b[151] and upregulation of miR-519d, miR-200 and miR-192 family members[152,153].
Mature microRNAs are biologically synthesized through multi-step processes consisting of transcription, excision, and nuclear transport as already extensively reviewed elsewhere[154,155]. Alterations of microRNA biogenesis can contribute to carcinogenesis[156,157]. Disruption of Dicer1 in a conditional knock-out mouse model revealed the critical roles of Dicer1 and microRNAs for hepatocyte survival, metabolism, and tumor suppression. Loss of Dicer1 in addition to other oncogenic stimuli can induce hepatocarcinogenesis[158]. Expression analysis of genes involved in microRNA biogenesis in primary HCC specimens has demonstrated a significant decrease in DCGR8, p68, p72, DICER1, AGO3, AGO4, and PIWIL4 expression compared to the adjacent liver tissues. Down-regulation of those genes correlated significantly with etiological factors and shorter HCC survival[159].
Aberrant microRNA expression in cancer has also been associated with epigenetic regulation such as DNA methylation and histone modifications. Since the first report about aberrant DNA methylation in a microRNA locus[6], several epigenetically silenced microRNAs have been reported across different type of cancers (reviewed in[160,161]). It is estimated that transcription of 10% of all microRNA species is controlled by DNA methylation[162]. However, a greater proportion of microRNAs silenced by DNA methylation has been suggested since 14.3% (218/1523) are located within 500 bp downstream of a CpG island[163]. Approaches frequently used for identification of epigenetically deregulated microRNAs in HCC cells are the treatment of HCC cell lines by epigenetic drugs such as de-methylating agents (5-aza-cytidine and 5-aza-deoxy-cytidine) and HDAC inhibitors (such as Trichostatin) as well as knockdown of DNMT family members followed by expression profiling. MicroRNAs repressed by DNA methylation in primary HCC specimens are summarized at Table 2. Among those, miR-9 and miR-124 seem to be commonly hypermethylated not only in HCC but also in other tumors[7,161].
Table 2.
MicroRNA genes targeted by DNA hypermethylation in hepatocellular carcinoma
| MicroRNA | Validated gene targets in HCC | Biological functions | Ref. |
| miR-1 | FoxP1, MET, HDAC4 | Regulates cell growth, replication and clonogenic survival | [164-166] |
| miR-10a | HOXB3, HOXA3, HOXA1, HOXD10, USF2, HOXD4 | Regulates embryonic development and cell differentiation | [167] |
| miR-124 | CDK6, VIM, SMYD3, E2F6, IQGAP1 | Regulates cell cycle progression (G1-S checkpoint), apoptosis, and metastasis | [166,168] |
| miR-1247 | ADAM15, CIT, MMP24 | Regulates cell proliferation and migration | [166] |
| miR-125b | PIGF, MMP2, MMP9, SIRT7, LIN28B | Regulates cell proliferation, anchorage-independent growth, cell migration, invasion, and angiogenesis | [169] |
| miR-129-2 | SOX4, VCP, IκBα | Regulates apoptosis | [166,170] |
| miR-132 | AKT1, CTNNB1, CCND1 | Regulates cell proliferation | [171] |
| miR-203 | ABCE1, CDK6 | Regulates cell growth and cell cycle progression | [168] |
| miR-320 | NRP1, CTNNB1 | Regulates cell migration, proliferation, and metasatasis | [167] |
| miR-335 | ROCK1, MAPK1, LRG1, MYCN | Regulates migration and cell proliferation | [172] |
| miR-596 | LGALS3BP, FOXP1, IGF2BP2 | Regulates cell growth and induces apoptosis | [166] |
| miR-663 | JUNB, JUND, | Regulates cell proliferation | [165] |
| miR-9 | MTHFD2, HOXD1, MMP14 | Regulates cell proliferation, invasion, metastasis, and angiogenesis | [166] |
HCC: Hepatocellular carcinoma.
Hsa-mir-1-1 is the first microRNA gene reported to be targeted by aberrant DNA methylation in primary HCC specimens[164]. Hypermethylation of hsa-mir-1-1 leads to overexpression of its target genes, FOXP1 and MET[164]. Transcriptional regulator FOXP1 protein plays a dual role as tumor promoting or suppressing protein depending on the tissue type[173]. FOXP1 is commonly upregulated in leukemia but donwregulated in kidney and colon cancer. A contribution to carcinogenesis is suggested by fusion with ABL1 and PAX5 in B-ALL and ETV1 in prostate cancer (reviewed in[174]). Elevated FOXP1 expression in HCC correlates with aggressive malignant phenotypes and poor survival[175]. In addition, miR-1 also targets MET, a tyrosine kinase that interacts with hepatocyte growth factor (HGF) upon external stimuli to subsequently activate Ras-mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)-AKT signaling pathways. The canonical c-MET/HGF pathway is an important player in many physiological functions including cell proliferation, growth, migration and angiogenesis and has been conveyed in liver development and regeneration as well as in hepatocarcingenesis[176]. Overexpression of c-MET that is observed in almost 80% of HCCs correlates significantly with worse clinical outcome[177,178]. Accumulating evidence shows the efficacy of c-MET inhibitors as alternative targeted therapy in HCC in which the clinical trials are still ongoing (reviewed in[178]).
Hypermethylation of hsa-mir-10a in HCC has also been recently reported. It is accompanied miR-10a downregulation and elevated expression of its host gene, HOXB4[167]. HOXB4 is widely represented in leukemogenesis acting as a transcription factor that promotes stem cell renewal[179] and in cervical cancer as a marker for non-differentiated cells[180]. However, overexpression of miR-10a was also reported in dysplastic nodules and HCC samples[181]. A recent study shows an interesting result since upregulation of miR-10a can promote cell migration and invasion in vitro but inhibit hepatocellular carcinoma metastasis in vivo[182]. It is suggested that in vivo, miR-10a can restrain cell-matrix adhesion directing its ability to further suppress cell invasion and metastasis[182].
The three genetic loci in the human genome that encode identical mature miR-124 (Hsa-mir-124-1, Hsa-mir-124-2, and Hsa-mir-124-3) are surrounded by CpG islands and are frequently targeted by DNA hypermethylation in HCC. CDK6 and E2F6 are confirmed as miR-124 gene targets[168]. Furthermore, an important component of mammalian target of rapamycin (mTOR) signaling pathway, PIK3CA, has also been reported as a novel target of miR-124 in HCC. Downregulation of miR-124 leads to over-activation of PI3K/Akt and mTOR signaling resulting in increased cell proliferation, survival, and metastasis[183]. MiR-124 is involved in feedback loop mechanism of liver inflammation mediated by hepatic nuclear factor-α and introduction of miR-124 systemically can inhibit liver carcinogenesis without significant side effects[184]. MiR 124 also regulates epithelial-mesenchymal transition (EMT) by directly targeting ROCK2 and EZH2 genes. Therefore, low expression of miR-124 in HCC correlates significantly with more aggressive behavior and shorter survival[185].
Hsa-miR-1247 is located at one of the largest microRNA clusters within the imprinted DLK1-MEG3 locus. However, apart from the other microRNAs located in that cluster, miR-1247 is the only one that is transcribed from 3’ to 5’ therefore it might not be affected by the imprint regulation. Aberrant methylation was first described in colorectal cancer[186]. In liver tumor, hypermethylation was reported in 37% of total HCC cases[166]. ADAM15, CIT, and MMP24 are putative miR-1247 target genes indicating miR-1247’s role in cellular migration and metastasis[186].
Downregulation of miR-125b in HCC through DNA methylation has been recently described[151,169]. MiR-125b regulates cell proliferation, anchorage-independent growth, cell migration, metastasis, and angiogenesis by targeting placenta growth factor, matrix metalloproteinase (MMP)2, and MMP9. Sirtuin7 (SIRT7) that functions as inhibitor of p21WAF1/Cip1 is also negatively regulated by miR-125b in HCC[151]. Tumor suppressive roles of miR-125b during liver carcinogenesis are also mediated through inhibition of LIN28B[187] and SUV39H1[188]. Downregulation of miR-125b is observed in around 70% of HCC primary samples and inversely correlated with expression of Ki-67, a cell proliferation index[187,188].
Hypermethylation at the upstream CpG island of hsa-mir-129-2 is reported in 60%-90% of primary HCC samples[166,170]. MiR-129 exerts tumor suppressive effects in HCC by inhibiting vaccinia virus complement control protein that forms a complex with p97 for stabilization of IκBα. Reduced miR-129 expression in HCC cells can inhibit apoptosis and stimulate cell migration[189]. In addition, DNA methylation at hsa-mir-129-2 is detected in plasma samples from 85% of stage I HCC patients rendering its potential for alternative surrogate marker for early diagnosis. In comparison, alpha-fetoprotein (AFP) that is widely used as a marker in liver cancer can be detected only in 10% of stage I HCC[170].
Repression of miR-132 expression by DNA methylation has also been reported in HCC. The hypermethylation appears to be mediated by interaction with hepatitis B virus x protein[171]. Meta-analysis of 38 microRNA profiling studies revealed widespread downregulation of miR-132 in various human cancers[190]. MiR-132 functions as an inhibitor of Akt-signaling pathway. As a result, inactivation of miR-132 caused induction of cell proliferation as well as colony formation in HCC cells[171]. Silencing of miR-132 by DNA methylation has also been documented in prostate[191] and pancreatic cancer[192]. Epigenetic silencing of miR-203 in HCC was reported by Furuta et al[168] CDK6 and ABCE1 were shown as direct target genes of miR-203 supporting the role of miR-203 as tumor suppressor. Hypermethylation was also reported in hematological malignancies[193] but not in esophageal squamous cell carcinoma[194]. However, a recent report showed no differential methylation between HCC tumors and the adjacent liver tissues[166]. Differences in methodology and the exact location of the CpG sites under study might explain the discrepancies.
Significant increased DNA methylation at hsa-miR-320 gene has also been reported in HCC tumors[167]. Expression of miR-320 is modulated upon HCV infection leading to alterations of cellular structures and malignant transformation[195]. Downregulation of miR-320 was reported in intrahepatic cholangiocarcinoma and contributed to neoplastic transformation by targeting the oncogenes MCL1 and BCL2[196]. Reduced miR-320 expression was linked with shortened recurrence free survival in colorectal cancer[197]. Loss of miR-320 functions in stromal fibroblasts was shown to cause oncogenic secretome release and reprogramming of the microenvironment in favor of tumor growth[198]. Furthermore, frequent downregulation of miR-320 in prostate cancer was associated with activation of Wnt/β-catenin pathway and stem-cell like properties[199].
Located at the intron of MEST gene, miR-335 is downregulated in 78% of HCC tumors by aberrant DNA hypermethylation[172]. Loss of miR-335 function is accompanied by dysregulation of the host gene MEST. Lower miR-335 expression correlates significantly with distant HCC metastasis[172]. Tumor suppressive effects of miR-335 were also demonstrated in breast cancer by targeting the BRCA1 pathway and downregulation was observed in distant metastasic cases[200,201]. Frequent downregulation was also found in prostate cancer and re-introduction of miR-335 in cell lines repressed cell proliferation, invasion, and migration[202].
Aberrant methylation of the hsa-mir-596 gene in HCC was identified by our group[165,166]. The tumor suppressing effects of miR-596 is mediated in cancer cells through negative regulation of LGALS3BP, FOXP1 and IGF2BP2 genes. MiR-596 is located at the short arm of chromosome 8 that is often affected by focal break points in cancer. A large deletion involving miR-596 was found in urothelial carcinomas[203]. Hypermethylation of miR-596 promoter was also found in oral cell squamous carcinoma lines and primary tissues. Ectopic expression of miR-596 caused apoptosis and reduced cell growth[204].
MiR-663 is also frequently targeted by DNA hypermethylation in HCC[165]. Proto-oncogenes JUNB and JUND are putative target genes of miR-663. The functions of miR-663 as an effective suppressor of tumor growth was shown in gastric cancer cell lines by Pan et al[205] Transient re-expression of miR-663 altered DNA content, induced cellular morphology changes and proliferative blockage[205]. However, functional analysis of miR-663 downregulation in HCC remains to be elucidated.
Mature miR-9 is encoded from 3 independent genomic loci in the human genome, i.e., hsa-mir-9-1, hsa-mir-9-2, and hsa-mir-9-3. Simultaneous hypermethylation was frequently found in different cancers including primary HCC[165,166]. However, how miR-9 contributes to carcinogenesis remains controversial. Upregulation of miR-9 induced by c-Myc was shown to prime breast cancer cells for epithelial-mesenchymal transition by directly inhibiting E-cadherin[206]. MiR-9 overexpression in HCC cells (SK-Hep-1) led to induction of cell migration through E-cadherin suppression[207]. Differential expression of miR-9 was reported in distant metastatic breast cancer[208]. On the other hand, Selcuklu et al[209] reported that miR-9 was repressed in primary breast cancer specimens and ectopic miR-9 expression in MCF-7 cells induced anti-proliferative and pro-apoptotic effects. In addition, hypermethylation of miR-9 promoters is observed as a potential diagnostic or prognostic parameter in head and neck cancer[210], lung cancer[211], bladder cancer[212], gastric cancer[213] and colorectal cancer[214]. Our group showed aberrant miR-9 methylation in HCC using quantitative methylation analysis[165] and demonstrated their correlation with clinical outcomes[166]. Figure 1 summarizes gene targets and key pathways affected by aberrant DNA methylation of microRNA genes in HCC.
Figure 1.
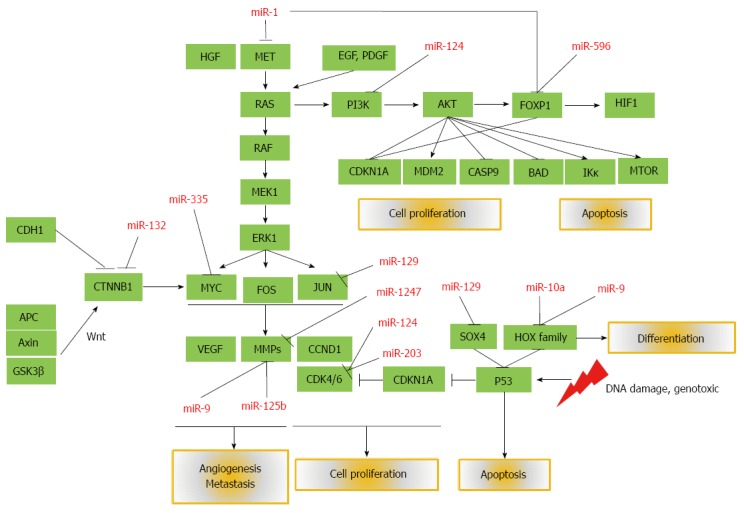
Mature microRNAs silenced by aberrant DNA methylation and their affected target genes and pathways that are important in the development and progression of hepatocellular carcinoma. PI3K: Phosphatidylinositol-3-kinase; MAPK: Mitogen-activated protein kinase; EGF: Epidermal growth factor; PDGF: Platelet-derived growth factor; HGF: Hepatocyte growth factor; mTOR: Mammalian target of rapamycin; VEGF: Vascular endothelial growth factor; MMP: Matrix metalloproteinase; APC: Activated protein C; CTNNB1: Beta-catenin.
GENOME-WIDE STUDIES OF DNA METHYLATION IN HUMAN HCC
The development of new technologies for DNA methylation analysis have greatly contributed to the advance of epigenomic studies in liver cancer especially with the recent application of more comprehensive, high-resolution genome-wide methods. We identified 10 studies implementing genome-wide methylation analysis in primary HCC specimens: a study used methylated CpG island amplification coupled with CpG island microarray (MCAM, n = 17)[215], two studies used MeDIP (n = 6[216,217], n = 11[217]), 6 studies employed Illumina’s Infinium Human Methylation27 (n = 23[218], n = 66[219], n = 13[220], n = 13[221], n = 63[89], and n = 62[222]), and two studies utilized Illumina’s Infinium HumanMethylation450 (n = 27[61] and n = 66[223]). Among these studies, however, only Shen et al[167] specifically addressed aberrant DNA methylation of microRNA genes. The 27k Bead array from Illumina contains 254 assays covering 110 intragenic microRNAs located within 64 host genes. Using a panel of microRNA gene methylation assays in the 27k array, HCC tumors can be differentiated from the corresponding adjacent liver tissues. More than 20% of the 254 CpG sites showed significant differential methylation affecting 27 genes. The newly released Illumina 450k has a greater coverage with almost 99% of RefSeq genes and 96% of CpG islands are included. Using this platform, Song et al[61] found 10775 CpG sites located within or adjacent to gene promoters were differentially methylated in HCC tumors. Of these CpG sites 493 are associated with microRNA genes. Using the same array platform, Shen et al[223] reported 28017 CpG sites (5.8%) to be hypermethylated in HCC tumors. These data indicate that many more microRNA genes are likely to be targeted by DNA hypermethylation in HCC and need to be further studied.
MICRORNA AND DNA METHYLATION IN LIVER CANCER STEM CELLS
In addition to the traditional view that cancer emerges through accumulation of genetic changes in a clonal population, another theory postulates a hierarchical organization of tumor cells. The latter suggests that cancer develops from cells with particular capabilities to self-renew, produce differentiated progeny, and initiate new focal tumors. These cells are known as cancer stem cells[224]. However, rather than representing independent mechanisms, the two models are believed to be complementary and substantially contribute to the inter- and intra-tumoral heterogeneity. The features of stemness and pluripotency are ultimately formed by unique epigenetic signatures[225,226]. Hepatic cancer stem cells can be distinguished from well differentiated tumor cells through their functional properties and specific surface markers such as CD133, CG90, EpCAM, and ALDH. Expression of CD133 is regulated by DNA methylation through modulation of TGFβ/Smad pathway[227]. Side population cells that represent cancer stem cells in HCC display differential gene expression[228] and unique DNA methylation patterns[229] that regulate pathways involved in pluripotentcy and self-renewal such as WNT/β-catenin, Hedgehog, MYC, TGFβ/Smad, Notch, MET, and BMI1[229].
The involvement of microRNAs in the regulation of cancer stem cells during hepatocarcinogenesis has also been suggested. MiR-181 is upregulated in EpCAM+ hepatic cancer stem cells through Wnt/β-catenin transcriptional regulation[230]. Differentiation-promoting genes, CDX2 and GATA6, are revealed as miR-181 gene targets. Cairo et al[150] reported MYC-dependent microRNAs that featured cell renewal and stemness in HCC. Other microRNAs, miR-216a/217 were shown to increase stem-like properties by targeting PTEN and SMAD7. Consistently, miR-216a/217 upregulation in HCC tissues correlated with EMT phenotypes, early recurrence, and shorter disease-free survival[144]. In addition, overexpression of microRNA clusters at the DLK1-DIO3 imprinted locus correlated with HCC stem cell markers, high AFP levels, and poor survival in HCC patients[120]. Since imprinting is regulated by DNA methylation, the cross-talk between DNA methylation and microRNAs during cancer stem cell de-differentiation in HCC may also be present. Some epigenetically-silenced microRNAs also target transcription factors and signaling pathways that are involved in cell-renewal and stemness phenotypes[164,168,199].
ENRICHED BIOLOGICAL PATHWAYS OF ABERRANTLY METHYLATED MICRORNA GENES IN HCC
MicroRNA target genes from the experimentally validated targets (TarBase or miRTarBase) and predicted algorithms (DIANA, miRDB, and TargetScan) of all epigenetically silenced microRNAs in HCC were used for enrichment biological pathway analysis using Kyoto Encyclopedia of Genes and Genomes and Panther. Metabolic, PIK3-Akt, MAPK, Wnt, inflammation, angiogenesis, epidermal growth factor receptor, Cadherin, and TGFβ are the most frequent pathways targeted by these microRNAs (Table 3 lists pathways and molecular functions regulated by methylation-silenced microRNA gene targets). Several biological functions such as metabolic, immune system, cell adhesion, cell communication, and developmental processes as well as molecular and cellular functions including protein binding, transcriptional regulator, catalytic and receptor activity are greatly enriched suggesting the importance of these epigenetically silenced microRNAs in the development of cancer.
Table 3.
Pathways and molecular functions regulated by methylation-silenced microRNA gene targets
| No. | Panther | Genes | Gene | Contrib |
| GO molecular function1 | ||||
| 1 | Binding (GO:0005488) | 2799 | 40.60% | 33.80% |
| 2 | Catalytic activity (GO:0003824) | 2126 | 30.80% | 25.70% |
| 3 | Transcription regulator activity (GO:0030528) | 920 | 13.30% | 11.10% |
| 4 | Receptor activity (GO:0004872) | 696 | 10.10% | 8.40% |
| 5 | Structural molecule activity (GO:0005198) | 558 | 8.10% | 6.70% |
| 6 | Enzyme regulator activity (GO:0030234) | 494 | 7.20% | 6.00% |
| 7 | Transporter activity (GO:0005215) | 418 | 6.10% | 5.00% |
| 8 | Ion channel activity (GO:0005216) | 152 | 2.20% | 1.80% |
| 9 | Translation regulator activity (GO:0045182) | 57 | 0.80% | 0.70% |
| 10 | Motor activity (GO:0003774) | 53 | 0.80% | 0.60% |
| GO Biological process2 | ||||
| 1 | Metabolic process (GO:0008152) | 3277 | 47.50% | 20.80% |
| 2 | Cellular process (GO:0009987) | 2564 | 37.20% | 16.30% |
| 3 | Cell communication (GO:0007154) | 1793 | 26.00% | 11.40% |
| 4 | Developmental process (GO:0032502) | 1315 | 19.10% | 8.40% |
| 5 | Transport (GO:0006810) | 1190 | 17.30% | 7.60% |
| 6 | Immune system process (GO:0002376) | 956 | 13.90% | 6.10% |
| 7 | System process (GO:0003008) | 927 | 13.40% | 5.90% |
| 8 | Cell cycle (GO:0007049) | 797 | 11.60% | 5.10% |
| 9 | Cellular component organization (GO:0016043) | 630 | 9.10% | 4.00% |
| 10 | Response to stimulus (GO:0050896) | 613 | 8.90% | 3.90% |
| GO cellular component3 | ||||
| 1 | Intracellular (GO:0005622) | 486 | 7.10% | 50.40% |
| 2 | Extracellular region (GO:0005576) | 233 | 3.40% | 24.10% |
| 3 | Plasma membrane (GO:0005886) | 86 | 1.20% | 8.90% |
| 4 | Ribonucleoprotein complex (GO:0030529) | 84 | 1.20% | 8.70% |
| 5 | Protein complex (GO:0043234) | 76 | 1.10% | 7.90% |
| GO protein class4 | ||||
| 1 | Nucleic acid binding (PC00171) | 1144 | 16.60% | 12.60% |
| 2 | Transcription factor (PC00218) | 920 | 13.30% | 10.20% |
| 3 | Hydrolase (PC00121) | 691 | 10.00% | 7.60% |
| 4 | Receptor (PC00197) | 690 | 10.00% | 7.60% |
| 5 | Transferase (PC00220) | 662 | 9.60% | 7.30% |
| 6 | Enzyme modulator (PC00095) | 634 | 9.20% | 7.00% |
| 7 | Signaling molecule (PC00207) | 481 | 7.00% | 5.30% |
| 8 | Transporter (PC00227) | 457 | 6.60% | 5.00% |
| 9 | Cytoskeletal protein (PC00085) | 402 | 5.80% | 4.40% |
| 10 | Kinase (PC00137) | 316 | 4.60% | 3.50% |
| Pathway5 | ||||
| 1 | Gonadotropin releasing hormone receptor pathway (P06664) | 159 | 2.30% | 4.80% |
| 2 | Wnt signaling pathway (P00057) | 156 | 2.30% | 4.70% |
| 3 | Inflammation mediated by chemokine and cytokine signaling pathway (P00031) | 134 | 1.90% | 4.00% |
| 4 | Integrin signalling pathway (P00034) | 116 | 1.70% | 3.50% |
| 5 | Angiogenesis (P00005) | 98 | 1.40% | 3.00% |
| 6 | EGF receptor signaling pathway (P00018) | 84 | 1.20% | 2.50% |
| 7 | Cadherin signaling pathway (P00012) | 80 | 1.20% | 2.40% |
| 8 | Huntington disease (P00029) | 77 | 1.10% | 2.30% |
| 9 | TGF-beta signaling pathway (P00052) | 73 | 1.10% | 2.20% |
| 10 | PDGF signaling pathway (P00047) | 74 | 1.10% | 2.20% |
| KEGG | ||||
| hsa01100 metabolic pathways (434) | ||||
| hsa05200 pathways in cancer (189) | ||||
| hsa04151 PI3K-Akt signaling pathway (162) | ||||
| hsa04010 MAPK signaling pathway (137) | ||||
| hsa05205 proteoglycans in cancer (134) | ||||
| hsa05166 HTLV-I infection (123) | ||||
| hsa04510 Focal adhesion (116) | ||||
| hsa04810 regulation of actin cytoskeleton (115) | ||||
| hsa05202 transcriptional misregulation in cancer (100) | ||||
| hsa04144 endocytosis (100) | ||||
Total genes n = 6893, total function hits n = 8282;
Total genes n = 6893, total process hits n = 15740;
Total genes n = 6893, total component hits n = 965;
Total genes n = 6893, total protein classess n = 9050;
Total genes n = 6893, total pathway hits n = 3313. Contrib: Contribution; PI3K: Phosphatidylinositol-3-kinase; MAPK: Mitogen-activated protein kinase; HTLV: Human T-lymphotropic virus 1; EGF: Epidermal growth factor; PDGF: Platelet-derived growth factor; TGF-β: Transforming growth factor beta.
DIAGNOSTIC, PROGNOSTIC, AND CLINICAL RELEVANCE OF DNA METHYLATION AT MICRORNA GENES IN HCC
Hepatic neoplasm consists of a range of benign and malignant tumors that differ in histopathology, etiology, disease progression, and clinical behavior. Aberrant DNA methylation that occurs during tumor initiation and development is relatively stable in tissues as well as serum/plasma. Additionally, dysregulation of DNA methylation and microRNA expression that are involved in the regulation of cell differentiation and developmental cell lineage is associated with poorly differentiated cancer and worse outcomes. Therefore, aberrant DNA methylation at microRNA genes is potentially a useful parameter for diagnosis as well as classification of human cancers including HCC. Single locus hypermethylation of miR-129-2 has been shown as a highly specific marker for distinguishing HCC from chronic hepatitis and healthy liver tissues[170].
Our recent study showed that DNA methylation at microRNA genes was a specific event detectable only in malignant liver cells and tissue samples but not in adjacent liver tissue, benign liver tumors, healthy liver cells, or in hepatocyte lines. These results indicated that methylation of microRNA genes might represent a new biomarker for specific detection of malignant liver tumors. In addition, concordant DNA methylation at certain microRNA loci correlated with poor HCC survival rendering its potential to be used as prognostic marker in HCC[166]. Differential methylation at these loci appears not to be a random event but highly organized during initiation and progression of HCC. In addition, several microRNAs affected by DNA methylation in HCC are suggested to modulate therapeutic responses upon conventional chemotherapy and or treatment with sorafenib. However, using a panel of DNA methylation aberrations in microRNA genes as a new marker for CIMP in HCC is a great challenge for future research. To evaluate this, quantitative DNA methylation analysis using diverse samples of liver diseases including adenoma, chronic hepatitis, cirrhosis, early and late stage of HCC in a setting of retrospective and prospective studies involving multi-center collaboration are needed. Genome-wide DNA methylation analysis will provide extensive information not only for microRNA loci but also other regions potentially important as a marker for CIMP. Correlating methylation status with clinocopathologic and molecular profiles from large HCC cohort will also strengthen the use of CIMP as a new classifier for HCC samples.
The involvement of aberrant DNA methylation at microRNA genes in the pathogenesis and progression of HCC also provide new insight into the molecular mechanisms of the disease and emerge as a candidate for novel alternative adjuvant therapy in HCC. This is of special importance since many clinical trials for molecular targeted therapies in HCC are not yet successful[231]. Systemic administration of microRNAs that are frequently silenced by DNA methylation such as miR-124 can inhibit the HCC progression in animal models. Moreover, a therapy using microRNA mimics is relatively effective and safe without any observed side effect[184]. In addition, miR-1274 that is commonly targeted by DNA hypermethylation in HCC has been demonstrated as an important regulator in response to therapy with sorafenib[232].
Demethylating agents such as 5-azacytidine and 5-aza-2’-deoxycytidine can induce re-expression of microRNA genes silenced by methylation. So far, these demethylating agents have been approved as adjuvant therapy in myelodysplastic syndrome[233]. Therapeutic effects of demethylating drugs for HCC have been established in vitro[234,235]. 5-azacytidine exerts anti-tumor effects not only by reversing epigenetic aberrations but also by re-sensitizing cells to apoptotic inducing therapy such as TRAIL[236]. However, both drugs are integrated into the replicating DNA and bind to DNMT enzymes irreversibly leading to unspecific biological side effects. A second generation demethylating agent, Zebularine, provides reversible binding and exhibit less toxicity. Late stage HCC patients usually with high degree of methylation levels are predicted to benefit from zebularine therapy[218]. However, most of demethylating agents cause some adverse events including liver dysfunction[237] that need to be carefully addressed especially in HCC patients. Adjustment in dose and administration schedule is therefore required for these agents to provide optimal results in cancer therapy[238]. Another epigenetic drug, belinostat, is a potent HDAC inhibit that has already been in phase I/II clinical trial for treatment of inoperable HCCs[239].
CONCLUSION
The discovery of aberrant DNA methylation at microRNA genes during liver carcinogenesis has contributed significantly not only to an understanding of the molecular pathogenesis of the disease but also provided potential new markers for diagnosis, prognosis, and prediction in HCC. Diverse major pathways such as Wnt, mTOR, MAPK, and nuclear factor kappa B signaling seem to be affected simultaneously by hypermethylation of those tumor suppressive microRNAs. Therefore, manipulating DNA methylation at those microRNA genes and/or their expression may provide a promising strategy for alternative adjuvant therapy in HCC. In addition, DNA methylation is relatively stable in tumor tissues and body fluids under many conditions suggesting it as a new promising biomarker for diagnosis and prognosis in HCC. However, confirmation with large multicenter HCC cohorts and using robust techniques for DNA methylation analysis of microRNA genes are warranted before the application in clinical practice.
Footnotes
Supported by Grant from the German Research Council (DFG), SFB-TRR77 “Liver cancer” (Project B1)
P- Reviewers: Carter WG, Du Z, Yao DF S- Editor: Gou SX L- Editor: A E- Editor: Wang CH
References
- 1.You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knudson AG. Cancer genetics. Am J Med Genet. 2002;111:96–102. doi: 10.1002/ajmg.10320. [DOI] [PubMed] [Google Scholar]
- 3.Rodríguez-Paredes M, Esteller M. Cancer epigenetics reaches mainstream oncology. Nat Med. 2011;17:330–339. doi: 10.1038/nm.2305. [DOI] [PubMed] [Google Scholar]
- 4.Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nat Rev Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ambros V. microRNAs: tiny regulators with great potential. Cell. 2001;107:823–826. doi: 10.1016/s0092-8674(01)00616-x. [DOI] [PubMed] [Google Scholar]
- 6.Saito Y, Liang G, Egger G, Friedman JM, Chuang JC, Coetzee GA, Jones PA. Specific activation of microRNA-127 with downregulation of the proto-oncogene BCL6 by chromatin-modifying drugs in human cancer cells. Cancer Cell. 2006;9:435–443. doi: 10.1016/j.ccr.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann U, Hasemeier B, Christgen M, Müller M, Römermann D, Länger F, Kreipe H. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 8.Szabo G, Bala S. MicroRNAs in liver disease. Nat Rev Gastroenterol Hepatol. 2013;10:542–552. doi: 10.1038/nrgastro.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Um TH, Kim H, Oh BK, Kim MS, Kim KS, Jung G, Park YN. Aberrant CpG island hypermethylation in dysplastic nodules and early HCC of hepatitis B virus-related human multistep hepatocarcinogenesis. J Hepatol. 2011;54:939–947. doi: 10.1016/j.jhep.2010.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 11.Yang JD, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Altekruse SF, McGlynn KA, Reichman ME. Hepatocellular carcinoma incidence, mortality, and survival trends in the United States from 1975 to 2005. J Clin Oncol. 2009;27:1485–1491. doi: 10.1200/JCO.2008.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pogribny IP, Rusyn I. Role of epigenetic aberrations in the development and progression of human hepatocellular carcinoma. Cancer Lett. 2014;342:223–230. doi: 10.1016/j.canlet.2012.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perret C. Methylation profile as a new tool for classification of hepatocellular carcinoma. J Hepatol. 2011;54:602–603. doi: 10.1016/j.jhep.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 15.Giordano S, Columbano A. MicroRNAs: new tools for diagnosis, prognosis, and therapy in hepatocellular carcinoma? Hepatology. 2013;57:840–847. doi: 10.1002/hep.26095. [DOI] [PubMed] [Google Scholar]
- 16.Borel F, Konstantinova P, Jansen PL. Diagnostic and therapeutic potential of miRNA signatures in patients with hepatocellular carcinoma. J Hepatol. 2012;56:1371–1383. doi: 10.1016/j.jhep.2011.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 18.Bird AP. CpG-rich islands and the function of DNA methylation. Nature. 1986;321:209–213. doi: 10.1038/321209a0. [DOI] [PubMed] [Google Scholar]
- 19.Saxonov S, Berg P, Brutlag DL. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci USA. 2006;103:1412–1417. doi: 10.1073/pnas.0510310103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suzuki MM, Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat Rev Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 21.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 22.Lister R, Pelizzola M, Dowen RH, Hawkins RD, Hon G, Tonti-Filippini J, Nery JR, Lee L, Ye Z, Ngo QM, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–322. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lister R, Mukamel EA, Nery JR, Urich M, Puddifoot CA, Johnson ND, Lucero J, Huang Y, Dwork AJ, Schultz MD, et al. Global epigenomic reconfiguration during mammalian brain development. Science. 2013;341:1237905. doi: 10.1126/science.1237905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feinberg AP, Vogelstein B. Hypomethylation of ras oncogenes in primary human cancers. Biochem Biophys Res Commun. 1983;111:47–54. doi: 10.1016/s0006-291x(83)80115-6. [DOI] [PubMed] [Google Scholar]
- 25.Fernandez AF, Assenov Y, Martin-Subero JI, Balint B, Siebert R, Taniguchi H, Yamamoto H, Hidalgo M, Tan AC, Galm O, et al. A DNA methylation fingerprint of 1628 human samples. Genome Res. 2012;22:407–419. doi: 10.1101/gr.119867.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berman BP, Weisenberger DJ, Aman JF, Hinoue T, Ramjan Z, Liu Y, Noushmehr H, Lange CP, van Dijk CM, Tollenaar RA, et al. Regions of focal DNA hypermethylation and long-range hypomethylation in colorectal cancer coincide with nuclear lamina-associated domains. Nat Genet. 2012;44:40–46. doi: 10.1038/ng.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.Sunami E, de Maat M, Vu A, Turner RR, Hoon DS. LINE-1 hypomethylation during primary colon cancer progression. PLoS One. 2011;6:e18884. doi: 10.1371/journal.pone.0018884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hur K, Cejas P, Feliu J, Moreno-Rubio J, Burgos E, Boland CR, Goel A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut. 2014;63:635–646. doi: 10.1136/gutjnl-2012-304219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazazian HH, Goodier JL. LINE drive. retrotransposition and genome instability. Cell. 2002;110:277–280. doi: 10.1016/s0092-8674(02)00868-1. [DOI] [PubMed] [Google Scholar]
- 31.Symer DE, Connelly C, Szak ST, Caputo EM, Cost GJ, Parmigiani G, Boeke JD. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–338. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 32.Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–165. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 33.Kongruttanachok N, Phuangphairoj C, Thongnak A, Ponyeam W, Rattanatanyong P, Pornthanakasem W, Mutirangura A. Replication independent DNA double-strand break retention may prevent genomic instability. Mol Cancer. 2010;9:70. doi: 10.1186/1476-4598-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee HS, Kim BH, Cho NY, Yoo EJ, Choi M, Shin SH, Jang JJ, Suh KS, Kim YS, Kang GH. Prognostic implications of and relationship between CpG island hypermethylation and repetitive DNA hypomethylation in hepatocellular carcinoma. Clin Cancer Res. 2009;15:812–820. doi: 10.1158/1078-0432.CCR-08-0266. [DOI] [PubMed] [Google Scholar]
- 35.Kim BH, Cho NY, Shin SH, Kwon HJ, Jang JJ, Kang GH. CpG island hypermethylation and repetitive DNA hypomethylation in premalignant lesion of extrahepatic cholangiocarcinoma. Virchows Arch. 2009;455:343–351. doi: 10.1007/s00428-009-0829-4. [DOI] [PubMed] [Google Scholar]
- 36.Zhang YJ, Wu HC, Yazici H, Yu MW, Lee PH, Santella RM. Global hypomethylation in hepatocellular carcinoma and its relationship to aflatoxin B(1) exposure. World J Hepatol. 2012;4:169–175. doi: 10.4254/wjh.v4.i5.169.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Calvisi DF, Ladu S, Gorden A, Farina M, Lee JS, Conner EA, Schroeder I, Factor VM, Thorgeirsson SS. Mechanistic and prognostic significance of aberrant methylation in the molecular pathogenesis of human hepatocellular carcinoma. J Clin Invest. 2007;117:2713–2722. doi: 10.1172/JCI31457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tangkijvanich P, Hourpai N, Rattanatanyong P, Wisedopas N, Mahachai V, Mutirangura A. Serum LINE-1 hypomethylation as a potential prognostic marker for hepatocellular carcinoma. Clin Chim Acta. 2007;379:127–133. doi: 10.1016/j.cca.2006.12.029. [DOI] [PubMed] [Google Scholar]
- 39.Wu HC, Wang Q, Yang HI, Tsai WY, Chen CJ, Santella RM. Global DNA methylation levels in white blood cells as a biomarker for hepatocellular carcinoma risk: a nested case-control study. Carcinogenesis. 2012;33:1340–1345. doi: 10.1093/carcin/bgs160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shukla R, Upton KR, Muñoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cedar H, Bergman Y. Programming of DNA methylation patterns. Annu Rev Biochem. 2012;81:97–117. doi: 10.1146/annurev-biochem-052610-091920. [DOI] [PubMed] [Google Scholar]
- 42.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhutani N, Burns DM, Blau HM. DNA demethylation dynamics. Cell. 2011;146:866–872. doi: 10.1016/j.cell.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park HJ, Yu E, Shim YH. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006;233:271–278. doi: 10.1016/j.canlet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 45.Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002;21:5427–5440. doi: 10.1038/sj.onc.1205600. [DOI] [PubMed] [Google Scholar]
- 46.Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 47.Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y, et al. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 sigma gene in human hepatocellular carcinoma. Oncogene. 2000;19:5298–5302. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 48.Lehmann U, Berg-Ribbe I, Wingen LU, Brakensiek K, Becker T, Klempnauer J, Schlegelberger B, Kreipe H, Flemming P. Distinct methylation patterns of benign and malignant liver tumors revealed by quantitative methylation profiling. Clin Cancer Res. 2005;11:3654–3660. doi: 10.1158/1078-0432.CCR-04-2462. [DOI] [PubMed] [Google Scholar]
- 49.Csepregi A, Röcken C, Hoffmann J, Gu P, Saliger S, Müller O, Schneider-Stock R, Kutzner N, Roessner A, Malfertheiner P, et al. APC promoter methylation and protein expression in hepatocellular carcinoma. J Cancer Res Clin Oncol. 2008;134:579–589. doi: 10.1007/s00432-007-0321-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moribe T, Iizuka N, Miura T, Kimura N, Tamatsukuri S, Ishitsuka H, Hamamoto Y, Sakamoto K, Tamesa T, Oka M. Methylation of multiple genes as molecular markers for diagnosis of a small, well-differentiated hepatocellular carcinoma. Int J Cancer. 2009;125:388–397. doi: 10.1002/ijc.24394. [DOI] [PubMed] [Google Scholar]
- 51.Hua D, Hu Y, Wu YY, Cheng ZH, Yu J, Du X, Huang ZH. Quantitative methylation analysis of multiple genes using methylation-sensitive restriction enzyme-based quantitative PCR for the detection of hepatocellular carcinoma. Exp Mol Pathol. 2011;91:455–460. doi: 10.1016/j.yexmp.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 52.Liu JB, Zhang YX, Zhou SH, Shi MX, Cai J, Liu Y, Chen KP, Qiang FL. CpG island methylator phenotype in plasma is associated with hepatocellular carcinoma prognosis. World J Gastroenterol. 2011;17:4718–4724. doi: 10.3748/wjg.v17.i42.4718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang J, Lin Y, Li L, Qing D, Teng XM, Zhang YL, Hu X, Hu Y, Yang P, Han ZG. ARHI, as a novel suppressor of cell growth and downregulated in human hepatocellular carcinoma, could contribute to hepatocarcinogenesis. Mol Carcinog. 2009;48:130–140. doi: 10.1002/mc.20461. [DOI] [PubMed] [Google Scholar]
- 54.Wu L, Li L, Meng S, Qi R, Mao Z, Lin M. Expression of argininosuccinate synthetase in patients with hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:365–368. doi: 10.1111/jgh.12043. [DOI] [PubMed] [Google Scholar]
- 55.Tsunedomi R, Ogawa Y, Iizuka N, Sakamoto K, Tamesa T, Moribe T, Oka M. The assessment of methylated BASP1 and SRD5A2 levels in the detection of early hepatocellular carcinoma. Int J Oncol. 2010;36:205–212. [PubMed] [Google Scholar]
- 56.Zhang X, Li HM, Liu Z, Zhou G, Zhang Q, Zhang T, Zhang J, Zhang C. Loss of heterozygosity and methylation of multiple tumor suppressor genes on chromosome 3 in hepatocellular carcinoma. J Gastroenterol. 2013;48:132–143. doi: 10.1007/s00535-012-0621-0. [DOI] [PubMed] [Google Scholar]
- 57.Zhang W, Zhou L, Ding SM, Xie HY, Xu X, Wu J, Chen QX, Zhang F, Wei BJ, Eldin AT, et al. Aberrant methylation of the CADM1 promoter is associated with poor prognosis in hepatocellular carcinoma treated with liver transplantation. Oncol Rep. 2011;25:1053–1062. doi: 10.3892/or.2011.1159. [DOI] [PubMed] [Google Scholar]
- 58.Cho S, Lee JH, Cho SB, Yoon KW, Park SY, Lee WS, Park CH, Joo YE, Kim HS, Choi SK, et al. Epigenetic methylation and expression of caspase 8 and survivin in hepatocellular carcinoma. Pathol Int. 2010;60:203–211. doi: 10.1111/j.1440-1827.2009.02507.x. [DOI] [PubMed] [Google Scholar]
- 59.Hirasawa Y, Arai M, Imazeki F, Tada M, Mikata R, Fukai K, Miyazaki M, Ochiai T, Saisho H, Yokosuka O. Methylation status of genes upregulated by demethylating agent 5-aza-2’-deoxycytidine in hepatocellular carcinoma. Oncology. 2006;71:77–85. doi: 10.1159/000100475. [DOI] [PubMed] [Google Scholar]
- 60.Harder J, Opitz OG, Brabender J, Olschewski M, Blum HE, Nomoto S, Usadel H. Quantitative promoter methylation analysis of hepatocellular carcinoma, cirrhotic and normal liver. Int J Cancer. 2008;122:2800–2804. doi: 10.1002/ijc.23433. [DOI] [PubMed] [Google Scholar]
- 61.Song MA, Tiirikainen M, Kwee S, Okimoto G, Yu H, Wong LL. Elucidating the landscape of aberrant DNA methylation in hepatocellular carcinoma. PLoS One. 2013;8:e55761. doi: 10.1371/journal.pone.0055761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding S, Gong BD, Yu J, Gu J, Zhang HY, Shang ZB, Fei Q, Wang P, Zhu JD. Methylation profile of the promoter CpG islands of 14 “drug-resistance” genes in hepatocellular carcinoma. World J Gastroenterol. 2004;10:3433–3440. doi: 10.3748/wjg.v10.i23.3433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakai M, Hibi K, Kanazumi N, Nomoto S, Inoue S, Takeda S, Nakao A. Aberrant methylation of the CHFR gene in advanced hepatocellular carcinoma. Hepatogastroenterology. 2005;52:1854–1857. [PubMed] [Google Scholar]
- 64.Lee S, Lee HJ, Kim JH, Lee HS, Jang JJ, Kang GH. Aberrant CpG island hypermethylation along multistep hepatocarcinogenesis. Am J Pathol. 2003;163:1371–1378. doi: 10.1016/S0002-9440(10)63495-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ding Z, Qian YB, Zhu LX, Xiong QR. Promoter methylation and mRNA expression of DKK-3 and WIF-1 in hepatocellular carcinoma. World J Gastroenterol. 2009;15:2595–2601. doi: 10.3748/wjg.15.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wong CM, Lee JM, Ching YP, Jin DY, Ng IO. Genetic and epigenetic alterations of DLC-1 gene in hepatocellular carcinoma. Cancer Res. 2003;63:7646–7651. [PubMed] [Google Scholar]
- 67.Qiu GH, Salto-Tellez M, Ross JA, Yeo W, Cui Y, Wheelhouse N, Chen GG, Harrison D, Lai P, Tao Q, et al. The tumor suppressor gene DLEC1 is frequently silenced by DNA methylation in hepatocellular carcinoma and induces G1 arrest in cell cycle. J Hepatol. 2008;48:433–441. doi: 10.1016/j.jhep.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Zhang C, Guo X, Jiang G, Zhang L, Yang Y, Shen F, Wu M, Wei L. CpG island methylator phenotype association with upregulated telomerase activity in hepatocellular carcinoma. Int J Cancer. 2008;123:998–1004. doi: 10.1002/ijc.23650. [DOI] [PubMed] [Google Scholar]
- 69.Xu X, Liu RF, Wan BB, Xing WM, Huang J, Han ZG. Expression of a novel gene FAM43B repressing cell proliferation is regulated by DNA methylation in hepatocellular carcinoma cell lines. Mol Cell Biochem. 2011;354:11–20. doi: 10.1007/s11010-011-0800-y. [DOI] [PubMed] [Google Scholar]
- 70.Kanda M, Nomoto S, Okamura Y, Hayashi M, Hishida M, Fujii T, Nishikawa Y, Sugimoto H, Takeda S, Nakao A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol Carcinog. 2011;50:571–579. doi: 10.1002/mc.20735. [DOI] [PubMed] [Google Scholar]
- 71.Li B, Liu W, Wang L, Li M, Wang J, Huang L, Huang P, Yuan Y. CpG island methylator phenotype associated with tumor recurrence in tumor-node-metastasis stage I hepatocellular carcinoma. Ann Surg Oncol. 2010;17:1917–1926. doi: 10.1245/s10434-010-0921-7. [DOI] [PubMed] [Google Scholar]
- 72.Fukai K, Yokosuka O, Chiba T, Hirasawa Y, Tada M, Imazeki F, Kataoka H, Saisho H. Hepatocyte growth factor activator inhibitor 2/placental bikunin (HAI-2/PB) gene is frequently hypermethylated in human hepatocellular carcinoma. Cancer Res. 2003;63:8674–8679. [PubMed] [Google Scholar]
- 73.Yau TO, Chan CY, Chan KL, Lee MF, Wong CM, Fan ST, Ng IO. HDPR1, a novel inhibitor of the WNT/beta-catenin signaling, is frequently downregulated in hepatocellular carcinoma: involvement of methylation-mediated gene silencing. Oncogene. 2005;24:1607–1614. doi: 10.1038/sj.onc.1208340. [DOI] [PubMed] [Google Scholar]
- 74.Zhang YJ, Li H, Wu HC, Shen J, Wang L, Yu MW, Lee PH, Bernard Weinstein I, Santella RM. Silencing of Hint1, a novel tumor suppressor gene, by promoter hypermethylation in hepatocellular carcinoma. Cancer Lett. 2009;275:277–284. doi: 10.1016/j.canlet.2008.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanafusa T, Yumoto Y, Nouso K, Nakatsukasa H, Onishi T, Fujikawa T, Taniyama M, Nakamura S, Uemura M, Takuma Y, et al. Reduced expression of insulin-like growth factor binding protein-3 and its promoter hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2002;176:149–158. doi: 10.1016/s0304-3835(01)00736-4. [DOI] [PubMed] [Google Scholar]
- 76.Xie B, Zhou J, Yuan L, Ren F, Liu DC, Li Q, Shu G. Epigenetic silencing of Klotho expression correlates with poor prognosis of human hepatocellular carcinoma. Hum Pathol. 2013;44:795–801. doi: 10.1016/j.humpath.2012.07.023. [DOI] [PubMed] [Google Scholar]
- 77.Lu CY, Hsieh SY, Lu YJ, Wu CS, Chen LC, Lo SJ, Wu CT, Chou MY, Huang TH, Chang YS. Aberrant DNA methylation profile and frequent methylation of KLK10 and OXGR1 genes in hepatocellular carcinoma. Genes Chromosomes Cancer. 2009;48:1057–1068. doi: 10.1002/gcc.20708. [DOI] [PubMed] [Google Scholar]
- 78.Okamura Y, Nomoto S, Kanda M, Li Q, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Leukemia inhibitory factor receptor (LIFR) is detected as a novel suppressor gene of hepatocellular carcinoma using double-combination array. Cancer Lett. 2010;289:170–177. doi: 10.1016/j.canlet.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 79.Qiu G, Fang J, He Y. 5’ CpG island methylation analysis identifies the MAGE-A1 and MAGE-A3 genes as potential markers of HCC. Clin Biochem. 2006;39:259–266. doi: 10.1016/j.clinbiochem.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 80.Zhang J, Gong C, Bing Y, Li T, Liu Z, Liu Q. Hypermethylation-repressed methionine adenosyltransferase 1A as a potential biomarker for hepatocellular carcinoma. Hepatol Res. 2013;43:374–383. doi: 10.1111/j.1872-034X.2012.01099.x. [DOI] [PubMed] [Google Scholar]
- 81.Kanda M, Nomoto S, Okamura Y, Nishikawa Y, Sugimoto H, Kanazumi N, Takeda S, Nakao A. Detection of metallothionein 1G as a methylated tumor suppressor gene in human hepatocellular carcinoma using a novel method of double combination array analysis. Int J Oncol. 2009;35:477–483. doi: 10.3892/ijo_00000359. [DOI] [PubMed] [Google Scholar]
- 82.Mao J, Yu H, Wang C, Sun L, Jiang W, Zhang P, Xiao Q, Han D, Saiyin H, Zhu J, et al. Metallothionein MT1M is a tumor suppressor of human hepatocellular carcinomas. Carcinogenesis. 2012;33:2568–2577. doi: 10.1093/carcin/bgs287. [DOI] [PubMed] [Google Scholar]
- 83.Fan H, Chen L, Zhang F, Quan Y, Su X, Qiu X, Zhao Z, Kong KL, Dong S, Song Y, et al. MTSS1, a novel target of DNA methyltransferase 3B, functions as a tumor suppressor in hepatocellular carcinoma. Oncogene. 2012;31:2298–2308. doi: 10.1038/onc.2011.411. [DOI] [PubMed] [Google Scholar]
- 84.Ling Y, Zhu J, Gao L, Liu Y, Zhu C, Li R, Wei L, Zhang C. The silence of MUC2 mRNA induced by promoter hypermethylation associated with HBV in Hepatocellular Carcinoma. BMC Med Genet. 2013;14:14. doi: 10.1186/1471-2350-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Macheiner D, Heller G, Kappel S, Bichler C, Stättner S, Ziegler B, Kandioler D, Wrba F, Schulte-Hermann R, Zöchbauer-Müller S, et al. NORE1B, a candidate tumor suppressor, is epigenetically silenced in human hepatocellular carcinoma. J Hepatol. 2006;45:81–89. doi: 10.1016/j.jhep.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 86.Tada M, Yokosuka O, Fukai K, Chiba T, Imazeki F, Tokuhisa T, Saisho H. Hypermethylation of NAD(P)H: quinone oxidoreductase 1 (NQO1) gene in human hepatocellular carcinoma. J Hepatol. 2005;42:511–519. doi: 10.1016/j.jhep.2004.11.024. [DOI] [PubMed] [Google Scholar]
- 87.Hayashi T, Tamori A, Nishikawa M, Morikawa H, Enomoto M, Sakaguchi H, Habu D, Kawada N, Kubo S, Nishiguchi S, et al. Differences in molecular alterations of hepatocellular carcinoma between patients with a sustained virological response and those with hepatitis C virus infection. Liver Int. 2009;29:126–132. doi: 10.1111/j.1478-3231.2008.01772.x. [DOI] [PubMed] [Google Scholar]
- 88.Fang S, Huang SF, Cao J, Wen YA, Zhang LP, Ren GS. Silencing of PCDH10 in hepatocellular carcinoma via de novo DNA methylation independent of HBV infection or HBX expression. Clin Exp Med. 2013;13:127–134. doi: 10.1007/s10238-012-0182-9. [DOI] [PubMed] [Google Scholar]
- 89.Neumann O, Kesselmeier M, Geffers R, Pellegrino R, Radlwimmer B, Hoffmann K, Ehemann V, Schemmer P, Schirmacher P, Lorenzo Bermejo J, et al. Methylome analysis and integrative profiling of human HCCs identify novel protumorigenic factors. Hepatology. 2012;56:1817–1827. doi: 10.1002/hep.25870. [DOI] [PubMed] [Google Scholar]
- 90.Nishida N, Kudo M, Nagasaka T, Ikai I, Goel A. Characteristic patterns of altered DNA methylation predict emergence of human hepatocellular carcinoma. Hepatology. 2012;56:994–1003. doi: 10.1002/hep.25706. [DOI] [PubMed] [Google Scholar]
- 91.Wang L, Wang WL, Zhang Y, Guo SP, Zhang J, Li QL. Epigenetic and genetic alterations of PTEN in hepatocellular carcinoma. Hepatol Res. 2007;37:389–396. doi: 10.1111/j.1872-034X.2007.00042.x. [DOI] [PubMed] [Google Scholar]
- 92.Zhang C, Ling Y, Zhang C, Xu Y, Gao L, Li R, Zhu J, Fan L, Wei L. The silencing of RECK gene is associated with promoter hypermethylation and poor survival in hepatocellular carcinoma. Int J Biol Sci. 2012;8:451–458. doi: 10.7150/ijbs.4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Okamura Y, Nomoto S, Kanda M, Hayashi M, Nishikawa Y, Fujii T, Sugimoto H, Takeda S, Nakao A. Reduced expression of reelin (RELN) gene is associated with high recurrence rate of hepatocellular carcinoma. Ann Surg Oncol. 2011;18:572–579. doi: 10.1245/s10434-010-1273-z. [DOI] [PubMed] [Google Scholar]
- 94.Zhang C, Li H, Wang Y, Liu W, Zhang Q, Zhang T, Zhang X, Han B, Zhou G. Epigenetic inactivation of the tumor suppressor gene RIZ1 in hepatocellular carcinoma involves both DNA methylation and histone modifications. J Hepatol. 2010;53:889–895. doi: 10.1016/j.jhep.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 95.Piao GH, Piao WH, He Y, Zhang HH, Wang GQ, Piao Z. Hyper-methylation of RIZ1 tumor suppressor gene is involved in the early tumorigenesis of hepatocellular carcinoma. Histol Histopathol. 2008;23:1171–1175. doi: 10.14670/HH-23.1171. [DOI] [PubMed] [Google Scholar]
- 96.Shih YL, Hsieh CB, Lai HC, Yan MD, Hsieh TY, Chao YC, Lin YW. SFRP1 suppressed hepatoma cells growth through Wnt canonical signaling pathway. Int J Cancer. 2007;121:1028–1035. doi: 10.1002/ijc.22750. [DOI] [PubMed] [Google Scholar]
- 97.Takagi H, Sasaki S, Suzuki H, Toyota M, Maruyama R, Nojima M, Yamamoto H, Omata M, Tokino T, Imai K, et al. Frequent epigenetic inactivation of SFRP genes in hepatocellular carcinoma. J Gastroenterol. 2008;43:378–389. doi: 10.1007/s00535-008-2170-0. [DOI] [PubMed] [Google Scholar]
- 98.Jin J, You H, Yu B, Deng Y, Tang N, Yao G, Shu H, Yang S, Qin W. Epigenetic inactivation of SLIT2 in human hepatocellular carcinomas. Biochem Biophys Res Commun. 2009;379:86–91. doi: 10.1016/j.bbrc.2008.12.022. [DOI] [PubMed] [Google Scholar]
- 99.Yoshikawa H, Matsubara K, Qian GS, Jackson P, Groopman JD, Manning JE, Harris CC, Herman JG. SOCS-1, a negative regulator of the JAK/STAT pathway, is silenced by methylation in human hepatocellular carcinoma and shows growth-suppression activity. Nat Genet. 2001;28:29–35. doi: 10.1038/ng0501-29. [DOI] [PubMed] [Google Scholar]
- 100.Shih YL, Hsieh CB, Yan MD, Tsao CM, Hsieh TY, Liu CH, Lin YW. Frequent concomitant epigenetic silencing of SOX1 and secreted frizzled-related proteins (SFRPs) in human hepatocellular carcinoma. J Gastroenterol Hepatol. 2013;28:551–559. doi: 10.1111/jgh.12078. [DOI] [PubMed] [Google Scholar]
- 101.Jia Y, Yang Y, Liu S, Herman JG, Lu F, Guo M. SOX17 antagonizes WNT/β-catenin signaling pathway in hepatocellular carcinoma. Epigenetics. 2010;5:743–749. doi: 10.4161/epi.5.8.13104. [DOI] [PubMed] [Google Scholar]
- 102.Zhang Y, Yang B, Du Z, Bai T, Gao YT, Wang YJ, Lou C, Wang FM, Bai Y. Aberrant methylation of SPARC in human hepatocellular carcinoma and its clinical implication. World J Gastroenterol. 2012;18:2043–2052. doi: 10.3748/wjg.v18.i17.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wong CM, Ng YL, Lee JM, Wong CC, Cheung OF, Chan CY, Tung EK, Ching YP, Ng IO. Tissue factor pathway inhibitor-2 as a frequently silenced tumor suppressor gene in hepatocellular carcinoma. Hepatology. 2007;45:1129–1138. doi: 10.1002/hep.21578. [DOI] [PubMed] [Google Scholar]
- 104.Lu B, Ma Y, Wu G, Tong X, Guo H, Liang A, Cong W, Liu C, Wang H, Wu M, et al. Methylation of Tip30 promoter is associated with poor prognosis in human hepatocellular carcinoma. Clin Cancer Res. 2008;14:7405–7412. doi: 10.1158/1078-0432.CCR-08-0409. [DOI] [PubMed] [Google Scholar]
- 105.Yu J, Tao Q, Cheung KF, Jin H, Poon FF, Wang X, Li H, Cheng YY, Röcken C, Ebert MP, et al. Epigenetic identification of ubiquitin carboxyl-terminal hydrolase L1 as a functional tumor suppressor and biomarker for hepatocellular carcinoma and other digestive tumors. Hepatology. 2008;48:508–518. doi: 10.1002/hep.22343. [DOI] [PubMed] [Google Scholar]
- 106.Hibi K, Sakuraba K, Shirahata A, Goto T, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y. Methylation of the UNC5C gene is frequently detected in hepatocellular carcinoma. Hepatogastroenterology. 2012;59:2573–2575. doi: 10.5754/hge10616. [DOI] [PubMed] [Google Scholar]
- 107.Kitamura Y, Shirahata A, Sakuraba K, Goto T, Mizukami H, Saito M, Ishibashi K, Kigawa G, Nemoto H, Sanada Y, et al. Aberrant methylation of the Vimentin gene in hepatocellular carcinoma. Anticancer Res. 2011;31:1289–1291. [PubMed] [Google Scholar]
- 108.Huang L, Li MX, Wang L, Li BK, Chen GH, He LR, Xu L, Yuan YF. Prognostic value of Wnt inhibitory factor-1 expression in hepatocellular carcinoma that is independent of gene methylation. Tumour Biol. 2011;32:233–240. doi: 10.1007/s13277-010-0117-6. [DOI] [PubMed] [Google Scholar]
- 109.Lv Z, Zhang M, Bi J, Xu F, Hu S, Wen J. Promoter hypermethylation of a novel gene, ZHX2, in hepatocellular carcinoma. Am J Clin Pathol. 2006;125:740–746. doi: 10.1309/09B4-52V7-R76K-7D6K. [DOI] [PubMed] [Google Scholar]
- 110.Shim YH, Yoon GS, Choi HJ, Chung YH, Yu E. p16 Hypermethylation in the early stage of hepatitis B virus-associated hepatocarcinogenesis. Cancer Lett. 2003;190:213–219. doi: 10.1016/s0304-3835(02)00613-4. [DOI] [PubMed] [Google Scholar]
- 111.Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–993. doi: 10.1038/nrc1507. [DOI] [PubMed] [Google Scholar]
- 112.Shen L, Ahuja N, Shen Y, Habib NA, Toyota M, Rashid A, Issa JP. DNA methylation and environmental exposures in human hepatocellular carcinoma. J Natl Cancer Inst. 2002;94:755–761. doi: 10.1093/jnci/94.10.755. [DOI] [PubMed] [Google Scholar]
- 113.Zhang C, Li Z, Cheng Y, Jia F, Li R, Wu M, Li K, Wei L. CpG island methylator phenotype association with elevated serum alpha-fetoprotein level in hepatocellular carcinoma. Clin Cancer Res. 2007;13:944–952. doi: 10.1158/1078-0432.CCR-06-2268. [DOI] [PubMed] [Google Scholar]
- 114.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 115.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 116.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 117.Yates LA, Norbury CJ, Gilbert RJ. The long and short of microRNA. Cell. 2013;153:516–519. doi: 10.1016/j.cell.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 118.Wong CM, Wong CC, Lee JM, Fan DN, Au SL, Ng IO. Sequential alterations of microRNA expression in hepatocellular carcinoma development and venous metastasis. Hepatology. 2012;55:1453–1461. doi: 10.1002/hep.25512. [DOI] [PubMed] [Google Scholar]
- 119.Iorio MV, Croce CM. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–159. doi: 10.1002/emmm.201100209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Luk JM, Burchard J, Zhang C, Liu AM, Wong KF, Shek FH, Lee NP, Fan ST, Poon RT, Ivanovska I, et al. DLK1-DIO3 genomic imprinted microRNA cluster at 14q32.2 defines a stemlike subtype of hepatocellular carcinoma associated with poor survival. J Biol Chem. 2011;286:30706–30713. doi: 10.1074/jbc.M111.229831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marquardt JU, Factor VM, Thorgeirsson SS. Epigenetic regulation of cancer stem cells in liver cancer: current concepts and clinical implications. J Hepatol. 2010;53:568–577. doi: 10.1016/j.jhep.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li R, Qian N, Tao K, You N, Wang X, Dou K. MicroRNAs involved in neoplastic transformation of liver cancer stem cells. J Exp Clin Cancer Res. 2010;29:169. doi: 10.1186/1756-9966-29-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Huang S, He X. The role of microRNAs in liver cancer progression. Br J Cancer. 2011;104:235–240. doi: 10.1038/sj.bjc.6606010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Law PT, Wong N. Emerging roles of microRNA in the intracellular signaling networks of hepatocellular carcinoma. J Gastroenterol Hepatol. 2011;26:437–449. doi: 10.1111/j.1440-1746.2010.06512.x. [DOI] [PubMed] [Google Scholar]
- 125.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ladeiro Y, Couchy G, Balabaud C, Bioulac-Sage P, Pelletier L, Rebouissou S, Zucman-Rossi J. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 127.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat Rev Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Law PT, Qin H, Ching AK, Lai KP, Co NN, He M, Lung RW, Chan AW, Chan TF, Wong N. Deep sequencing of small RNA transcriptome reveals novel non-coding RNAs in hepatocellular carcinoma. J Hepatol. 2013;58:1165–1173. doi: 10.1016/j.jhep.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 129.Hou J, Lin L, Zhou W, Wang Z, Ding G, Dong Q, Qin L, Wu X, Zheng Y, Yang Y, et al. Identification of miRNomes in human liver and hepatocellular carcinoma reveals miR-199a/b-3p as therapeutic target for hepatocellular carcinoma. Cancer Cell. 2011;19:232–243. doi: 10.1016/j.ccr.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 130.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 131.Budhu A, Jia HL, Forgues M, Liu CG, Goldstein D, Lam A, Zanetti KA, Ye QH, Qin LX, Croce CM, et al. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- 132.Liu S, Guo W, Shi J, Li N, Yu X, Xue J, Fu X, Chu K, Lu C, Zhao J, et al. MicroRNA-135a contributes to the development of portal vein tumor thrombus by promoting metastasis in hepatocellular carcinoma. J Hepatol. 2012;56:389–396. doi: 10.1016/j.jhep.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 133.Ji J, Shi J, Budhu A, Yu Z, Forgues M, Roessler S, Ambs S, Chen Y, Meltzer PS, Croce CM, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–1447. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Han ZB, Zhong L, Teng MJ, Fan JW, Tang HM, Wu JY, Chen HY, Wang ZW, Qiu GQ, Peng ZH. Identification of recurrence-related microRNAs in hepatocellular carcinoma following liver transplantation. Mol Oncol. 2012;6:445–457. doi: 10.1016/j.molonc.2012.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Augello C, Vaira V, Caruso L, Destro A, Maggioni M, Park YN, Montorsi M, Santambrogio R, Roncalli M, Bosari S. MicroRNA profiling of hepatocarcinogenesis identifies C19MC cluster as a novel prognostic biomarker in hepatocellular carcinoma. Liver Int. 2012;32:772–782. doi: 10.1111/j.1478-3231.2012.02795.x. [DOI] [PubMed] [Google Scholar]
- 136.Tomimaru Y, Eguchi H, Nagano H, Wada H, Tomokuni A, Kobayashi S, Marubashi S, Takeda Y, Tanemura M, Umeshita K, et al. MicroRNA-21 induces resistance to the anti-tumour effect of interferon-α/5-fluorouracil in hepatocellular carcinoma cells. Br J Cancer. 2010;103:1617–1626. doi: 10.1038/sj.bjc.6605958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tomokuni A, Eguchi H, Tomimaru Y, Wada H, Kawamoto K, Kobayashi S, Marubashi S, Tanemura M, Nagano H, Mori M, et al. miR-146a suppresses the sensitivity to interferon-α in hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2011;414:675–680. doi: 10.1016/j.bbrc.2011.09.124. [DOI] [PubMed] [Google Scholar]
- 138.Fornari F, Milazzo M, Chieco P, Negrini M, Calin GA, Grazi GL, Pollutri D, Croce CM, Bolondi L, Gramantieri L. MiR-199a-3p regulates mTOR and c-Met to influence the doxorubicin sensitivity of human hepatocarcinoma cells. Cancer Res. 2010;70:5184–5193. doi: 10.1158/0008-5472.CAN-10-0145. [DOI] [PubMed] [Google Scholar]
- 139.Zhang J, Wang Y, Zhen P, Luo X, Zhang C, Zhou L, Lu Y, Yang Y, Zhang W, Wan J. Genome-wide analysis of miRNA signature differentially expressed in doxorubicin-resistant and parental human hepatocellular carcinoma cell lines. PLoS One. 2013;8:e54111. doi: 10.1371/journal.pone.0054111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Xu Y, Xia F, Ma L, Shan J, Shen J, Yang Z, Liu J, Cui Y, Bian X, Bie P, et al. MicroRNA-122 sensitizes HCC cancer cells to adriamycin and vincristine through modulating expression of MDR and inducing cell cycle arrest. Cancer Lett. 2011;310:160–169. doi: 10.1016/j.canlet.2011.06.027. [DOI] [PubMed] [Google Scholar]
- 141.Ma K, He Y, Zhang H, Fei Q, Niu D, Wang D, Ding X, Xu H, Chen X, Zhu J. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem. 2012;287:5639–5649. doi: 10.1074/jbc.M111.291229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Bai S, Nasser MW, Wang B, Hsu SH, Datta J, Kutay H, Yadav A, Nuovo G, Kumar P, Ghoshal K. MicroRNA-122 inhibits tumorigenic properties of hepatocellular carcinoma cells and sensitizes these cells to sorafenib. J Biol Chem. 2009;284:32015–32027. doi: 10.1074/jbc.M109.016774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Shimizu S, Takehara T, Hikita H, Kodama T, Miyagi T, Hosui A, Tatsumi T, Ishida H, Noda T, Nagano H, et al. The let-7 family of microRNAs inhibits Bcl-xL expression and potentiates sorafenib-induced apoptosis in human hepatocellular carcinoma. J Hepatol. 2010;52:698–704. doi: 10.1016/j.jhep.2009.12.024. [DOI] [PubMed] [Google Scholar]
- 144.Xia H, Ooi LL, Hui KM. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 145.Tsang TY, Tang WY, Chan JY, Co NN, Au Yeung CL, Yau PL, Kong SK, Fung KP, Kwok TT. P-glycoprotein enhances radiation-induced apoptotic cell death through the regulation of miR-16 and Bcl-2 expressions in hepatocellular carcinoma cells. Apoptosis. 2011;16:524–535. doi: 10.1007/s10495-011-0581-5. [DOI] [PubMed] [Google Scholar]
- 146.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, Iorio MV, Visone R, Sever NI, Fabbri M, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 147.Starczynowski DT, Morin R, McPherson A, Lam J, Chari R, Wegrzyn J, Kuchenbauer F, Hirst M, Tohyama K, Humphries RK, et al. Genome-wide identification of human microRNAs located in leukemia-associated genomic alterations. Blood. 2011;117:595–607. doi: 10.1182/blood-2010-03-277012. [DOI] [PubMed] [Google Scholar]
- 148.O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 149.Chang TC, Yu D, Lee YS, Wentzel EA, Arking DE, West KM, Dang CV, Thomas-Tikhonenko A, Mendell JT. Widespread microRNA repression by Myc contributes to tumorigenesis. Nat Genet. 2008;40:43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Cairo S, Wang Y, de Reyniès A, Duroure K, Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM, et al. Stem cell-like micro-RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci USA. 2010;107:20471–20476. doi: 10.1073/pnas.1009009107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ, Kim MG, Chang YG, Shen Q, Park WS, Lee JY, et al. Sirtuin7 oncogenic potential in human hepatocellular carcinoma and its regulation by the tumor suppressors MiR-125a-5p and MiR-125b. Hepatology. 2013;57:1055–1067. doi: 10.1002/hep.26101. [DOI] [PubMed] [Google Scholar]
- 152.Fornari F, Milazzo M, Chieco P, Negrini M, Marasco E, Capranico G, Mantovani V, Marinello J, Sabbioni S, Callegari E, et al. In hepatocellular carcinoma miR-519d is up-regulated by p53 and DNA hypomethylation and targets CDKN1A/p21, PTEN, AKT3 and TIMP2. J Pathol. 2012;227:275–285. doi: 10.1002/path.3995. [DOI] [PubMed] [Google Scholar]
- 153.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, et al. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 155.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11:597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 156.Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, et al. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Nakamura T, Canaani E, Croce CM. Oncogenic All1 fusion proteins target Drosha-mediated microRNA processing. Proc Natl Acad Sci USA. 2007;104:10980–10985. doi: 10.1073/pnas.0704559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Sekine S, Ogawa R, Ito R, Hiraoka N, McManus MT, Kanai Y, Hebrok M. Disruption of Dicer1 induces dysregulated fetal gene expression and promotes hepatocarcinogenesis. Gastroenterology. 2009;136:2304–2315.e1-4. doi: 10.1053/j.gastro.2009.02.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kitagawa N, Ojima H, Shirakihara T, Shimizu H, Kokubu A, Urushidate T, Totoki Y, Kosuge T, Miyagawa S, Shibata T. Downregulation of the microRNA biogenesis components and its association with poor prognosis in hepatocellular carcinoma. Cancer Sci. 2013;104:543–551. doi: 10.1111/cas.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Kunej T, Godnic I, Horvat S, Zorc M, Calin GA. Cross talk between microRNA and coding cancer genes. Cancer J. 2012;18:223–231. doi: 10.1097/PPO.0b013e318258b771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Han L, Witmer PD, Casey E, Valle D, Sukumar S. DNA methylation regulates MicroRNA expression. Cancer Biol Ther. 2007;6:1284–1288. doi: 10.4161/cbt.6.8.4486. [DOI] [PubMed] [Google Scholar]
- 163.Kozaki K, Inazawa J. Tumor-suppressive microRNA silenced by tumor-specific DNA hypermethylation in cancer cells. Cancer Sci. 2012;103:837–845. doi: 10.1111/j.1349-7006.2012.02236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Datta J, Kutay H, Nasser MW, Nuovo GJ, Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et al. Methylation mediated silencing of MicroRNA-1 gene and its role in hepatocellular carcinogenesis. Cancer Res. 2008;68:5049–5058. doi: 10.1158/0008-5472.CAN-07-6655. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 165.Potapova A, Albat C, Hasemeier B, Haeussler K, Lamprecht S, Suerbaum S, Kreipe H, Lehmann U. Systematic cross-validation of 454 sequencing and pyrosequencing for the exact quantification of DNA methylation patterns with single CpG resolution. BMC Biotechnol. 2011;11:6. doi: 10.1186/1472-6750-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Anwar SL, Albat C, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Lehmann U. Concordant hypermethylation of intergenic microRNA genes in human hepatocellular carcinoma as new diagnostic and prognostic marker. Int J Cancer. 2013;133:660–670. doi: 10.1002/ijc.28068. [DOI] [PubMed] [Google Scholar]
- 167.Shen J, Wang S, Zhang YJ, Kappil MA, Chen Wu H, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, et al. Genome-wide aberrant DNA methylation of microRNA host genes in hepatocellular carcinoma. Epigenetics. 2012;7:1230–1237. doi: 10.4161/epi.22140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Furuta M, Kozaki KI, Tanaka S, Arii S, Imoto I, Inazawa J. miR-124 and miR-203 are epigenetically silenced tumor-suppressive microRNAs in hepatocellular carcinoma. Carcinogenesis. 2010;31:766–776. doi: 10.1093/carcin/bgp250. [DOI] [PubMed] [Google Scholar]
- 169.Alpini G, Glaser SS, Zhang JP, Francis H, Han Y, Gong J, Stokes A, Francis T, Hughart N, Hubble L, et al. Regulation of placenta growth factor by microRNA-125b in hepatocellular cancer. J Hepatol. 2011;55:1339–1345. doi: 10.1016/j.jhep.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer. 2013;52:636–643. doi: 10.1002/gcc.22059. [DOI] [PubMed] [Google Scholar]
- 171.Wei X, Tan C, Tang C, Ren G, Xiang T, Qiu Z, Liu R, Wu Z. Epigenetic repression of miR-132 expression by the hepatitis B virus x protein in hepatitis B virus-related hepatocellular carcinoma. Cell Signal. 2013;25:1037–1043. doi: 10.1016/j.cellsig.2013.01.019. [DOI] [PubMed] [Google Scholar]
- 172.Dohi O, Yasui K, Gen Y, Takada H, Endo M, Tsuji K, Konishi C, Yamada N, Mitsuyoshi H, Yagi N, et al. Epigenetic silencing of miR-335 and its host gene MEST in hepatocellular carcinoma. Int J Oncol. 2013;42:411–418. doi: 10.3892/ijo.2012.1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Katoh M, Igarashi M, Fukuda H, Nakagama H, Katoh M. Cancer genetics and genomics of human FOX family genes. Cancer Lett. 2013;328:198–206. doi: 10.1016/j.canlet.2012.09.017. [DOI] [PubMed] [Google Scholar]
- 175.Zhang Y, Zhang S, Wang X, Liu J, Yang L, He S, Chen L, Huang J. Prognostic significance of FOXP1 as an oncogene in hepatocellular carcinoma. J Clin Pathol. 2012;65:528–533. doi: 10.1136/jclinpath-2011-200547. [DOI] [PubMed] [Google Scholar]
- 176.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. doi: 10.1038/nrc3205. [DOI] [PubMed] [Google Scholar]
- 177.Chu JS, Ge FJ, Zhang B, Wang Y, Silvestris N, Liu LJ, Zhao CH, Lin L, Brunetti AE, Fu YL, et al. Expression and prognostic value of VEGFR-2, PDGFR-β, and c-Met in advanced hepatocellular carcinoma. J Exp Clin Cancer Res. 2013;32:16. doi: 10.1186/1756-9966-32-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Goyal L, Muzumdar MD, Zhu AX. Targeting the HGF/c-MET pathway in hepatocellular carcinoma. Clin Cancer Res. 2013;19:2310–2318. doi: 10.1158/1078-0432.CCR-12-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Buske C, Feuring-Buske M, Abramovich C, Spiekermann K, Eaves CJ, Coulombel L, Sauvageau G, Hogge DE, Humphries RK. Deregulated expression of HOXB4 enhances the primitive growth activity of human hematopoietic cells. Blood. 2002;100:862–868. doi: 10.1182/blood-2002-01-0220. [DOI] [PubMed] [Google Scholar]
- 180.Barba-de la Rosa AP, Briones-Cerecero E, Lugo-Melchor O, De León-Rodríguez A, Santos L, Castelo-Ruelas J, Valdivia A, Piña P, Chagolla-López A, Hernandez-Cueto D, et al. Hox B4 as potential marker of non-differentiated cells in human cervical cancer cells. J Cancer Res Clin Oncol. 2012;138:293–300. doi: 10.1007/s00432-011-1081-2. [DOI] [PubMed] [Google Scholar]
- 181.Varnholt H, Drebber U, Schulze F, Wedemeyer I, Schirmacher P, Dienes HP, Odenthal M. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 182.Yan Y, Luo YC, Wan HY, Wang J, Zhang PP, Liu M, Li X, Li S, Tang H. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology. 2013;57:667–677. doi: 10.1002/hep.26071. [DOI] [PubMed] [Google Scholar]
- 183.Lang Q, Ling C. MiR-124 suppresses cell proliferation in hepatocellular carcinoma by targeting PIK3CA. Biochem Biophys Res Commun. 2012;426:247–252. doi: 10.1016/j.bbrc.2012.08.075. [DOI] [PubMed] [Google Scholar]
- 184.Hatziapostolou M, Polytarchou C, Aggelidou E, Drakaki A, Poultsides GA, Jaeger SA, Ogata H, Karin M, Struhl K, Hadzopoulou-Cladaras M, et al. An HNF4α-miRNA inflammatory feedback circuit regulates hepatocellular oncogenesis. Cell. 2011;147:1233–1247. doi: 10.1016/j.cell.2011.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zheng F, Liao YJ, Cai MY, Liu YH, Liu TH, Chen SP, Bian XW, Guan XY, Lin MC, Zeng YX, et al. The putative tumour suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut. 2012;61:278–289. doi: 10.1136/gut.2011.239145. [DOI] [PubMed] [Google Scholar]
- 186.Yan H, Choi AJ, Lee BH, Ting AH. Identification and functional analysis of epigenetically silenced microRNAs in colorectal cancer cells. PLoS One. 2011;6:e20628. doi: 10.1371/journal.pone.0020628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Liang L, Wong CM, Ying Q, Fan DN, Huang S, Ding J, Yao J, Yan M, Li J, Yao M, et al. MicroRNA-125b suppressesed human liver cancer cell proliferation and metastasis by directly targeting oncogene LIN28B2. Hepatology. 2010;52:1731–1740. doi: 10.1002/hep.23904. [DOI] [PubMed] [Google Scholar]
- 188.Fan DN, Tsang FH, Tam AH, Au SL, Wong CC, Wei L, Lee JM, He X, Ng IO, Wong CM. Histone lysine methyltransferase, suppressor of variegation 3-9 homolog 1, promotes hepatocellular carcinoma progression and is negatively regulated by microRNA-125b. Hepatology. 2013;57:637–647. doi: 10.1002/hep.26083. [DOI] [PubMed] [Google Scholar]
- 189.Liu Y, Hei Y, Shu Q, Dong J, Gao Y, Fu H, Zheng X, Yang G. VCP/p97, down-regulated by microRNA-129-5p, could regulate the progression of hepatocellular carcinoma. PLoS One. 2012;7:e35800. doi: 10.1371/journal.pone.0035800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang W, Peng B, Wang D, Ma X, Jiang D, Zhao J, Yu L. Human tumor microRNA signatures derived from large-scale oligonucleotide microarray datasets. Int J Cancer. 2011;129:1624–1634. doi: 10.1002/ijc.25818. [DOI] [PubMed] [Google Scholar]
- 191.Formosa A, Lena AM, Markert EK, Cortelli S, Miano R, Mauriello A, Croce N, Vandesompele J, Mestdagh P, Finazzi-Agrò E, et al. DNA methylation silences miR-132 in prostate cancer. Oncogene. 2013;32:127–134. doi: 10.1038/onc.2012.14. [DOI] [PubMed] [Google Scholar]
- 192.Zhang S, Hao J, Xie F, Hu X, Liu C, Tong J, Zhou J, Wu J, Shao C. Downregulation of miR-132 by promoter methylation contributes to pancreatic cancer development. Carcinogenesis. 2011;32:1183–1189. doi: 10.1093/carcin/bgr105. [DOI] [PubMed] [Google Scholar]
- 193.Bueno MJ, Pérez de Castro I, Gómez de Cedrón M, Santos J, Calin GA, Cigudosa JC, Croce CM, Fernández-Piqueras J, Malumbres M. Genetic and epigenetic silencing of microRNA-203 enhances ABL1 and BCR-ABL1 oncogene expression. Cancer Cell. 2008;13:496–506. doi: 10.1016/j.ccr.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 194.Chen X, Hu H, Guan X, Xiong G, Wang Y, Wang K, Li J, Xu X, Yang K, Bai Y. CpG island methylation status of miRNAs in esophageal squamous cell carcinoma. Int J Cancer. 2012;130:1607–1613. doi: 10.1002/ijc.26171. [DOI] [PubMed] [Google Scholar]
- 195.Ishida H, Tatsumi T, Hosui A, Nawa T, Kodama T, Shimizu S, Hikita H, Hiramatsu N, Kanto T, Hayashi N, et al. Alterations in microRNA expression profile in HCV-infected hepatoma cells: involvement of miR-491 in regulation of HCV replication via the PI3 kinase/Akt pathway. Biochem Biophys Res Commun. 2011;412:92–97. doi: 10.1016/j.bbrc.2011.07.049. [DOI] [PubMed] [Google Scholar]
- 196.Chen L, Yan HX, Yang W, Hu L, Yu LX, Liu Q, Li L, Huang DD, Ding J, Shen F, et al. The role of microRNA expression pattern in human intrahepatic cholangiocarcinoma. J Hepatol. 2009;50:358–369. doi: 10.1016/j.jhep.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 197.Schepeler T, Reinert JT, Ostenfeld MS, Christensen LL, Silahtaroglu AN, Dyrskjøt L, Wiuf C, Sørensen FJ, Kruhøffer M, Laurberg S, et al. Diagnostic and prognostic microRNAs in stage II colon cancer. Cancer Res. 2008;68:6416–6424. doi: 10.1158/0008-5472.CAN-07-6110. [DOI] [PubMed] [Google Scholar]
- 198.Bronisz A, Godlewski J, Wallace JA, Merchant AS, Nowicki MO, Mathsyaraja H, Srinivasan R, Trimboli AJ, Martin CK, Li F, et al. Reprogramming of the tumour microenvironment by stromal PTEN-regulated miR-320. Nat Cell Biol. 2012;14:159–167. doi: 10.1038/ncb2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Hsieh IS, Chang KC, Tsai YT, Ke JY, Lu PJ, Lee KH, Yeh SD, Hong TM, Chen YL. MicroRNA-320 suppresses the stem cell-like characteristics of prostate cancer cells by downregulating the Wnt/beta-catenin signaling pathway. Carcinogenesis. 2013;34:530–538. doi: 10.1093/carcin/bgs371. [DOI] [PubMed] [Google Scholar]
- 200.Heyn H, Engelmann M, Schreek S, Ahrens P, Lehmann U, Kreipe H, Schlegelberger B, Beger C. MicroRNA miR-335 is crucial for the BRCA1 regulatory cascade in breast cancer development. Int J Cancer. 2011;129:2797–2806. doi: 10.1002/ijc.25962. [DOI] [PubMed] [Google Scholar]
- 201.Png KJ, Yoshida M, Zhang XH, Shu W, Lee H, Rimner A, Chan TA, Comen E, Andrade VP, Kim SW, et al. MicroRNA-335 inhibits tumor reinitiation and is silenced through genetic and epigenetic mechanisms in human breast cancer. Genes Dev. 2011;25:226–231. doi: 10.1101/gad.1974211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Xiong SW, Lin TX, Xu KW, Dong W, Ling XH, Jiang FN, Chen G, Zhong WD, Huang J. MicroRNA-335 acts as a candidate tumor suppressor in prostate cancer. Pathol Oncol Res. 2013;19:529–537. doi: 10.1007/s12253-013-9613-5. [DOI] [PubMed] [Google Scholar]
- 203.Williams SV, Platt FM, Hurst CD, Aveyard JS, Taylor CF, Pole JC, Garcia MJ, Knowles MA. High-resolution analysis of genomic alteration on chromosome arm 8p in urothelial carcinoma. Genes Chromosomes Cancer. 2010;49:642–659. doi: 10.1002/gcc.20775. [DOI] [PubMed] [Google Scholar]
- 204.Endo H, Muramatsu T, Furuta M, Uzawa N, Pimkhaokham A, Amagasa T, Inazawa J, Kozaki K. Potential of tumor-suppressive miR-596 targeting LGALS3BP as a therapeutic agent in oral cancer. Carcinogenesis. 2013;34:560–569. doi: 10.1093/carcin/bgs376. [DOI] [PubMed] [Google Scholar]
- 205.Pan J, Hu H, Zhou Z, Sun L, Peng L, Yu L, Sun L, Liu J, Yang Z, Ran Y. Tumor-suppressive mir-663 gene induces mitotic catastrophe growth arrest in human gastric cancer cells. Oncol Rep. 2010;24:105–112. doi: 10.3892/or_00000834. [DOI] [PubMed] [Google Scholar]
- 206.Ma L, Young J, Prabhala H, Pan E, Mestdagh P, Muth D, Teruya-Feldstein J, Reinhardt F, Onder TT, Valastyan S, et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat Cell Biol. 2010;12:247–256. doi: 10.1038/ncb2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tan HX, Wang Q, Chen LZ, Huang XH, Chen JS, Fu XH, Cao LQ, Chen XL, Li W, Zhang LJ. MicroRNA-9 reduces cell invasion and E-cadherin secretion in SK-Hep-1 cell. Med Oncol. 2010;27:654–660. doi: 10.1007/s12032-009-9264-2. [DOI] [PubMed] [Google Scholar]
- 208.Gravgaard KH, Lyng MB, Laenkholm AV, Søkilde R, Nielsen BS, Litman T, Ditzel HJ. The miRNA-200 family and miRNA-9 exhibit differential expression in primary versus corresponding metastatic tissue in breast cancer. Breast Cancer Res Treat. 2012;134:207–217. doi: 10.1007/s10549-012-1969-9. [DOI] [PubMed] [Google Scholar]
- 209.Selcuklu SD, Donoghue MT, Rehmet K, de Souza Gomes M, Fort A, Kovvuru P, Muniyappa MK, Kerin MJ, Enright AJ, Spillane C. MicroRNA-9 inhibition of cell proliferation and identification of novel miR-9 targets by transcriptome profiling in breast cancer cells. J Biol Chem. 2012;287:29516–29528. doi: 10.1074/jbc.M111.335943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Minor J, Wang X, Zhang F, Song J, Jimeno A, Wang XJ, Lu X, Gross N, Kulesz-Martin M, Wang D, et al. Methylation of microRNA-9 is a specific and sensitive biomarker for oral and oropharyngeal squamous cell carcinomas. Oral Oncol. 2012;48:73–78. doi: 10.1016/j.oraloncology.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C, Minichsdorfer C, Lang G, Döme B, End-Pfützenreuter A, et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non-small cell lung cancers. Clin Cancer Res. 2012;18:1619–1629. doi: 10.1158/1078-0432.CCR-11-2450. [DOI] [PubMed] [Google Scholar]
- 212.Shimizu T, Suzuki H, Nojima M, Kitamura H, Yamamoto E, Maruyama R, Ashida M, Hatahira T, Kai M, Masumori N, et al. Methylation of a panel of microRNA genes is a novel biomarker for detection of bladder cancer. Eur Urol. 2013;63:1091–1100. doi: 10.1016/j.eururo.2012.11.030. [DOI] [PubMed] [Google Scholar]
- 213.Du Y, Liu Z, Gu L, Zhou J, Zhu BD, Ji J, Deng D. Characterization of human gastric carcinoma-related methylation of 9 miR CpG islands and repression of their expressions in vitro and in vivo. BMC Cancer. 2012;12:249. doi: 10.1186/1471-2407-12-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 215.Shitani M, Sasaki S, Akutsu N, Takagi H, Suzuki H, Nojima M, Yamamoto H, Tokino T, Hirata K, Imai K, et al. Genome-wide analysis of DNA methylation identifies novel cancer-related genes in hepatocellular carcinoma. Tumour Biol. 2012;33:1307–1317. doi: 10.1007/s13277-012-0378-3. [DOI] [PubMed] [Google Scholar]
- 216.Deng YB, Nagae G, Midorikawa Y, Yagi K, Tsutsumi S, Yamamoto S, Hasegawa K, Kokudo N, Aburatani H, Kaneda A. Identification of genes preferentially methylated in hepatitis C virus-related hepatocellular carcinoma. Cancer Sci. 2010;101:1501–1510. doi: 10.1111/j.1349-7006.2010.01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Stefanska B, Huang J, Bhattacharyya B, Suderman M, Hallett M, Han ZG, Szyf M. Definition of the landscape of promoter DNA hypomethylation in liver cancer. Cancer Res. 2011;71:5891–5903. doi: 10.1158/0008-5472.CAN-10-3823. [DOI] [PubMed] [Google Scholar]
- 218.Andersen JB, Factor VM, Marquardt JU, Raggi C, Lee YH, Seo D, Conner EA, Thorgeirsson SS. An integrated genomic and epigenomic approach predicts therapeutic response to zebularine in human liver cancer. Sci Transl Med. 2010;2:54ra77. doi: 10.1126/scitranslmed.3001338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Yang JD, Seol SY, Leem SH, Kim YH, Sun Z, Lee JS, Thorgeirsson SS, Chu IS, Roberts LR, Kang KJ. Genes associated with recurrence of hepatocellular carcinoma: integrated analysis by gene expression and methylation profiling. J Korean Med Sci. 2011;26:1428–1438. doi: 10.3346/jkms.2011.26.11.1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tao R, Li J, Xin J, Wu J, Guo J, Zhang L, Jiang L, Zhang W, Yang Z, Li L. Methylation profile of single hepatocytes derived from hepatitis B virus-related hepatocellular carcinoma. PLoS One. 2011;6:e19862. doi: 10.1371/journal.pone.0019862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Ammerpohl O, Pratschke J, Schafmayer C, Haake A, Faber W, von Kampen O, Brosch M, Sipos B, von Schönfels W, Balschun K, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130:1319–1328. doi: 10.1002/ijc.26136. [DOI] [PubMed] [Google Scholar]
- 222.Shen J, Wang S, Zhang YJ, Kappil M, Wu HC, Kibriya MG, Wang Q, Jasmine F, Ahsan H, Lee PH, et al. Genome-wide DNA methylation profiles in hepatocellular carcinoma. Hepatology. 2012;55:1799–1808. doi: 10.1002/hep.25569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Shen J, Wang S, Zhang YJ, Wu HC, Kibriya MG, Jasmine F, Ahsan H, Wu DP, Siegel AB, Remotti H, et al. Exploring genome-wide DNA methylation profiles altered in hepatocellular carcinoma using Infinium HumanMethylation 450 BeadChips. Epigenetics. 2013;8:34–43. doi: 10.4161/epi.23062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Nguyen LV, Vanner R, Dirks P, Eaves CJ. Cancer stem cells: an evolving concept. Nat Rev Cancer. 2012;12:133–143. doi: 10.1038/nrc3184. [DOI] [PubMed] [Google Scholar]
- 225.Spivakov M, Fisher AG. Epigenetic signatures of stem-cell identity. Nat Rev Genet. 2007;8:263–271. doi: 10.1038/nrg2046. [DOI] [PubMed] [Google Scholar]
- 226.Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.You H, Ding W, Rountree CB. Epigenetic regulation of cancer stem cell marker CD133 by transforming growth factor-beta. Hepatology. 2010;51:1635–1644. doi: 10.1002/hep.23544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Chiba T, Kita K, Zheng YW, Yokosuka O, Saisho H, Iwama A, Nakauchi H, Taniguchi H. Side population purified from hepatocellular carcinoma cells harbors cancer stem cell-like properties. Hepatology. 2006;44:240–251. doi: 10.1002/hep.21227. [DOI] [PubMed] [Google Scholar]
- 229.Zhai JM, Yin XY, Hou X, Hao XY, Cai JP, Liang LJ, Zhang LJ. Analysis of the genome-wide DNA methylation profile of side population cells in hepatocellular carcinoma. Dig Dis Sci. 2013;58:1934–1947. doi: 10.1007/s10620-013-2663-4. [DOI] [PubMed] [Google Scholar]
- 230.Ji J, Yamashita T, Budhu A, Forgues M, Jia HL, Li C, Deng C, Wauthier E, Reid LM, Ye QH, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Villanueva A. Rethinking future development of molecular therapies in hepatocellular carcinoma: a bottom-up approach. J Hepatol. 2013;59:392–395. doi: 10.1016/j.jhep.2013.03.025. [DOI] [PubMed] [Google Scholar]
- 232.Zhou C, Liu J, Li Y, Liu L, Zhang X, Ma CY, Hua SC, Yang M, Yuan Q. microRNA-1274a, a modulator of sorafenib induced a disintegrin and metalloproteinase 9 (ADAM9) down-regulation in hepatocellular carcinoma. FEBS Lett. 2011;585:1828–1834. doi: 10.1016/j.febslet.2011.04.040. [DOI] [PubMed] [Google Scholar]
- 233.Kaminskas E, Farrell A, Abraham S, Baird A, Hsieh LS, Lee SL, Leighton JK, Patel H, Rahman A, Sridhara R, et al. Approval summary: azacitidine for treatment of myelodysplastic syndrome subtypes. Clin Cancer Res. 2005;11:3604–3608. doi: 10.1158/1078-0432.CCR-04-2135. [DOI] [PubMed] [Google Scholar]
- 234.Kanda T, Tada M, Imazeki F, Yokosuka O, Nagao K, Saisho H. 5-aza-2’-deoxycytidine sensitizes hepatoma and pancreatic cancer cell lines. Oncol Rep. 2005;14:975–979. [PubMed] [Google Scholar]
- 235.Venturelli S, Armeanu S, Pathil A, Hsieh CJ, Weiss TS, Vonthein R, Wehrmann M, Gregor M, Lauer UM, Bitzer M. Epigenetic combination therapy as a tumor-selective treatment approach for hepatocellular carcinoma. Cancer. 2007;109:2132–2141. doi: 10.1002/cncr.22652. [DOI] [PubMed] [Google Scholar]
- 236.Venturelli S, Berger A, Weiland T, Zimmermann M, Häcker S, Peter C, Wesselborg S, Königsrainer A, Weiss TS, Gregor M, et al. Dual antitumour effect of 5-azacytidine by inducing a breakdown of resistance-mediating factors and epigenetic modulation. Gut. 2011;60:156–165. doi: 10.1136/gut.2010.208041. [DOI] [PubMed] [Google Scholar]
- 237.Stewart DJ, Issa JP, Kurzrock R, Nunez MI, Jelinek J, Hong D, Oki Y, Guo Z, Gupta S, Wistuba II. Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res. 2009;15:3881–3888. doi: 10.1158/1078-0432.CCR-08-2196. [DOI] [PubMed] [Google Scholar]
- 238.Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Yeo W, Chung HC, Chan SL, Wang LZ, Lim R, Picus J, Boyer M, Mo FK, Koh J, Rha SY, et al. Epigenetic therapy using belinostat for patients with unresectable hepatocellular carcinoma: a multicenter phase I/II study with biomarker and pharmacokinetic analysis of tumors from patients in the Mayo Phase II Consortium and the Cancer Therapeutics Research Group. J Clin Oncol. 2012;30:3361–3367. doi: 10.1200/JCO.2011.41.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]


