Abstract
Free-living amoebae (FLA) include opportunistic pathogens such as Naegleria fowleri, Balamuthia mandrillaris, and the genera Sappinia and Acanthamoeba. In this study, a survey was conducted in order to evaluate the presence of potentially pathogenic amoebic strains in water samples collected from wells located in the western part of Guinea-Bissau. The samples were left to precipitate for 48 hours and then the sediments were seeded on non-nutrient agar plates containing Escherichia coli spread and cultures were checked daily for the presence of FLA. Identification of FLA strains was based on the morphological and polymerase chain reaction (PCR) using the 18S rDNA or 16S mitochondrial rDNA genes in the case of Naegleria and Balamuthia genera, respectively. In the case of positive samples of Acanthamoeba, strains were further classified at the genotype level by sequencing the diagnostic fragment 3 (DF3) region located in the 18S rDNA gene as previously described. Sappinia sp. was not isolated during the study and thus, no molecular analysis was performed for this genus. The obtained results revealed the presence of Acanthamoeba (genotypes T3 and T4), Naegleria fowleri, and Balamuthia mandrillaris. To the best of our knowledge, this is the first report demonstrating the presence of FLA in water bodies from Guinea-Bissau and the first report on the isolation of Balamuthia mandrillaris from environmental sources in Africa.
Keywords: Acanthamoeba, Balamuthia mandrillaris, Naegleria, Well water, Genotype, Guinea-Bissau
Introduction
Free-living amoebae (FLA) are widely distributed protozoa in the environment that are able to survive in different habitats such as water, soil, dust and air sources.1,2 Among FLA, some species are causative agents of disease in humans and other animals, such as Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea.1,3 Specifically, N. fowleri, B. mandrillaris, and S. diploidea have been reported as causative agents of encephalitis. Moreover, Acanthamoeba pathogenic strains are the causative agents of a multifocal encephalitis called granulomatous amebic encephalitis, a chronic central nervous system disease of immunocompromised hosts, and various other system disease states, including Acanthamoeba keratitis (AK) and pneumonitis.2 Most studies have shown that 90% of Acanthamoeba isolates that produce infections belong to the T4 genotype, although other genotypes have been reported as causative agents of amoebic infections in humans and other animals, such as T1, T3, T5, T10, T11, T15, T17, and T18.2,4
The presence of FLA in water bodies may represent a health risk for both immunocompromised and immunocompetent individuals. Moreover, FLA are highly resistant to extreme conditions of temperature, pH, and exposure to chemicals. In addition to their pathogenicity, FLA serve as hosts for a large number of pathogenic bacteria and viruses of clinical relevance for humans and other animals.2
It is important to mention that there is a lack of studies in the African continent about the distribution of FLA in environmental sources, such as soil and water bodies. Nevertheless, studies on the presence of potentially pathogenic genotypes of Acanthamoeba, as well as Naegleria fowleri, in environmental sources has been reported by other authors in this continent (including countries such as Nigeria, Tunisia, Benin, and Egypt among others).5–9 Moreover, only a few clinical cases of amoebic encephalitis have been reported in Nigeria (N. fowleri),10–12 South Africa (N. fowleri),13 and Senegal (Acanthamoeba).14 Furthermore, up to five cases of AK have been informed in northern Africa (Tunisia).15,16 Up to date, Sappinia spp. or B. mandrillaris have not been reported in Africa. Nevertheless, the presence of B. mandrillaris in this continent is suspected since immunoreactivity towards this species in individuals from Ivory Coast was recently reported.17
Therefore, the aim of this study was to determine the presence of potentially pathogenic FLA in water samples from Guinea-Bissau. To the best of our knowledge, this is the first study of Acanthamoeba strains at the genotype level in this country and the first report on the isolation of Balamuthia mandrillaris from environmental sources in Africa.
Materials and Methods
Sample sites and culture of FLA
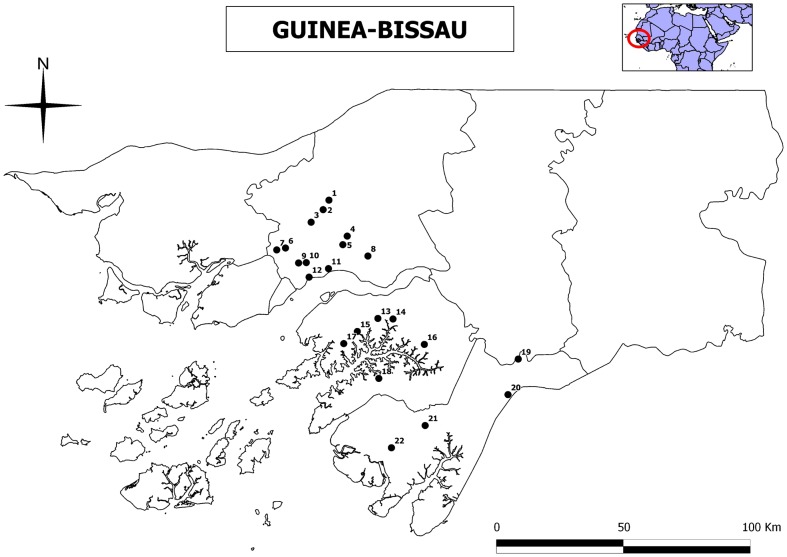
Twenty-two water samples were collected from wells during the wet season of 2011 at rural areas from three regions of the western part of Guinea-Bissau (Fig. 1). The two southern regions (Quínara and Tombalí) are undisturbed forest areas, whereas the northern one (Oio) has slightly human impacts by extensive agricultural and livestock uses. Water samples were collected from the wells most frequently used by the local population in 22 tabankas (small traditional villages). Average depth of wells was always less than 20 m. Values of pH, conductivity (µS/cm), and total hardness (mg/l CaCO3) of the collected water samples were also determined at collection. Conductivity and pH were measured in situ with a HANNA Model HI9829 portable conductivity/pH meter. Total hardness was determined with a Lovibond PC MultiDirect spectrophotometer. Three water replicates of 1 l were collected in each well. Samples were left 48 hours at room temperature so that precipitation could occur. After that, the sediment was carefully removed and transferred to 50 ml sterile polypropylene tubes. After that, 150 µl of each sediment sample were seeded onto 2% non-nutrient agar plates with heat killed E. coli and incubated at 25°C. The cultures were monitored every 24 hours, for up to 7 days. Positive isolates for FLA (identified under the inverted microscope) were cloned by dilution and transferred to new culture plates for further molecular analyses as previously described.7 In the case of positive samples for Acanthamoeba, amoebae were transferred into axenic cultures by placing them in PYG medium [0.75% Proteose peptone (w/v), 0.75% yeast extract (w/v), and 1.5% glucose (w/v)]. A type strain of Acanthamoeba (Acanthamoeba castellanii Neff ATCC 30010) from the American Type Culture Collection (ATCC) was grown without shaking under the same conditions. Other genera of FLA were not axenified.
Figure 1.
Sampling points surveyed during 2011 in three regions of Guinea-Bissau. Oio Region: 1, Culcunhe; 2, Watine; 3, Ufle; 4, Braia; 5, Clack N’djassé; 6, Bera; 7, Palche Iala; 8, Wedequeia; 9, Saw; 10, Djugudul Com; 11, Thugue; 12, Oco Grande. Quínara Region: 13, Ganjetra; 14, Fulacunda; 15, Brandao; 16, Madina Hatche; 17, Nova Sintra; 18, Empada. Tombalí Region: 19, Madina Contabane; 20, Afia Bunhio; 21, Botchi Minde; 22, Mato Forroba.
DNA extraction
DNA from cultures identified as positive for FLA by microscopy was extracted by placing 1–2 ml of amoebic cultures directly into the Maxwell® 16 Tissue DNA Purification Kit sample cartridge (Promega, Madrid, Spain). Amoebic genomic DNA was purified using the Maxwell 16 Instrument as described in the Maxwell 16 DNA Purification Kits Technical Manual #TM284 (Promega). DNA yield and purity were determined using the NanoDrop® 1000 spectrophotometer (Fisher Scientific, Madrid, Spain).
Polymerase chain reaction (PCR) reactions and molecular characterization of FLA isolates
For the identification and genotyping of Acanthamoeba isolates, the diagnostic fragment 3 (DF3) region of Acanthamoeba 18S rDNA gene was amplified as previously described using the JDP1/JDP2 primer pair.18,19 This PCR amplifies a 495 bp fragment of the 18S rDNA gene that allows genotype identification after sequencing.
For Vahlkampfiidae of the genus Naegleria, the complete ITS region (ITS1, 5.8S, and ITS2) was amplified using the NFITSFW and NFITSRV primer pair for N. fowleri and the ITSFW and ITSRV primers for other Naegleria species as previously described,20,21 which amplify a fragment of around 300–400 bp depending on the species of Naegleria. In the case of other Vahlkamphfiidae, the JITSFW/JITSRV primer pair was used as previously described.21 Moreover, the 18S rDNA gene of these FLA was also amplified using the universal eukaryotic P2 and P3r primer pair, which amplifies a fragment of around 500 bp in FLA and allows identification of FLA at the genus level after sequencing.22 Naegleria canariensis CCAP 1518/27 DNA was used a positive control in the PCR reactions.
For the identification of Balamuthia mandrillaris, the 16S rDNA gene was amplified and sequenced as previously described,19 using the Balspec16S primer pair which amplifies a product of 1075 bp. Balamuthia mandrillaris CDC:V416 strain (ATCC®PRA290TM) DNA was used as a positive control.
The obtained PCR products were purified using the Qiaquick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using a MEGABACE 1000 automatic sequencer (Healthcare Biosciences, Barcelona, Spain) in the University of La Laguna Sequencing Service (Servicio de Secuenciación SEGAI, University of La Laguna). Sequences were obtained twice from both strands. The obtained sequences were aligned using Mega 5.0 software program.23 Moreover, nucleotide similarity search was performed by BLAST search (Basic Local Alignment search tool) of the sequenced amplicons against amoeba species.
Genotype identification of Acanthamoeba was carried out based on sequence analysis of DF3 region as previously described by comparison to the available Acanthamoeba DNA sequences in Genbank database.18,19 Acanthamoeba castellanii Neff ATCC 30010 (genotype T4) DNA was used as a positive control in the PCR reactions. Phylogenetic analyses for Acanthamoeba strains was carried out using maximum parsimony, minimum evolution and maximum likelihood optimality criteria, implemented in Mega 5.0.23 Transition/transversion ratios were estimated by maximum likelihood heuristic searches. Estimates of node support were obtained by performing 1000 bootstrap replicates. Obtained sequences were compared to sequences available in GenBank database.
The sequences for the new FLA isolates are deposited in the Genbank database under the accession numbers KJ000391–KJ000399 and are shown in Table 1.
Table 1. Free-living amoebae isolated in the analysed 22 water samples from Guinea-Bissau (Fig. 1). Values of pH, conductivity (μS/cm), and hardness (mg/l), and Genbank accession numbers for the new FLA strains are also shown.
| Sampling point | Location coordinates | Isolated amoebae | pH | Conductivity | Hardness | Accession # | |
| (1) Culcunhe | N12°14′36.3″ | W15°25′30.0″ | Negative | 4.08 | 346 | 190 | |
| (2) Watine | N12°12′21.7″ | W15°26′52.0″ | Naegleria fowleri, Tetramitus sp. | 4.06 | 275 | 170 | KJ000395 |
| (3) Ufle | N12°09′23.3″ | W15°29′42.1″ | Negative | 4.15 | 355 | 90 | |
| (4) Braia | N12°06′06.4″ | W15°21′10.5″ | Negative | 2.20 | 43 | 90 | |
| (5) Clack N’djassé | N12°04′05.0″ | W15°22′12.9″ | Acanthamoeba sp. T4 (GBW-1), Willaertia sp. | 2.25 | 392 | 230 | KJ000391 |
| (6) Bera | N12°03′17.4″ | W15°35′45.7″ | Willaertia sp. | 4.17 | 306 | 200 | |
| (7) Palche Iala | N12°02′50.7″ | W15°37′50.0″ | Naegleria fowleri, Tetramitus sp. | 4.70 | 213 | 90 | KJ000396 |
| (8) Wedequeia | N12°01'23.9″ | W15°16'17.3″ | Negative | 2.23 | … | 350 | |
| (9) Saw | N11°59′45.3″ | W15°32′40.0″ | Willaertia sp. | 2.14 | 102 | 80 | |
| (10) Djugudul Com | N11°59′50.8″ | W15°30′51.1″ | Negative | 2.12 | 197 | 110 | |
| (11) Thugue | N11°58′ 25.1″ | W15°25′36.8″ | Tetramitus sp. | 2.16 | 216 | 160 | |
| (12) Oco Grande | N11°56′24.4″ | W15°30′11.2″ | Tetramitus sp. | 2.22 | 524 | 300 | |
| (13) Ganjetra | N11°46′38.7″ | W15°13′56.0″ | Willaertia sp. | 6,00 | 44 | 15 | |
| (14) Fulacunda | N11°46′30.9″ | W15°10′21.9″ | Naegleria sp., Willaertia sp. | 5.10 | 328 | 96 | |
| (15) Brandao | N11°43′34.0″ | W15°18′47.5″ | Negative | 5.10 | 78 | 18 | |
| (16) Madina Hatche | N11°40′30.4″ | W15°02′57.3″ | Acanthamoeba sp. T3 (GBW-4), Willaertia sp | 5.20 | 49 | 14 | KJ000394 |
| (17) Nova Sintra | N11°40′41.8″ | W15°21′59.9″ | Balamuthia mandrillaris | 5.50 | 100 | 42 | KJ000399 |
| (18) Empada | N11°32′27.9″ | W15°13′43.2″ | Naegleria fowleri, Tetramitus sp | 5.10 | 71 | 7 | KJ000397 |
| (19) Madina Contabane | N11°37′02.6″ | W14°40′45.8″ | Acanthamoeba sp. T4 (GBW-2), Willaertia sp | 5.50 | 111 | 10 | KJ000392 |
| (20) Afia Bunhio | N11°28′38.2″ | W14°43′11.8″ | Acanthamoeba sp. T4 (GBW-3) | 6.10 | 241 | 150 | KJ000393 |
| (21) Botchi Minde | N11°17′05.6″ | W15°02′23.6″ | Negative | 5.10 | 283 | 10 | |
| (22) Mato Forroba | N11°16′04.7″ | W15°10′43.8″ | Naegleria fowleri, Tetramitus sp. | 5.50 | 100 | 39 | KJ000398 |
Results and Discussion
Regarding the collected water samples, the mineral content was highly variable within the studied area, with pH values ranging from 2.12 to 6.00, and conductivity values varying between 43 and 524 μS/cm. Furthermore, total hardness varied between 7 and 350 mg/l (Table 1). However, no correlation could be found between the concentration of the different parameters and the presence of potentially pathogenic FLA. Mainly because of the high resistance of FLA against harsh environmental conditions as it has been reported before.2
FLA were isolated in 15 of the 22 samples (68.2%) included in this study (Table 1). However, Sappinia spp. was not identified within these samples. Members of the Vahlkampfidae family were isolated in 13 of the 15 positive samples (86.6%). However, most of the isolated strains belonged to Willaertia or Tetramitus genera (10 samples), which are non-pathogenic FLA and are widely distributed worldwide in water related habitats. Moreover, Naegleria genus was also observed in five of the 13 samples that were Vahlkampfidae-positive. Interestingly, sequencing of the FLA 18S rDNA gene and ITS region revealed the presence of Naegleria fowleri in four of the five positive samples for the genus Naegleria (98–99% homology with other N. fowleri strains available in Genbank). Previous studies have reported the isolation of N. fowleri from dust and soil from Nigeria5,12 and water bodies in Egypt.6 Interestingly, human cases of encephalitis due to N. fowleri have been reported in Nigeria (2)10,11 and South Africa (1).13 In all the reported cases, water and/or dust sources were suspected to be the source of infection.
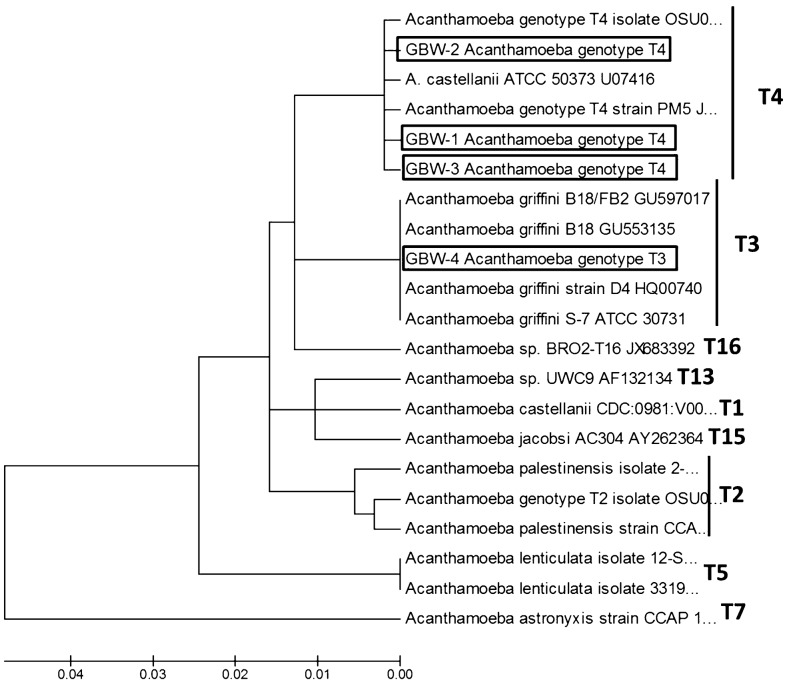
Regarding Acanthamoeba genus, four of the 15 positive samples (27.6%) were positive for Acanthamoeba strains belonging to genotype T3 (one sample, 25%) and T4 (three samples, 75%) (Fig. 2). Previous environmental studies in different countries in Africa have reported the presence of potentially pathogenic Acanthamoeba strains mainly in water sources.6–9 Furthermore, only three clinical related studies regarding Acanthamoeba genus have been reported in Africa: one case of encephalitis due to Acanthamoeba in Dakar, Senegal,14 an epidemiological study to determine the presence of FLA in the nasal passages of a population in Zaria, Nigeria24 and the report of three cases of AK in patients from Tunisia.16 Another case of encephalitis due to a hartmannellid amoeba was reported in Zambia;25 however, it is possible that the identification of this strain was not carried out correctly.
Figure 2.
18S rDNA DF3 linearized neighbour-joining tree obtained by using the Kimura two-parameter distance algorithm, produced in MEGA 5.0. The isolates obtained in the present study are identified in the tree (boxes). The type sequences were taken from GenBank and are presented under the following numbers: A. astronyxis strain CCAP 1534/1 #AF239293, A. castellanii strain CDC:0981:V006 Accession #U07400, A. castellanii ATCC 50373 Accession # U07416, A. griffini S-7 ATCC 30731 Accession #U07412, A. griffini B-18 FB2 Acession #GU597017, A. griffini B-18 Acession #GU553135, A. griffini strain D4 Acession #HQ00740, A. lenticulata isolate 12-SO #KC694184, A. lenticulata isolate 33195463 Accession #KC438381, Acanthamoeba sp. isolate OSU09-002 Accession # JQ669657, Acanthamoeba sp. genotype T2 Isolate OSU09-006 #JQ669661, Acanthamoeba palestinensis isolate TW-2 Accession #KC694193, A. palestinensis strain CCAP 1547-1 #AF239296, Acanthamoeba sp. isolate BRO2-T16 Accession #JX683392, Acanthamoeba sp. UWC9 Accession #AF132134, Acanthamoeba sp. PM5 Accession #JX494395, Acanthamoeba jacobsi AC304 Accession #AY262364.
Finally, one sample was positive for B. mandrillaris. Interestingly, the region where this water sample was collected is often flooded by high tides during the rainy season and showed high values of salinity. Nevertheless, and to the best of our knowledge, this is the first report of B. mandrillaris isolation from the environment in the African continent. Although this study may be the first to identify B. mandrillaris from the environment in Africa, it should be noted that its presence on the continent was suspected because of the prevalence of immunoreactivity towards this species in individuals in Ivory Coast (Western Africa).17 Despite all the recent advances in the study of B. mandrillaris, several questions about this organism remain unanswered. Although the number of infections due to B. mandrillaris is relatively low, the estimated frequency of reported cases so far is likely to be underestimated.26 However, even in the present study, environmental isolation of B. mandrillaris seems to be a low frequency occurrence. In addition to this study, researchers have only been able to isolate B. mandrillaris from the environment on five occasions. The development of improved culture techniques to allow a fast, reliable, and repetitive approach to isolate and grown these amoebae in axenic conditions is urgently required.
Nevertheless, the present study reports the presence of potentially pathogenic FLA strains of Acanthamoeba genus and Naegleria fowleri and Balamuthia mandrillaris species in water sources from Guinea-Bissau. The isolation of these potential pathogens from the studied water sources in this country should raise awareness within clinicians and public health professionals. Moreover, knowledge of the prevalence of FLA in this country may help clinicians to diagnose and treat either healthy or immunocompromised individuals. Presence of potentially pathogenic FLA in the studied wells should lead to take precautions in order to avoid potential health risks to individuals inhabiting these areas.
Disclaimer Statements
Contributors RB, JGE and GGN collected the water samples, worked on the initial isolation of amoebae and drafted the manuscript. JLM, MRB, CMMN an ALA worked on the initial isolation of amoebae, morphological and molecular characterization and drafted the manuscript. BV, EMC and JEP participated on the amoebae characterization and drafted parts of the manuscript.
Funding This work was supported by the grants RICET (project no. RD12/0018/0012 of the programme of Redes Temáticas de Investigación Cooperativa, FIS), Spanish Ministry of Health, Madrid, Spain, and the Project FIS PI10/01298 ‘Protozoosis emergentes por amebas de vida libre: aislamiento y caracterización molecular, identificación de cepas transportadoras de otros agentes patógenos y búsqueda de quimioterapias efectivas’, PI13/00490 ‘Protozoosis Emergentes por Amebas de Vida Libre: Aislamiento, Caracterización, Nuevas Aproximaciones Terapéuticas y Traslación Clínica de los Resultados’ from the Instituto de Salud Carlos III, and Project ref. AGUA3 ‘Amebas de Vida Libre como Marcadores de Calidad del Agua’ from CajaCanarias Fundación.
ALA was funded by a grant ‘Ayudas del Programa de Formación de Personal Investigador, para la realización de Tesis Doctorales’ from the Agencia Canaria de Investigación, Innovación y Sociedad de la Información from the Canary Islands Government. MRB was funded by CEI Canarias, Campus Atlántico Internacional, and Becas Fundación Cajacanarias para Postgraduados 2014. RABN and GGN were supported by ‘II y III Convocatoria de Ayudas en Cooperación para el Desarrollo para profesores’ and ‘V Programa de Prácticas y Proyectos de Cooperación al Desarrollo de la UCLM’, of University of Castilla-La Mancha. JLM was supported by the Ramón y Cajal Subprogramme from the Spanish Ministry of Economy and Competitivity RYC-2011-08863.
Conflicts of interest The authors declare no conflicts of interest.
Ethics approval No applicable.
References
- 1.Siddiqui R, Khan NA. Biology and pathogenesis of Acanthamoeba. Parasit Vectors. 2012;5:6. doi: 10.1186/1756-3305-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lorenzo-Morales J, Martín-Navarro CM, López-Arencibia A, Arnalich-Montiel F, Piñero JE, Valladares B. Acanthamoeba keratitis: an emerging disease gathering importance worldwide? Trends Parasitol. 2013;29:181–7. doi: 10.1016/j.pt.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Visvesvara GS, Moura H, Schuster FL. Pathogenic and opportunistic free-living amoebae: Acanthamoeba spp., Balamuthia mandrillaris, Naegleria fowleri, and Sappinia diploidea. FEMS Immunol Med Microbiol. 2007;50:1–26. doi: 10.1111/j.1574-695X.2007.00232.x. [DOI] [PubMed] [Google Scholar]
- 4.Qvarnstrom Y, Nerad TA, Visvesvara GS. Characterization of a new pathogenic Acanthamoeba species, A. byersi n. sp., isolated from a human with fatal amoebic encephalitis. J Eukaryot Microbiol. 2013;60:626–33. doi: 10.1111/jeu.12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lawande RV. Recovery of soil amoebae from the air during the harmattan in Zaria, Nigeria. Ann Trop Med Parasitol. 1983;77(1):45–9. doi: 10.1080/00034983.1983.11811671. [DOI] [PubMed] [Google Scholar]
- 6.Sadaka HA, el-Nassery SF, abou Samra LM, Awadalla HN. Isolation and identification of free-living amoebae from some water sources in Alexandria. J Egypt Soc Parasitol. 1994;2:247–57. [PubMed] [Google Scholar]
- 7.Lorenzo-Morales J, Ortega-Rivas A, Martínez E, Khoubbane M, Artigas P, Periago MV, et al. Acanthamoeba isolates belonging to T1, T2, T3, T4 and T7 genotypes from environmental freshwater samples in the Nile Delta region, Egypt. Acta Trop. 2006;100:63–9. doi: 10.1016/j.actatropica.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 8.Eddyani M, de Jonckheere JF, Durnez L, Suykerbuyk P, Leirs H, Portaels F. Occurrence of free-living amoebae in communities of low and high endemicity for Buruli ulcer in southern Benin. Appl Environ Microbiol. 2008;74(21):6547–53. doi: 10.1128/AEM.01066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trabelsi H, Sellami A, Dendena F, Sellami H, Cheikh-Rouhou F, Makni F, et al. Free-living Amoebae (FLA): morphological and molecular identification of Acanthamoeba in dental unit water. Parasite. 2010;1:67–70. doi: 10.1051/parasite/2010171067. [DOI] [PubMed] [Google Scholar]
- 10.Lawande RV, John I, Dobbs RH, Egler LJ. A case of primary amebic meningoencephalitis in Zaria, Nigeria. Am J Clin Pathol. 1979;71(5):591–4. doi: 10.1093/ajcp/71.5.591. [DOI] [PubMed] [Google Scholar]
- 11.Lawande RV, Macfarlane JT, Weir WR, Awunor-Renner C. A case of primary amebic meningoencephalitis in a Nigerian farmer. Am J Trop Med Hyg. 1980;29(1):21–5. doi: 10.4269/ajtmh.1980.29.21. [DOI] [PubMed] [Google Scholar]
- 12.Ugonabo JA, Gugnani HC. Nasal carriage of Naegleria fowleri and its environmental occurrence in Borno State, Nigeria. J Commun Dis. 1989;21(2):111–3. [PubMed] [Google Scholar]
- 13.Schoeman CJ, van der Vyver AE, Visvesvara GS. Primary amoebic meningoencephalitis in southern Africa. J Infect. 1993;26:211–4. doi: 10.1016/0163-4453(93)93085-i. [DOI] [PubMed] [Google Scholar]
- 14.Ndiaye M, Diop AG, Dieng Y, Seydi M, Diouf FS, Diop BM, et al. A case of meningoencephalitis caused by Acanthamoeba sp. in Dakar. Med Trop (Mars). 2005;1:67–8. [PubMed] [Google Scholar]
- 15.Ben Salah S, Makni F, Cheikrouhou F, Ben Zina Z, Mlik M, Feki J, et al. Acanthamoeba keratitis: about the first two Tunisian cases. Bull Soc Pathol Exot. 2007;100(1):41–2. [PubMed] [Google Scholar]
- 16.Fathallah A, Ben Rayana N, Knani L, Meksi SG, Saghrouni F, Ghorbel M, et al. Acanthamoeba keratitis. Report of 3 cases diagnosed in central Tunisia. Tunis Med. 2010;88(2):111–5. [PubMed] [Google Scholar]
- 17.Kiderlen AF, Radam E, Schuster FL, Adjogoua EV, Akoua-Koffi C, Leendertz FH. Balamuthia and Acanthamoeba-binding antibodies in West African human sera. Exp Parasitol. 2010;126(1):28–32. doi: 10.1016/j.exppara.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 18.Booton GC, Visvesvara GS, Byers TJ, Kelly DJ, Fuerst PA. Identification and distribution of Acanthamoeba species genotypes associated with nonkeratitis infections. J Clin Microbiol. 2005;43:1689–93. doi: 10.1128/JCM.43.4.1689-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niyyati M, Lorenzo-Morales J, Rezaie S, Rahimi F, Mohebali M, Maghsood AH, et al. Genotyping of Acanthamoeba isolates from clinical and environmental specimens in Iran. Exp Parasitol. 2009;121:242–5. doi: 10.1016/j.exppara.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 20.de Jonckheere JF, Brown S. The identification of vahlkampfiid amoebae by ITS sequencing. Protist. 2005;1:89–96. doi: 10.1016/j.protis.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 21.Moussa M, de Jonckheere JF, Guerlotté J, Richard V, Bastaraud A, Romana M, et al. Survey of Naegleria fowleri in geothermal recreational waters of Guadeloupe (French West Indies). PLoS One. 2013;8(1):e54414. doi: 10.1371/journal.pone.0054414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mulec J, Vaupotič J, Walochnik J. Prokaryotic and eukaryotic airborne microorganisms as tracers of microclimatic changes in the underground (Postojna Cave, Slovenia). Microb Ecol. 2012;64(3):654–67. doi: 10.1007/s00248-012-0059-1. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abraham SN, Lawande RV. Incidence of free-living amoebae in the nasal passages of local population in Zaria, Nigeria. J Trop Med Hyg. 1982;5:217–22. [PubMed] [Google Scholar]
- 25.Bhagwandeen SB, Carter RF, Naik KG, Levitt D. A case of hartmannellid amebic meningoencephalitis in Zambia. Am J Clin Pathol. 1975;4:483–92. doi: 10.1093/ajcp/63.4.483. [DOI] [PubMed] [Google Scholar]
- 26.Lorenzo-Morales J, Cabello-Vílchez AM, Martín-Navarro CM, Martínez-Carretero E, Piñero JE, Valladares B. Is Balamuthia mandrillaris a public health concern worldwide? Trends Parasitol. 2013;29(10):483–8. doi: 10.1016/j.pt.2013.07.009. [DOI] [PubMed] [Google Scholar]