Abstract
Bacterioplankton of the SAR11 clade are the most abundant microorganisms in marine systems, usually representing 25% or more of the total bacterial cells in seawater worldwide. SAR11 is divided into subclades with distinct spatiotemporal distributions (ecotypes), some of which appear to be specific to deep water. Here we examine the genomic basis for deep ocean distribution of one SAR11 bathytype (depth-specific ecotype), subclade Ic. Four single-cell Ic genomes, with estimated completeness of 55%–86%, were isolated from 770 m at station ALOHA and compared with eight SAR11 surface genomes and metagenomic datasets. Subclade Ic genomes dominated metagenomic fragment recruitment below the euphotic zone. They had similar COG distributions, high local synteny and shared a large number (69%) of orthologous clusters with SAR11 surface genomes, yet were distinct at the 16S rRNA gene and amino-acid level, and formed a separate, monophyletic group in phylogenetic trees. Subclade Ic genomes were enriched in genes associated with membrane/cell wall/envelope biosynthesis and showed evidence of unique phage defenses. The majority of subclade Ic-specfic genes were hypothetical, and some were highly abundant in deep ocean metagenomic data, potentially masking mechanisms for niche differentiation. However, the evidence suggests these organisms have a similar metabolism to their surface counterparts, and that subclade Ic adaptations to the deep ocean do not involve large variations in gene content, but rather more subtle differences previously observed deep ocean genomic data, like preferential amino-acid substitutions, larger coding regions among SAR11 clade orthologs, larger intergenic regions and larger estimated average genome size.
Keywords: bathytype, ecotype, metagenomics, SAR11, single-cell genomics, deep ocean
Introduction
Characterized by darkness, average temperatures of approximately 2–4 °C, increased hydrostatic pressure and general oligotrophy, the relatively extreme environment of the deep ocean is also the largest biome on the Earth. The mesopelagic (200–1000 m) and bathypelagic (1000–4000 m) zones contain >70% of marine microbial biomass (Arıstegui et al., 2009) and these organisms have vital roles in global cycling of carbon, nitrogen and other biogeochemical processes (Nagata et al., 2010; Robinson et al., 2010). In addition to microorganisms necessarily being adapted to cold and increased pressure there, the deep sea also contains more recalcitrant forms of carbon than at the surface (Arıstegui et al., 2009; Nagata et al., 2010; Robinson et al., 2010). Cultivated isolates have revealed some microbial adaptations associated with life at depth, including increased intergenic spacer regions, rRNA gene indels and higher abundances of membrane polyunsaturated fatty acids and surface-adhesion/motility genes (Simonato et al., 2006; Lauro and Bartlett, 2008; Wang et al., 2008; Nagata et al., 2010).
However, many of the most abundant bacterial groups from the deep ocean remain uncultivated, for example, the SAR202, SAR324 and SAR406 clades, which make up significant fractions of microbial communities at depth (Giovannoni et al., 1996; Gordon and Giovannoni, 1996; Wright et al., 1997; DeLong et al., 2006; Morris et al., 2006; Varela et al., 2008; Schattenhofer et al., 2009; Treusch et al., 2009; Morris et al., 2012). Thus, it remains uncertain how widespread the known adaptations of cultivated isolates are among deep ocean microorganisms. Metagenomic analyses have provided evidence for common genomic features in the deep ocean, such as increased proliferation of transposable elements and phage, amino-acid content changes, and increased average genome size (DeLong et al., 2006; Konstantinidis et al., 2009). Single-cell genomic analyses provide another powerful means to understand the metabolism and evolution of organisms eluding cultivation-based techniques (Stepanauskas, 2012; Blainey, 2013; Lasken, 2013; Rinke et al., 2013). This approach provided the first insight into the metabolism of several of these deep ocean clades, including SAR324, Arctic96BD-19 and Agg47, and made the important discovery that at least some of these organisms are capable of chemoautotrophy (Swan et al., 2011). The findings from single-cell genomics are consistent with widespread autotrophy genes in other dominant deep ocean microorganisms, such as the Thaumarchaea (Karner et al., 2001; Pester et al., 2011), and direct measurements of high levels of carbon fixation in the meso- and bathypelagic zones (Reinthaler et al., 2010).
Another abundant group of microorganisms that populates the deep ocean is SAR11. Bacterioplankton of the SAR11 clade are the most numerous in marine systems, typically comprising ∼25% of all prokaryotic cells (Morris et al., 2002; Schattenhofer et al., 2009). Although the majority of research has focused on the SAR11 clade in the euphotic and upper mesopelagic zones, multiple studies have demonstrated evidence of substantial SAR11 populations deeper in the mesopelagic, as well as in the bathy- and even hadopelagic (>6000 m) realms (Martin-Cuadrado et al., 2007; Konstantinidis et al., 2009; Schattenhofer et al., 2009; Quaiser et al., 2010; Swan et al., 2011; Eloe et al., 2011a, 2011b; King et al., 2013).
SAR11, or the ‘Pelagibacterales,' is a diverse group, spanning at least 18% 16S rRNA gene divergence, and is comprised of subclades with unique spatiotemporal distributions (ecotypes) that follow seasonal patterns (Field et al., 1997; Carlson et al., 2009; Giovannoni and Vergin, 2012; Grote et al., 2012; Vergin et al., 2013). All genome-sequenced representatives are characterized by small (1.3-1.4 Mbp), streamlined genomes with low GC content, few gene duplications and an obligately aerobic, heterotrophic metabolism generally focused on oxidation of low-molecular-weight carbon compounds, such as carboxylic and amino acids, osmolytes and methylated compounds (Schwalbach et al., 2010; Yilmaz et al., 2011; Carini et al., 2012; Grote et al., 2012). Representatives spanning the known subclade diversity have an unusually high level of core genome conservation and gene synteny, however, some subclade-specific genomic features have been identified (Grote et al., 2012). The subclade V representative, HIMB59, encodes a complete glycolysis pathway and a variety of predicted sugar transporters. As subclade V organisms bloom at the surface concurrently with the more numerically dominant subclade Ia ecotype (Vergin et al., 2013), genetic machinery for the oxidation of sugars may provide a means of niche differentiation.
A recent study has pointed toward a deep SAR11 bathytype (depth-specific ecotype (Lauro and Bartlett, 2008)), phylogenetically distinct from the currently cultivated strains. This ‘subclade Ic' was represented by a single 16S clone library sequence that preferentially recruited pyrosequencing reads from depths of 200 m and below at the Bermuda Atlantic Time-series Study site (BATS; Vergin et al., 2013), and formed a monophyletic group with 16S sequences from single-cell genomes collected at 770 m at Station ALOHA. Here we present a comparative analysis of subclade Ic utilizing four single-amplified genomes (SAGs), metagenomes from euphotic, meso-, bathy- and hadopelagic samples and eight pure-culture SAR11 genomes from three surface subclades. We tested the hypothesis that the subclade Ic genomes would have features that distinguish this bathytype from surface organisms to yield a better understanding of SAR11 adaptations to the ocean interior and of the genomic basis for SAR11 subclade differentiation by depth.
Materials and methods
Comparative genomics
Single-cell separation, multiple displacement amplification (MDA), quality control and SAG selection for sequencing based on MDA kinetics were all carried out as described previously (Swan et al., 2011). More detailed descriptions are available in Supplementary Methods. Sequencing and assembly of the SAGs were carried out by the DOE Joint Genome Institute as part of a Community Sequencing Program grant 2011-387. The SAG Whole Genome Shotgun projects have been deposited at DDBJ/EMBL/GenBank under the accession numbers: AZHR00000000 (AAA240-E13), AZHQ00000000 (AAA288-E13), AZYB00000000 (AAA288-G21) and AZYC00000000 (AAA288-N07). The versions described in this paper are versions AZHR01000000 (AAA240-E13), AZHQ01000000 (AAA288-E13), AZYB01000000 (AAA288-G21) and AZYC01000000 (AAA288-N07). Genome annotations can be accessed using the Integrated Microbial Genome (IMG) database (http://img.jgi.doe.gov).
SAG gene orthology with other SAR11 genomes was completed using the Hal pipeline (Robbertse et al., 2011) and a series of custom filters, described in detail in Supplementary Methods. Post assembly quality control was assisted by examination of gene conservation across SAR11 strains. SAG genome completion was evaluated based on 599 single-copy genes present in all eight pure culture SAR11 genomes. Overall, SAG genome completion percentage was based on the percentage of these orthologs found in the SAGs (Supplementary Table S1). Average amino-acid identity and local synteny between genomes were calculated with the scripts/methods of Yelton et al. (2011). Pairwise 16S rRNA gene identity was calculated with megablast using default settings. COG distribution among SAR11 genomes was part of data supplied by IMG (Supplementary Table S1). Patterns of amino-acid substitution between surface and deep-water strains of SAR11 were analyzed as described in Konstantinidis et al. (2009). Fold-change abundance of amino acids across similar and non-similar substitutions was calculated from all vs all BLASTP output within homologous clusters. Intergenic spacer regions were provided as part of the IMG annotation process. Distribution of intergenic regions was examined in R (http://www.R-project.org). Transposable elements were assessed using TBLASTN and the sequences collected by Brian Haas of the Broad Institute for the program TransposonPSI (http://transposonpsi.sourceforge.net). Clustered regularly interspaced short palindromic repeats (CRISPRs) were detected as part of the automated IMG annotation process. A search for cas genes was conducted using 78 hidden Markov models (HMMs) developed by Haft et al. (2005) and Makarova et al. (2011), and hmmsearch (Eddy, 2011) using default settings.
All phylogenetic analyses, with the exception of proteorhodopsin, were completed by aligning sequences with MUSCLE (Edgar, 2004) and computing trees with RAxML (Stamatakis, 2006; Stamatakis et al., 2008). Alignments for trees in Figures 1 and 5 were curated for poorly aligned sites using Gblocks (Castresana, 2000). ProtTest (Abascal et al., 2005) was utilized to optimize amino-acid substitution modeling for protein-coding trees. The concatenated protein phylogeny of the SAR11 clade was completed using the Hal pipeline (Robbertse et al., 2011). The proteorhodopsin tree was computed using the iterative Bayesian alignment/phylogeny program HandAlign (Westesson et al., 2012). Detailed methodology for every tree, unaligned fasta files for each of the single gene trees, and the super alignment and model file for the concatenated protein tree, are provided in the Supplementary Information.
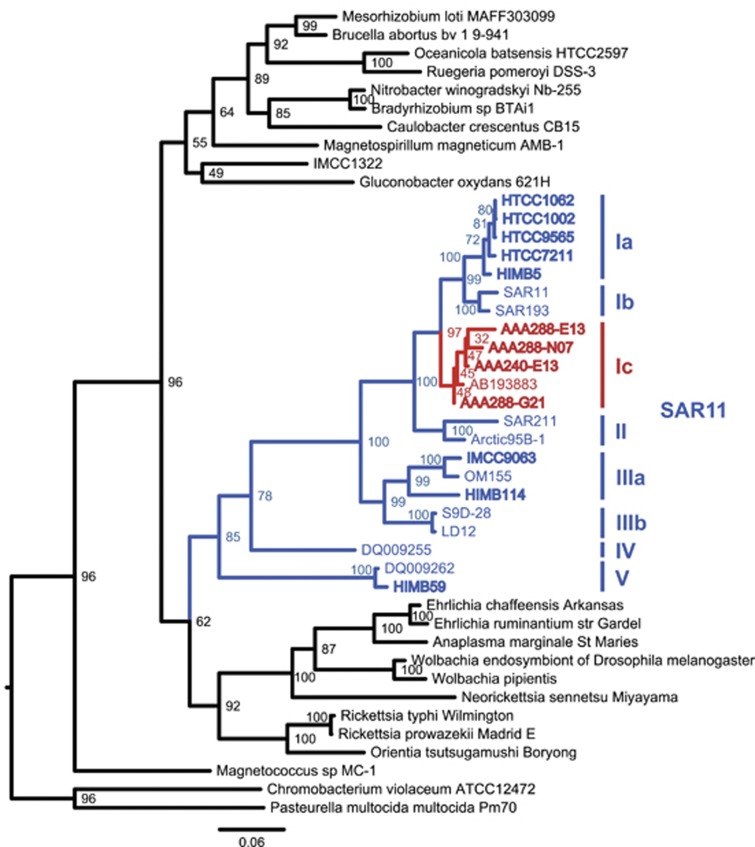
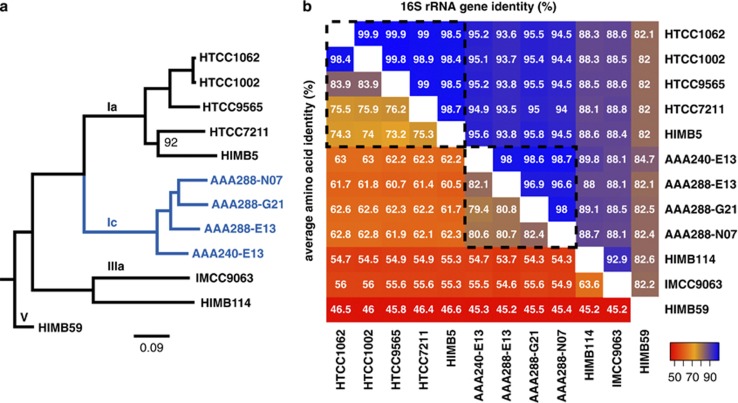
Figure 1.
Maximum-likelihood tree of the 16S rRNA gene for the SAR11 clade in the context of other Alphaproteobacteria. Genome sequenced strains are in bold, with subclade Ic sequences in red and other SAR11 sequences in blue. Bootstrap values (n=1000) are indicated at the nodes; scale bar represents changes per position.
Metagenomics
DNA was extracted from microbial biomass collected from BATS in August 2002 across a depth profile (0, 40, 80, 120, 160, 200 and 250 m) and sequenced using 454 pyrosequencing (GS-FLX, Roche, Basel, Switzerland). Data is available at CAMERA (https://portal.camera.calit2.net) under CAM_PROJ_BATS. Metagenomes from ALOHA were previously described in Shi et al. (2011). Data were also analyzed from 454 metagenomic sequences collected from Eastern Tropical South Pacific Oxygen Minimum Zone (Stewart et al., 2012), the Puerto Rico Trench (Eloe et al., 2011a), the Sea of Marmara (Quaiser et al., 2010) and the Matapan-Vavilov Deep in the Mediterranean Sea (Smedile et al., 2013). All raw data were trimmed of low-quality end sequences using Lucy (Chou and Holmes, 2001) and de-replicated using CDHIT-454 (Fu et al., 2012). Sanger-sequenced reads from 4000 m at ALOHA (Konstantinidis et al., 2009) were also analyzed but not compared with the 454 pyrosequenced reads. GOS (Venter et al., 2004; Rusch et al., 2007; Brown et al., 2012) surface sequences were analyzed for temperature dependence of subclade Ic abundance, but also not included in gene relative abundance normalizations (Supplementary Information).
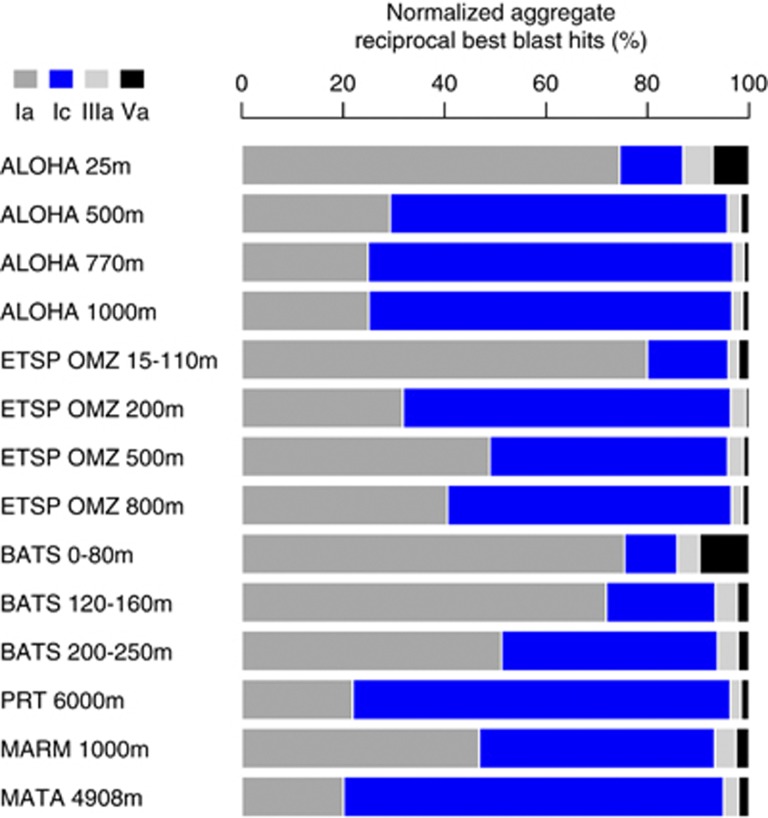
Comparative recruitment of metagenomic sequences was completed using a reciprocal best BLAST (rbb; for example, Wilhelm et al., 2007) of eight SAR11 isolate genomes (HTCC1062, HTCC1002, HTCC9565, HTCC7211, HIMB5, HIMB114, IMCC9063, HIMB59) and the four SAR11 SAGs. Each concatenated SAR11 genome sequence was searched against each metagenome database with BLASTN. All hits to SAR11 genomes were then searched against the entire IMG database (v400), containing the 12 SAR11 genome sequences using BLASTN. The best hits to each genome after this reciprocal blast were then normalized by gene length, the average number of sequences and relative abundance of SAR11 per sample. Taxonomic relative abundance for SAR11 and non-SAR11 organisms was estimated with metagenomic best-blast hits to whole-genome sequences in the IMG v400 database. The results presented in Figure 2 represent an aggregation of all normalized metagenomic recruitment for all genomes in a given subclade, divided by the total number of SAR11 hits in that sample.
Figure 2.
Relative abundance of SAR11 subclades based on reciprocal best blast recruitment of metagenomic sequences.
Gene clusters that may putatively have a role in depth adaptation in subclade 1c were identified as follows: metagenomic samples were classified as ‘surface' (<200 m) or ‘deep' (⩾200 m) and gene cluster abundance in surface and deep samples was determined by reciprocal best-BLAST. The R package DESeq (Anders and Huber, 2010) was used to identify genes that were statistically significantly enriched at depth and at the surface. Detailed workflows for the metagenomic analyses are available in Supplementary Information.
Results and discussion
Subclade Ic relative abundance in metagenomic datasets
Previous results demonstrated an abundance of upper mesopelagic 16S rRNA gene sequences phylogenetically affiliated with a single clone branching between SAR11 subclades Ia/Ib and subclades IIa/IIb, termed subclade Ic (Vergin et al., 2013; Figure 1). Phylogenetic evaluation of SAR11-type SAG 16S rRNA gene sequences demonstrated a congruent topology, with a monophyletic group of SAGs collected from mesopelagic samples corresponding to the subclade Ic position (Supplementary Figure S1). Four SAGs were selected to represent the breadth of the clade, determined by branch lengths (Supplementary Figure S1). The 16S rRNA gene sequences from the SAGs formed a monophyletic group with the subclade Ic clone from (Vergin et al., 2013) basal to subclades Ia/b (Figure 1). All four SAGs were isolated from a single station ALOHA sample taken at 770 m.
Recruitment of metagenomic 454 pyrosequences from Station ALOHA, the Eastern Subtropical Pacific oxygen minimum zone (ESTP OMZ) and BATS indicated a higher relative abundance of subclade Ic in the mesopelagic compared to the euphotic zone (Figure 2, Supplementary Figures S2–S4), and greater relative abundance in the 6000 m Puerto Rico Trench (PRT) metagenomic dataset compared with other subclades (Supplementary Figure S5). The Sea of Marmara (MARM) dataset showed similar distributions between subclade Ia (predominantly HTCC1062 type) and Ic (Supplementary Figure S6), and although the Matapan-Vavilov Deep (MATA) dataset had very little recruitment to any SAR11 genome (Supplementary Figure S7), consistent with the previous analysis (Smedile et al., 2013), those sequences that did recruit to SAR11 genomes were predominantly Ic-like. Longer Sanger shotgun-sequencing reads from 4000 m at Station ALOHA (Konstantinidis et al., 2009) also demonstrated increased recruitment to the SAGs relative to other genomes in deeper water (Supplementary Figure S8). We tested whether the increased abundance at depth might be due to temperature dependence. Recruitment from the GOS dataset (Venter et al., 2004; Rusch et al., 2007; Brown et al., 2012) consistently showed a dearth of subclade Ic abundance relative to Ia in surface waters around the globe, and did not support the conclusion that subclade Ic abundance at depth was driven by temperature (Supplementary Information).
Comparisons with surface SAR11 genomes
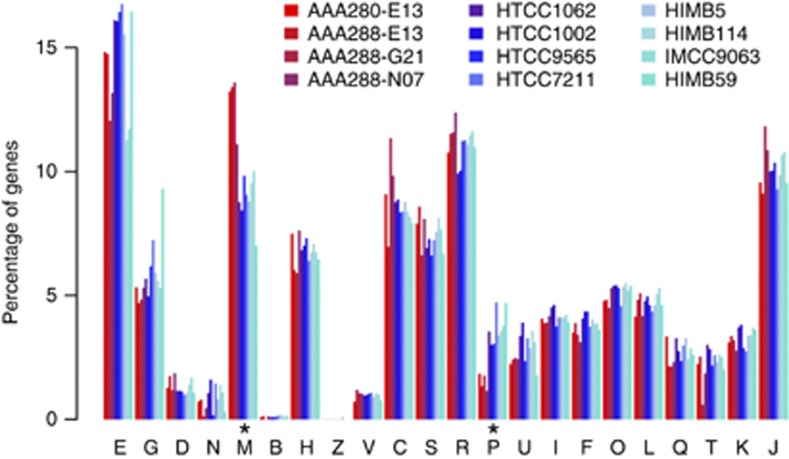
The SAGs had total assembly sizes between 0.81 and 1.40 Mbp spanning 81–151 scaffolds >500 bp, GC content between 29% and 30%, and coded for 948–1621 genes (Table 1). Estimated genome completeness, using 599 SAR11-specific single-copy orthologs (Supplementary Table S1), was between 55% and 86% with the corresponding estimated average genome size for the subclade Ic organisms at 1.49±0.09 Mbp. Protein-coding orthologous clusters for the SAGs and eight isolate SAR11 genomes were determined by all vs all BLASTP and Markov clustering using the automated pipeline Hal (Robbertse et al., 2011) and custom filters for length and synteny. Of the 3158 total orthologous clusters in the 12 SAR11 genomes, 1764 (56%) were present in at least one SAG, and 69% of the orthologous clusters found in the SAGs were shared with between one and eight other SAR11 genomes. COG distribution among the SAGs was generally the same as in surface genomes, except for categories M and P (Figure 3, Supplementary Figure S9, see below). The majority of Ic-specific genes were hypothetical (Supplementary Table S1), although several notable Ic-specific genes were present (see below). As would be expected from a low percentage of unique genes in the SAGs, much of the metabolism of these organisms appeared to be similar to that of the surface strains, particularly the subclade Ia organisms. Collectively, the Ic subclades were predicted to be obligate aerobic organisms, with cytochrome c oxidase as the sole predicted terminal reductatse, a complete tricarboxylic acid cycle, conserved lesions in several glycolytic pathways (Schwalbach et al., 2010), a reliance on reduced sulfur compounds (Tripp et al., 2008) and pathways for the metabolism and oxidation of small organic molecules such as amino/carboxylic acids and one-carbon and methylated compounds (Yilmaz et al., 2011; Grote et al., 2012; Carini et al., 2012; Supplementary Table S1).
Table 1. Subclade Ic SAG genome characteristics.
| Genome | AAA240-E13 | AAA288-E13 | AAA288-G21 | AAA288-N07 | Other SAR11a |
|---|---|---|---|---|---|
| Number of scaffolds | 151 | 106 | 139 | 81 | — |
| Assembly size (Mbp) | 1.40 | 0.81 | 0.91 | 0.95 | — |
| Est. genome completeness (%) | 86 | 55 | 64 | 66 | — |
| Est. genome size (Mbp) | 1.62 | 1.48 | 1.43 | 1.44 | 1.29–1.41* |
| GC content (%) | 29 | 29 | 30 | 29 | 29–32 |
| Number of genes | 1621 | 948 | 1103 | 1110 | 1357–1576 |
| Number of genes (prot. cod.) | 1581 | 923 | 1074 | 1083 | 1321–1541 |
Abbreviations: IMG, Integrated Microbial Genome; SAG, single-amplified genome.
Values from Grote et al. (2012) and IMG.
*Actual (not estimated) sizes.
Figure 3.
COG distribution as a percentage of total genes assigned to COGs. Y axis: percentage of genes, x axis: COG categories. Colors correspond to the genomes according to the key. Asterisks indicate categories with differential distribution in the SAGs relative to the isolate genomes. E, amino-acid metabolism and transport; G, carbohydrate metabolism and transport; D, cell division and chromosome partitioning; N, cell motility and secretion; M, cell wall/membrane/envelope biogenesis; B, chromatin structure and dynamics; H, coenzyme metabolism; Z, cytoskeleton; V-, C, energy production and conversion; S, unknown function; R, general function prediction only; P, inorganic ion transport and metabolism; U, intracellular trafficking and secretion; I, lipid metabolism; F, nucleotide transport and metabolism; O, posttranslational modification, protein turnover, chaperones; L, DNA replication, recombination and repair; Q, secondary metabolite biosynthesis, transport and catabolism; T, signal transduction mechanisms; K, transcription; J, translation.
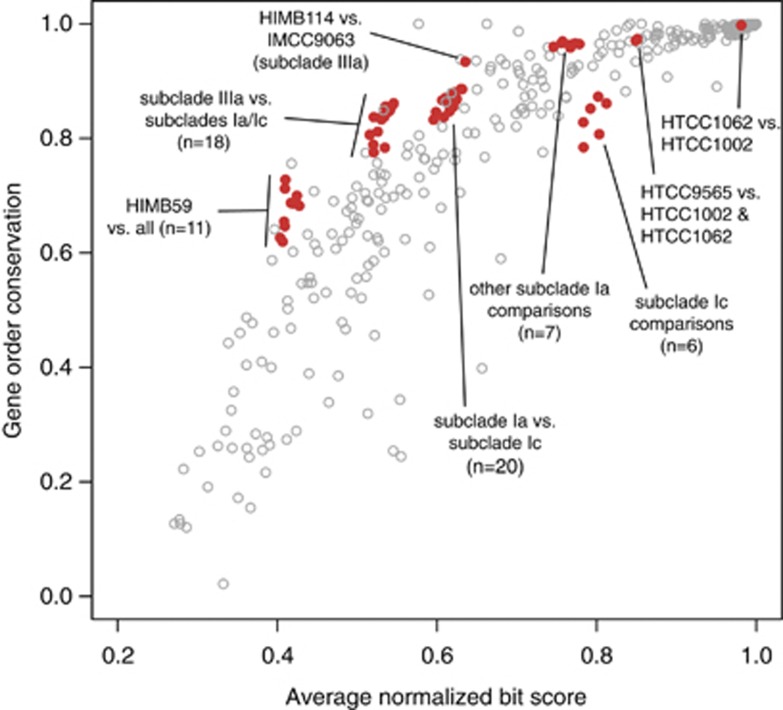
Also consistent with previous findings about the Pelagibacterales (Grote et al., 2012), the Ic SAGs had an unusually high conservation of local synteny among SAR11 genes (Figure 4). When compared among themselves, the Ic SAGs had less local synteny than most organisms at that level of 16S rRNA gene identity. However, we attributed this to the SAGs being incomplete and fragmented, because when the SAGs were compared with other SAR11 genomes, syntenic genes were a characteristically high percentage of the total shared genes. High amounts of local synteny may seem unlikely given predicted SAR11 recombination rates are among the highest measured for prokaryotes (Vergin et al., 2007; Vos and Didelot, 2009), however, it was shown previously that much of the rearrangement within genomes occurs at operon boundaries, and thus local synteny is not disrupted (Wilhelm et al., 2007). Further, the rates in Vergin et al. (2007) were restricted to closely related organisms within subclade Ia.
Figure 4.
Local synteny in SAR11 genomes. The percentage of genes in conserved order relative to the total number of shared genes (gene order conservation) vs average normalized bit score of the shared amino-acid content. Red dots are all pairwise comparisons of SAR11 genomes, the total in a given area indicated by n. Data are overlaid on that from Yelton et al. (2011; open gray circles).
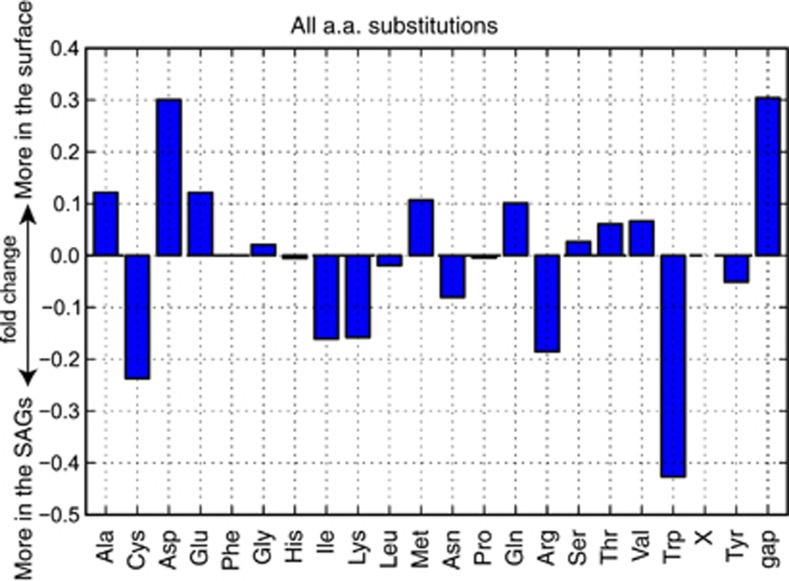
Although gene content and local gene order conservation between the isolate genomes and the SAGs was high, the SAGs were distinct at the amino-acid level. A concatenated protein phylogeny using 322 single-copy orthologs supported the 16S phylogeny, placing the subclade Ic SAGs as a monophyletic sister group to the subclade Ia surface strains (Figure 5a). The divergence from other strains and the depth of branching within the subclade Ic supported conceptualization of subclade Ic as a new genus of SAR11, separate from the subclade Ia, or Pelagibacter genus (Grote et al., 2012). Comparison of average amino-acid identity vs 16S rRNA gene identity was also in accordance with the metrics proposed by Konstantinidis and Tiedje (2007) for delineation of genera (66%–72% amino-acid identity; Grote et al., 2012; Figure 5b). Specific amino-acid substitution patterns among orthologs shared between the SAGs and the surface genomes showed relative increases in cysteine, isoleucine, lysine, asparagine, arginine and tryptophan in the predicted subclade Ic protein sequences at the expense of alanine, aspartatic acid, glutamic acid, methionine, glutamine, threonine and valine (Figure 6, Supplementary Figure S10).
Figure 5.
(a) Maximum likelihood tree of the SAR11 clade using 322 concatenated proteins. Subclade Ic highlighted in blue. All nodes had 100% bootstrap support unless otherwise indicated. Scale bar indicates changes per position. Root was inferred from Thrash et al. (2011) and Figure 1. (b) Average amino-acid identity vs 16S rRNA gene identity. Colors correspond to values in each cell according to the key. Dashed line indicates genus-level boundaries according to Konstantinidis and Tiedje (2007). Note, AAA240-E13 has only a partial 16S rRNA gene sequence, all others are full-length (see Supplementary Information).
Figure 6.
Fold change in amino-acid substitutions between the SAGs and the surface genomes. Pair-wise substitutions were quantified based on BLAST alignments of homologs between surface genomes and SAGs. X, unknown codons.
Many of the previously reported features associated with deep-ocean adaptation in microorganisms were not observed in the SAGs, such as rRNA gene insertions, increased transposable elements, or genes for chemoautotrophy (see Supplementary Information for detailed discussion). Nevertheless, there were still some distinguishing characteristics between subclade Ic and surface strains at the whole-genome level that were similar to or matching those previously observed in deep ocean metagenomic data sets (DeLong et al., 2006; Konstantinidis et al., 2009) and comparative genomics studies. The subclade Ic genomes had a small, but statistically significantly increase in intergenic space (Supplementary Figure S11) and a slightly (but statistically insignificant) higher estimated average genome size than that of current surface genomes (1.49±0.09 vs 1.33±0.07 Mbp, Supplementary Table S1). Also, consistent with Konstantinidis et al. (2009) and a general trend toward larger genomes in deeper samples, there were more gaps in the surface strain ortholog alignments (Supplementary Figure S10), indicating insertions and thus larger coding regions in the subclade Ic open reading frames. Unlike the surface strains, three of the four SAGs showed a statistically significant enrichment in category M, cell wall/membrane/envelope biogenesis (Figure 3 and Supplementary Figure S9). An increase in COG M genes was previously noted in the deep ocean Photobacterium profundum SS9 relative to mesophilic Vibrionaceae strains (Campanaro et al., 2008) and in a deep water ecotype of Alteromonas macleodii (Ivars-Martínez et al., 2008). COG M genes enriched in the SAGs include glycosyltransferases, methyltransferases, sugar epimerases, a sialic acid synthase, the cellular morphology gene ccmA (Hay et al., 1999) and polysaccharide export proteins (Supplementary Table S1). The SAGs also showed a significant reduction of COG P genes for inorganic ion transport and metabolism that may reflect increased reliance on organic N and P sources. In support of this hypothesis, none of the SAGs had homologs of the phosphate metabolism genes phoU, pstS, pstA or pstC, and although they had predicted ammonia permeases that clustered with ammonium transporters (clusters 150010.f.ok and 1500936.f.ok), none had genes annotated as an ammonium transporter. Furthermore, the SAGs had unique genes for purine degradation to ammonia (Supplementary Figure S12), including a 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline (OHCU) decarboxylase that was specific to, and conserved in, all four SAGs, possibly indicating a clade-specific nitrogen salvage pathway.
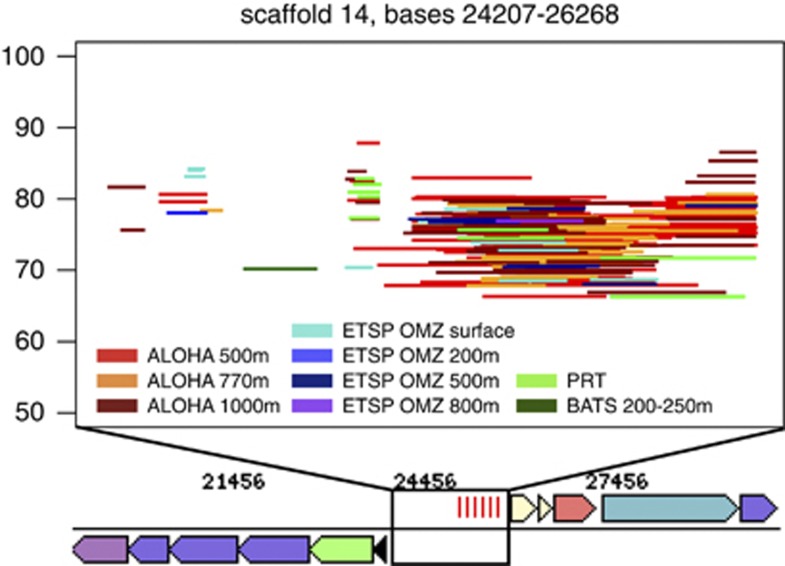
There were also indications of unique phage interactions and defense mechanisms in subclade Ic compared with the surface strains, consistent with previous studies showing enrichment of phage genes at depth (Martin-Cuadrado et al., 2007; Konstantinidis et al., 2009). The SAGs had unique phage integrases and phage protein D genes (Supplementary Table S1), and AAA240-E13 contained a predicted CRISPR region (Makarova et al., 2011) on scaffold 14 (Figure 7). A search for corresponding CRISPR-associated (cas) genes using HMMs developed by (Haft et al., 2005; Makarova et al., 2011) found some evidence for a cas4-like gene currently annotated as a hypothetical protein, conserved in three SAGs and HTCC9565 (Supplementary Table S1, cluster 15001317). In AAA240-E13, this cas4-like protein was on scaffold 18 and thus not located directly nearby the CRISPR. Widespread Pelagiphage that infect at least a subset of the known surface strains have been recently discovered (Zhao et al., 2013), but this is the only putative CRISPR locus identified so far in SAR11 genomes. Detailed analysis showed that this region had recruitment of metagenomic sequences mostly from the mesopelagic samples of station ALOHA, indicating that the CRISPR is relatively specific, geographically (Figure 7). The observed increase in subclade Ic COG M genes may also have a role in phage defense (Rodriguez-Valera et al., 2009).
Figure 7.
Recruitment of metagenomic sequences to the predicted CRISPR region. Upper box represents a magnification of the genomic region on scaffold 14 indicated in the title. Each line is a metagenomic sequence with reciprocal best hits (rbhs) to this region, organized by % identity (y axis) and sample (color). Those samples not appearing in the analysis either had only rbhs <50 bp or no rbhs.
Gene-specific relative abundance in metagenomic datasets
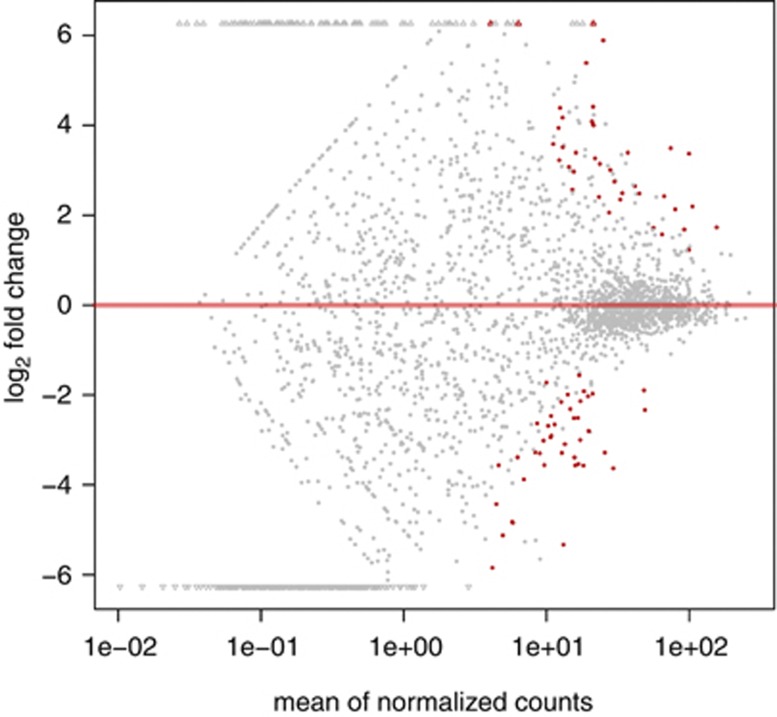
We used metagenomic data to evaluate the relative importance of SAG genes in situ, postulating that genes with little or no recruitment could be discounted as being present in fewer organisms, whereas those with high levels of recruitment could be inferred as being the most conserved, and therefore most important, to Ic-type organisms. Broadly, patterns of differential gene abundance between the SAR11 subclades could be identified across data sets. In most of the deep water samples, SAGs formed statistically significant grouping based on hierarchical clustering of recruitment profiles, indicating that these genomes are highly similar based on relative abundance of reciprocal best blast hits in deep-water environments (Supplementary Figure S13). The relative abundances of every gene for each SAG are reported in Supplementary Table S1 for all normalized datasets. Thirty-nine clusters showed significantly higher relative abundance of metagenomic sequence recruitment in deep water (those at 200 m and below) compared with surface datasets (Figure 8, Supplementary Information). Only two of these clusters did not contain SAG genes; whereas, of the 42 clusters that were significantly more abundant in surface samples, only two contained SAG genes and the rest were exclusive surface genomes. Half of the deep abundance clusters were exclusive to the SAGs, the other half had some shared distribution between the SAGs and surface genomes (Supplementary Table S1).
Figure 8.
Plot of normalized mean vs log-fold change for surface vs deep gene clusters.
Of the 19 of these clusters that were specific to subclade Ic, 9 were annotated as hypothetical proteins. A subclade Ic-specific cluster of putative Fe-S oxidoreductases contained multiple copies from each SAG, and all of the SAGs also had multiple copies of uncharacterized genes that clustered with single copies of predicted membrane occupation and recognition nexus (MORN) repeat genes from the subclade Ia genomes. The gene expansions for both these clusters suggested the proteins were important in the Ic subclade and in support of this hypothesis both were among the clusters significantly more abundant in deep metagenomic data sets (Supplementary Table S1). A predicted adenosine deaminase, unique to the SAGs, was highly abundant in deep samples. This gene works upstream of xanthine dehydrogenase (also significantly more abundant) in purine degradation, and although not statistically significant, other elements of the putative subclade Ic-specific purine degradation pathway, including the 2-oxo-4-hydroxy-4-carboxy-5-ureidoimidazoline decarboxylase, had high recruitment in deep samples compared with surface samples. Putative pillin assembly (pilF) genes, shared with other SAR11s, were also significantly more abundant in deep water samples, as were several methyltransferases, a Na+/proline symporter and a high-affinity Fe2+/Pb2+ permease.
Sulfite oxidase genes, conserved in three SAGs and shared only with HTCC9565, showed more recruitment in deep water samples, and were located directly adjacent to a cytochrome in the same configuration as the sorAB genes with proven sulfite oxidase activity in Starkeya novella ATCC 8083T (Kappler et al., 2000, 2012). The predicted AAA240-E13 sulfite oxidase had 33% identity with the S. novella SorA protein (blastp). Nearby were genes encoding for predicted Fe-S proteins, molybdopterin biosynthesis enzymes and molybdenum cofactor synthesis (Mo and heme are required cofactors (Kappler et al., 2000; Aguey-Zinsou et al., 2003)), which also appeared qualitatively more abundant in deep water samples. This may therefore indicate a mechanism for sulfur chemolithotrophy in subclade Ic and HTCC9565. Utilization of partially reduced sulfur compounds could also potentially explain the high abundance of SAR11 organisms and SAR11-type adenosine phosphosulfate reductase (aprAB) genes found in the ESTP OMZ, particularly at 200 m where dissolved oxygen is lowest and sulfur cycling has been identified (Figure 2; Canfield et al., 2010; Stewart et al., 2012). The aprAB genes were found in all subclade Ia and two of the subclade Ic genomes (Supplementary Table S1), and had high abundances in most of the deep water samples and higher abundance in deep vs shallow samples in datasets from the same water column. Given the lack of additional genes in the assimilatory sulfate reduction pathway in most SAR11 organisms (there was a predicted sat gene in HTCC9565 (Grote et al., 2012)), aprAB have been proposed to have a role in taurine metabolism (Williams et al., 2012), and may serve as a key sulfur cycling process for SAR11 in deep water as well. Our results indicate that the observed abundance of aprAB in the ESTP OMZ may be due to subclade Ic, rather than subclade Ia organisms.
Metagenomic relative abundance measurements allowed us to evaluate the potential importance of other notable genes found in the SAGs. Two, AAA288-G21 and AAA288-N07, contained predicted copies of proteorhodopsin, unexpected given the predominance of subclade Ic below the photic zone. The phylogeny of the proteorhodopsin genes generally matched the topology of the species tree (Supplementary Figure S14) and these loci showed modest recruitment in many of the samples for both strains (Supplementary Table S1), indicating that the subclade Ic may cycle to the euphotic zone with enough frequency, as a population, for the physiological benefits of retaining proteorhodopsin to be realized. Many of the unique or unexpected SAG genes with annotations were located in hypervariable regions (genomic islands), where there was little or no recruitment of metagenomic sequences (Coleman, 2006; Wilhelm et al., 2007; Tully et al., 2011; Grote et al., 2012; Supplementary Table S1). AAA240-E13 and AAA288-E13 had copies of predicted flagellar proteins, including a motor switch protein and a basal-body P-ring protein located together, and AAA240-E13 additionally had a putative flagellar biosynthesis/type III secretory pathway protein. However, the first two genes showed no recruitment in any of the metagenomic data sets, and the third had recruitment in only one, indicating that they were unlikely to be a common trait among subclade Ic strains (Supplementary Table S1). AAA240-E13 had the first mismatch repair (mutS) family homolog found in a SAR11 genome (Viklund et al., 2012), but it too was located in a hypervariable region.
Summary
The results of our metagenomic analyses from a variety of locations strongly support the conclusion that the subclade Ic organisms are autochthonous to the deep ocean. However, this raises the question, what are the depths to which they are best adapted? Are subclade Ic SAR11 truly piezophilic (growth rates increasing with pressure from 1 to 500 atm (Madigan and Martinko, 2006)), or are they primarily adapted to the shallower mesopelagic zone (piezotolerant)? Although the ALOHA 4000 m and PRT metagenomic analyses demonstrated subclade Ic organisms can be found in abysso- and hadopelagic realms, the lack of additional data from extreme deep water sites leaves the abundance of Pelagibacterales subclade Ic in such locations in question. Further, many previously identified features of both piezophilic isolates and deep ocean single-cell genomes (Simonato et al., 2006; Lauro and Bartlett, 2008; Nagata et al., 2010; Swan et al., 2011) are absent in the SAR11 SAGs. Although the incomplete state of the SAGs leaves open the possibility that these features may be contained in the unsequenced portion of the genomes, their absence in the nearly complete of AAA240-E13 SAG implies that even if present in some SAR11 Ic organisms, they are not universally conserved by the subclade. Alternatively, previously described features of deep ocean isolates may not be common to all piezophiles, and some piezophilic adaptations may not be directly observable at the level of nucleic acid or protein sequence variation. For example, many, but not all, piezophiles contain polyunsaturated acids, and cold or high pressure adaption can also be achieved by changing the ratio of unsaturated to saturated monounsaturated fatty acids in membrane lipids (DeLong and Yayanos, 1985). Such properties are not readily predictable from genomes. Finally, as these SAGs were isolated from 770 m, a depth that does not usually represent a piezophilic environment, the possibility exists that the Ic subclade may have further bathytype divisions, including true piezophiles that occupy the deeper realms.
The evidence herein suggests these are a piezotolerant subclade, with metabolism similar to that of surface subclades focused on aerobic oxidation of organic acids, amino acids, and C1 and methylated compounds, universal products of metabolism that are expected to be found in all biomes, and may contain mechanisms for nitrogen salvage and sulfur chemolithotrophy unusual in most surface SAR11 genomes. They also appear to have been evolving as an environmentally isolated subclade for long enough to show distinct signatures at the genome level. Thus, we can affirm our hypothesis: the subclade Ic SAGs did contain genomic features that distinguished them from the surface SAR11 genomes, although these features were generally more subtle than large-scale gene content variations. They had larger intergenic regions and larger coding regions in SAR11 clade orthologs, had a slightly larger estimated average genome size, were distinct phylogenetically and at the amino-acid content level, were enriched and depleted in COG M and P genes compared with other SAR11 genomes, respectively, and contained clade-specific hypothetical genes with increased relative abundances in deep water samples. Further examination of such hypothetical genes and cultivation successes with deep ocean SAR11 strains will help provide a mechanistic explanation for how the features described by this study contribute to the predominance of subclade Ic organisms in deeper water.
Acknowledgments
This work was supported by the Gordon and Betty Moore Foundation (SJG and EFD), the US Department of Energy Joint Genome Institute (JGI) Community Supported Program grant 2011-387 (RS, BKS, EFD, SJG), National Science Foundation (NSF) Science and Technology Center Award EF0424599 (EFD), NSF awards EF-826924 (RS), OCE-821374 (RS) and OCE-1232982 (RS and BKS), and is based on work supported by the NSF under Award no. DBI-1003269 (JCT). Sequencing was conducted by JGI and supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Christopher M Sullivan and the Oregon State University Center for Genome Research and Biocomputing, as well as the Louisiana State University Center for Computation and Technology for vital computational resources. We also thank Kelly C Wrighton and Laura A Hug for critical discussions about single-cell genomics, metagenomics and metabolic reconstruction.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aguey-Zinsou K-F, Bernhardt PV, Kappler U, McEwan AG. Direct electrochemistry of a bacterial sulfite dehydrogenase. J Am Chem Soc. 2003;125:530–535. doi: 10.1021/ja028293e. [DOI] [PubMed] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arıstegui J, Gasol JM, Duarte CM, Herndl GJ. Microbial oceanography of the dark ocean's pelagic realm. Limnol Oceanogr. 2009;54:1501–1529. [Google Scholar]
- Blainey PC. The future is now: single-cell genomics of bacteria and archaea. FEMS Microbiol Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MV, Lauro FM, DeMaere MZ, Les M, Wilkins D, Thomas T, et al. Global biogeography of SAR11 marine bacteria. Mol Sys Biol. 2012;8:1–13. doi: 10.1038/msb.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanaro S, Treu L, Valle G. Protein evolution in deep sea bacteria: an analysis of amino acids substitution rates. BMC Evol Biol. 2008;8:313. doi: 10.1186/1471-2148-8-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield DE, Stewart FJ, Thamdrup B, De Brabandere L, Dalsgaard T, DeLong EF, et al. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science. 2010;330:1375–1378. doi: 10.1126/science.1196889. [DOI] [PubMed] [Google Scholar]
- Carini P, Steindler L, Beszteri S, Giovannoni SJ. Nutrient requirements for growth of the extreme oligotroph 'Candidatus Pelagibacter ubique' HTCC1062 on a defined medium. ISME J. 2012;7:592–602. doi: 10.1038/ismej.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson CA, Morris R, Parsons R, Treusch AH, Giovannoni SJ, Vergin K. Seasonal dynamics of SAR11 populations in the euphotic and mesopelagic zones of the northwestern Sargasso Sea. ISME J. 2009;3:283–295. doi: 10.1038/ismej.2008.117. [DOI] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chou HH, Holmes MH. DNA sequence quality trimming and vector removal. Bioinformatics. 2001;17:1093–1104. doi: 10.1093/bioinformatics/17.12.1093. [DOI] [PubMed] [Google Scholar]
- Coleman ML. Genomic islands and the ecology and evolution of Prochlorococcus. Science. 2006;311:1768–1770. doi: 10.1126/science.1122050. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Yayanos AA. Adaptation of the membrane lipids of a deep-sea bacterium to changes in hydrostatic pressure. Science. 1985;228:1101–1103. doi: 10.1126/science.3992247. [DOI] [PubMed] [Google Scholar]
- DeLong EF, Preston CM, Mincer T, Rich V, Hallam SJ, Frigaard NU, et al. Community genomics among stratified microbial assemblages in the ocean's interior. Science. 2006;311:496. doi: 10.1126/science.1120250. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Accelerated Profile HMM Searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe EA, Fadrosh DW, Novotny M, Zeigler Allen L, Kim M, Lombardo M-J, et al. Going deeper: metagenome of a hadopelagic microbial community. PLoS One. 2011;6:e20388. doi: 10.1371/journal.pone.0020388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eloe EA, Shulse CN, Fadrosh DW, Williamson SJ, Allen EE, Bartlett DH. Compositional differences in particle-associated and free-living microbial assemblages from an extreme deep-ocean environment. Environ Microbiol Rep. 2011;3:449–458. doi: 10.1111/j.1758-2229.2010.00223.x. [DOI] [PubMed] [Google Scholar]
- Field K, Gordon D, Wright T, Rappe M, Urbach E, Vergin K, et al. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Rappe MS, Vergin KL, Adair NL. 16S rRNA genes reveal stratified open ocean bacterioplankton populations related to the green non-sulfur bacteria. Proc Natl Acad Sci USA. 1996;93:7979–7984. doi: 10.1073/pnas.93.15.7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Vergin KL. Seasonality in ocean microbial communities. Science. 2012;335:671–676. doi: 10.1126/science.1198078. [DOI] [PubMed] [Google Scholar]
- Gordon DA, Giovannoni SJ. Detection of stratified microbial populations related to Chlorobium and Fibrobacter species in the Atlantic and Pacific oceans. Appl Environ Microbiol. 1996;62:1171–1177. doi: 10.1128/aem.62.4.1171-1177.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote J, Thrash JC, Huggett MJ, Landry ZC, Carini P, Giovannoni SJ, et al. Streamlining and core genome conservation among highly divergent members of the SAR11 Clade. mBio. 2012;3:e00252–00212. doi: 10.1128/mBio.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLOS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay NA, Tipper DJ, Gygi D, Hughes C. A novel membrane protein influencing cell shape and multicellular swarming of Proteus mirabilis. J Bacteriol. 1999;181:2008–2016. doi: 10.1128/jb.181.7.2008-2016.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivars-Martínez E, Martin-Cuadrado A-B, D'Auria G, Mira A, Ferriera S, Johnson J, et al. Comparative genomics of two ecotypes of the marine planktonic copiotroph Alteromonas macleodii suggests alternative lifestyles associated with different kinds of particulate organic matter. ISME J. 2008;2:1194–1212. doi: 10.1038/ismej.2008.74. [DOI] [PubMed] [Google Scholar]
- Kappler U, Bennett B, Rethmeier J, Schwarz G, Deutzmann R, McEwan AG, et al. Sulfite: cytochrome c oxidoreductase from Thiobacillus novellus. J Biol Chem. 2000;275:13202–13212. doi: 10.1074/jbc.275.18.13202. [DOI] [PubMed] [Google Scholar]
- Kappler U, Davenport K, Beatson S, Lucas S, Lapidus A, Copeland A, et al. Complete genome sequence of the facultatively chemolithoautotrophic and methylotrophic alpha Proteobacterium Starkeya novella type strain (ATCC 8093(T)) Stand Genomic Sci. 2012;7:44–58. doi: 10.4056/sigs.3006378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature. 2001;409:507–510. doi: 10.1038/35054051. [DOI] [PubMed] [Google Scholar]
- King GM, Smith CB, Tolar B, Hollibaugh JT. Analysis of composition and structure of coastal to mesopelagic bacterioplankton communities in the northern gulf of Mexico. Front Microbiol. 2013;3:438. doi: 10.3389/fmicb.2012.00438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantinidis KT, Tiedje JM. Prokaryotic taxonomy and phylogeny in the genomic era: advancements and challenges ahead. Curr Opin Microbiol. 2007;10:504–509. doi: 10.1016/j.mib.2007.08.006. [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, Braff J, Karl DM, DeLong EF. Comparative metagenomic analysis of a microbial community residing at a depth of 4,000 meters at station ALOHA in the North Pacific subtropical gyre. Appl Environ Microbiol. 2009;75:5345–5355. doi: 10.1128/AEM.00473-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasken RS. Single-cell sequencing in its prime. Nat Biotechnol. 2013;31:211–212. doi: 10.1038/nbt.2523. [DOI] [PubMed] [Google Scholar]
- Lauro FM, Bartlett DH. Prokaryotic lifestyles in deep sea habitats. Extremophiles. 2008;12:15–25. doi: 10.1007/s00792-006-0059-5. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJJ, Charpentier E, Horvath P, et al. Evolution and classification of the CRISPR/Cas systems. Nat Rev Micro. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Cuadrado A-B, López-García P, Alba J-C, Moreira D, Monticelli L, Strittmatter A, et al. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One. 2007;2:e914. doi: 10.1371/journal.pone.0000914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madigan MT, Martinko JM.2006Brock Biology of Microorganisms,11th edn. Pearson Prentice Hall: Upper Saddle River, NJ, USA [Google Scholar]
- Morris R, Rappé M, Connon S, Vergin K, Siebold WA, Carlson CA, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- Morris RM, Longnecker K, Giovannoni SJ. Pirellula and OM43 are among the dominant lineages identified in an Oregon coast diatom bloom. Environ Microbiol. 2006;8:1361–1370. doi: 10.1111/j.1462-2920.2006.01029.x. [DOI] [PubMed] [Google Scholar]
- Morris RM, Frazar CD, Carlson CA. Basin-scale patterns in the abundance of SAR11 subclades, marine Actinobacteria (OM1), members of the Roseobacter clade and OCS116 in the South Atlantic. Environ Microbiol. 2012;14:1133–1144. doi: 10.1111/j.1462-2920.2011.02694.x. [DOI] [PubMed] [Google Scholar]
- Nagata T, Tamburini C, Arístegui J, Baltar F, Bochdansky AB, Fonda-Umani S, et al. Emerging concepts on microbial processes in the bathypelagic ocean – ecology, biogeochemistry, and genomics. Deep-Sea Res II. 2010;57:1519–1536. [Google Scholar]
- Pester M, Schleper C, Wagner M. The Thaumarchaeota: an emerging view of their phylogeny and ecophysiology. Curr Opin Microbiol. 2011;14:300–306. doi: 10.1016/j.mib.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaiser A, Zivanovic Y, Moreira D, López-García P. Comparative metagenomics of bathypelagic plankton and bottom sediment from the Sea of Marmara. ISME J. 2010;5:285–304. doi: 10.1038/ismej.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinthaler T, van Aken HM, Herndl GJ. Major contribution of autotrophy to microbial carbon cycling in the deep North Atlantic's interior. Deep-Sea Res II. 2010;57:1572–1580. [Google Scholar]
- Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, et al. Insights into the phylogeny and coding potential of microbial dark matter. Nature. 2013;499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- Robbertse B, Yoder RJ, Boyd A, Reeves J, Spatafora JW. Hal: an automated pipeline for phylogenetic analyses of genomic data. PLoS Curr Tree Life. 2011;3:RRN1213. doi: 10.1371/currents.RRN1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson C, Steinberg DK, Anderson TR, Arístegui J, Carlson CA, Frost JR, et al. Mesopelagic zone ecology and biogeochemistry–a synthesis. Deep-Sea Res II. 2010;57:1504–1518. [Google Scholar]
- Rodriguez-Valera F, Martin-Cuadrado A-B, Rodriguez-Brito B, Pašić L, Thingstad TF, Rohwer F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, et al. The Sorcerer II Global Ocean Sampling Expedition: Northwest Atlantic through Eastern Tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattenhofer M, Fuchs BM, Amann R, Zubkov MV, Tarran GA, Pernthaler J. Latitudinal distribution of prokaryotic picoplankton populations in the Atlantic Ocean. Environ Microbiol. 2009;11:2078–2093. doi: 10.1111/j.1462-2920.2009.01929.x. [DOI] [PubMed] [Google Scholar]
- Schwalbach MS, Tripp HJ, Steindler L, Smith DP, Giovannoni SJ. The presence of the glycolysis operon in SAR11 genomes is positively correlated with ocean productivity. Environ Microbiol. 2010;12:490–500. doi: 10.1111/j.1462-2920.2009.02092.x. [DOI] [PubMed] [Google Scholar]
- Shi Y, Tyson GW, Eppley JM, DeLong EF. Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J. 2011;5:999–1013. doi: 10.1038/ismej.2010.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonato F, Campanaro S, Lauro FM, Vezzi A, D'Angelo M, Vitulo N, et al. Piezophilic adaptation: a genomic point of view. J Biotechnol. 2006;126:11–25. doi: 10.1016/j.jbiotec.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Smedile F, Messina E, La Cono V, Tsoy O, Monticelli LS, Borghini M, et al. Metagenomic analysis of hadopelagic microbial assemblages thriving at the deepest part of Mediterranean Sea, Matapan-Vavilov Deep. Environ Microbiol. 2013;15:167–182. doi: 10.1111/j.1462-2920.2012.02827.x. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Stepanauskas R. Single cell genomics: an individual look at microbes. Curr Opin Microbiol. 2012;15:613–620. doi: 10.1016/j.mib.2012.09.001. [DOI] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol. 2012;14:23–40. doi: 10.1111/j.1462-2920.2010.02400.x. [DOI] [PubMed] [Google Scholar]
- Swan BK, Martinez-Garcia M, Preston CM, Sczyrba A, Woyke T, Lamy D, et al. Potential for chemolithoautotrophy among ubiquitous bacteria lineages in the Dark Ocean. Science. 2011;333:1296–1300. doi: 10.1126/science.1203690. [DOI] [PubMed] [Google Scholar]
- Thrash JC, Boyd A, Huggett MJ, Grote J, Carini P, Yoder RJ, et al. Phylogenomic evidence for a common ancestor of mitochondria and the SAR11 clade. Sci Rep. 2011;1:1–9. doi: 10.1038/srep00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treusch AH, Vergin KL, Finlay LA, Donatz MG, Burton RM, Carlson CA, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- Tripp HJ, Kitner JB, Schwalbach MS, Dacey JWH, Wilhelm LJ, Giovannoni SJ. SAR11 marine bacteria require exogenous reduced sulphur for growth. Nature. 2008;452:741–744. doi: 10.1038/nature06776. [DOI] [PubMed] [Google Scholar]
- Tully BJ, Nelson WC, Heidelberg JF. Metagenomic analysis of a complex marine planktonic thaumarchaeal community from the Gulf of Maine. Environ Microbiol. 2011;14:254–267. doi: 10.1111/j.1462-2920.2011.02628.x. [DOI] [PubMed] [Google Scholar]
- Varela MM, Van Aken HM, Herndl GJ. Abundance and activity of Chloroflexi-type SAR202 bacterioplankton in the meso- and bathypelagic waters of the (sub) tropical Atlantic. Environ Microbiol. 2008;10:1903–1911. doi: 10.1111/j.1462-2920.2008.01627.x. [DOI] [PubMed] [Google Scholar]
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, et al. Environmental genome shotgun sequencing of the Sargasso Sea. Science. 2004;304:66–74. doi: 10.1126/science.1093857. [DOI] [PubMed] [Google Scholar]
- Vergin KL, Tripp HJ, Wilhelm LJ, Denver DR, Rappé MS, Giovannoni SJ. High intraspecific recombination rate in a native population of Candidatus Pelagibacter ubique (SAR11) Environ Microbiol. 2007;9:2430–2440. doi: 10.1111/j.1462-2920.2007.01361.x. [DOI] [PubMed] [Google Scholar]
- Vergin KL, Beszteri B, Monier A, Thrash JC, Temperton B, Treusch AH, et al. High-resolution SAR11 ecotype dynamics at the Bermuda Atlantic Time-series Study site by phylogenetic placement of pyrosequences. ISME J. 2013;7:1322–1332. doi: 10.1038/ismej.2013.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viklund J, Ettema TJG, Andersson SGE. Independent genome reduction and phylogenetic reclassification of the oceanic SAR11 clade. Mol Biol Evol. 2012;29:599–615. doi: 10.1093/molbev/msr203. [DOI] [PubMed] [Google Scholar]
- Vos M, Didelot X. A comparison of homologous recombination rates in bacteria and archaea. ISME J. 2009;3:199–208. doi: 10.1038/ismej.2008.93. [DOI] [PubMed] [Google Scholar]
- Wang F, Wang J, Jian H, Zhang B, Li S, Wang F, et al. Environmental adaptation: genomic analysis of the piezotolerant and psychrotolerant deep-sea iron reducing bacterium Shewanella piezotolerans WP3. PLoS One. 2008;3:e1937. doi: 10.1371/journal.pone.0001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westesson O, Barquist L, Holmes I. HandAlign: Bayesian multiple sequence alignment, phylogeny and ancestral reconstruction. Bioinformatics. 2012;28:1170–1171. doi: 10.1093/bioinformatics/bts058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm LJ, Tripp HJ, Givan SA, Smith DP, Giovannoni SJ. Natural variation in SAR11 marine bacterioplankton genomes inferred from metagenomic data. Biol Direct. 2007;2:27. doi: 10.1186/1745-6150-2-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ, et al. A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J. 2012;6:1883–1900. doi: 10.1038/ismej.2012.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright TD, Vergin KL, Boyd PW, Giovannoni SJ. A novel delta-subdivision proteobacterial lineage from the lower ocean surface layer. Appl Environ Microbiol. 1997;63:1441–1448. doi: 10.1128/aem.63.4.1441-1448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yelton AP, Thomas BC, Simmons SL, Wilmes P, Zemla A, Thelen MP, et al. A semi-quantitative, synteny-based method to improve functional predictions for hypothetical and poorly annotated bacterial and archaeal genes. PLoS Comput Biol. 2011;7:e1002230. doi: 10.1371/journal.pcbi.1002230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz P, Gilbert JA, Knight R, Amaral-Zettler L, Karsch-Mizrachi I, Cochrane G, et al. The genomic standards consortium: bringing standards to life for microbial ecology. ISME J. 2011;5:1565–1567. doi: 10.1038/ismej.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Temperton B, Thrash JC, Schwalbach MS, Vergin KL, Landry ZC, et al. Abundant SAR11 viruses in the ocean. Nature. 2013;494:357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.