Abstract
Despite the frequent isolation of nitrate-respiring Epsilonproteobacteria from deep-sea hydrothermal vents, the genes coding for the nitrate reduction pathway in these organisms have not been investigated in depth. In this study we have shown that the gene cluster coding for the periplasmic nitrate reductase complex (nap) is highly conserved in chemolithoautotrophic, nitrate-reducing Epsilonproteobacteria from deep-sea hydrothermal vents. Furthermore, we have shown that the napA gene is expressed in pure cultures of vent Epsilonproteobacteria and it is highly conserved in microbial communities collected from deep-sea vents characterized by different temperature and redox regimes. The diversity of nitrate-reducing Epsilonproteobacteria was found to be higher in moderate temperature, diffuse flow vents than in high temperature black smokers or in low temperatures, substrate-associated communities. As NapA has a high affinity for nitrate compared with the membrane-bound enzyme, its occurrence in vent Epsilonproteobacteria may represent an adaptation of these organisms to the low nitrate concentrations typically found in vent fluids. Taken together, our findings indicate that nitrate reduction is widespread in vent Epsilonproteobacteria and provide insight on alternative energy metabolism in vent microorganisms. The occurrence of the nap cluster in vent, commensal and pathogenic Epsilonproteobacteria suggests that the ability of these bacteria to respire nitrate is important in habitats as different as the deep-sea vents and the human body.
Keywords: Epsilonproteobacteria, nitrate reduction, deep-sea vents, napA
Introduction
The contribution of deep-sea vent microorganisms to the sulfur cycle has been extensively investigated (Jannasch, 1995; Karl, 1995; Reysenbach et al., 2002). However, the isolation and characterization of a number of novel bacteria and archaea from deep-sea hydrothermal vents revealed that most of them can use nitrate in addition to sulfur as their terminal electron acceptor. During growth, these microorganisms reduce nitrate to dinitrogen (Takai et al., 2004; Nakagawa et al., 2005; Takai et al., 2006) or ammonium (Blochl et al., 1997; Vetriani et al., 2004; Voordeckers et al., 2008; Perez-Rodriguez et al., 2010, 2012). In most cases, nitrate reduction is coupled to the oxidation of molecular hydrogen (and less frequently of formate or thiosulfate), and these organisms have a shorter doubling time when nitrate, rather than sulfur, is supplemented as the terminal electron acceptor (Vetriani et al., 2004; Voordeckers et al., 2005; Perez-Rodriguez et al., 2010, 2012). In particular, most of the Epsilonproteobacteria isolated from deep-sea hydrothermal vents conserve energy by reducing nitrate (reviewed in Campbell et al., 2006). Epsilonproteobacteria have been found to be abundant in geographically distant deep-sea vent sites, and colonization experiments that were carried out in the vicinity of active deep-sea vents revealed that between 66 and 98% of the microorganisms associated with these substrates belonged to this group of bacteria (Lopez-Garcia et al., 2003; Takai et al., 2003; Alain et al., 2004). Overall, these observations indicate that Epsilonproteobacteria represent a large fraction of the primary producers at deep-sea hydrothermal vents at temperatures between 25 and 60 °C, and imply that the use of nitrate as a terminal electron acceptor in these environments may be more common than previously thought.
Data on the nitrogen chemistry of hydrothermal fluids indicate a depletion of both nitrate and nitrite in vent fluids. Nitrate, however, is present in bottom seawater (Millero, 1996) and it is therefore available to be used by vent bacteria both as an electron acceptor and as a nitrogen source. More recently, Bourbonnais et al. (2012a, 2012b) found that the rates for nitrate reduction to dinitrogen gas and ammonium in fluids from diffuse flow vents on the Juan de Fuca Ridge were up to ∼1000 and 150 nmol N l−1 per day, respectively, and that the isotopic composition of nitrate and ammonium suggested a role for vent microorganisms in nitrogen cycling.
Prokaryotes are able to synthesize three distinct types of nitrate reductases. All three enzymes reduce nitrate to nitrite, but they are involved in different physiological processes (reviewed in Lin and Stewart, 1998; Moreno-Vivian et al., 1999; Potter et al., 2001; Richardson et al., 2001). The Nas enzyme is located in the cytoplasm and participates in nitrogen assimilation, whereas the other two nitrate reductases (Nar and Nap) are both linked to respiratory electron transport systems. The Nar enzyme is a three-subunit complex attached to the cytoplasmic side of the membrane. The Nap enzyme is a two-subunit complex located in the periplasmic space of Gram-negative bacteria. All three types of nitrate reductases contain molybdenum bound to the bis-molybdopterin guanine dinucleotide cofactor at the active site. However, although the assimilatory enzyme (Nas) uses NAD(P)H or ferredoxin as reductant, the respiratory Nar and Nap enzymes ultimately take electrons from the membranous quinone pool.
The periplasmic nitrate reductase (NapA, encoded by the napA gene) has been described in several different organisms, and its physiological functions may vary in different bacteria (Moreno-Vivian et al., 1999; Potter et al., 2001; Richardson et al., 2001). Although in some bacteria NapA has a dissimilatory function to maintain redox balance during growth on highly reduced carbon sources (for example, in Rhodobacter and Paracoccus spp.; Richardson et al., 2001), in some enteric and rumen bacteria, such as Escherichia coli and Wolinella succinogenes, NapA is expressed during anaerobic growth and catalyzes the first step in the respiratory reduction of nitrate to ammonium (Grove et al., 1996; Simon et al., 2003). Within the Epsilonproteobacteria, the napA gene has been detected in the genomes of the rumen bacterium W. succinogenes (Simon et al., 2003); of the pathogens Helicobacter hepaticus (Suerbaum et al., 2003), Campylobacter jejuni (Parkhill et al., 2000) and several other Campylobacter spp. (Kern and Simon, 2009a); and of Arcobacter butzleri (Miller et al., 2007), Sulfurimonas denitrificans (Sievert et al., 2008) and several Epsilonproteobacteria isolated from marine geothermal environments (Inagaki et al., 2003; Voordeckers et al., 2005; Nakagawa et al., 2007; Campbell et al., 2009; Sikorski et al., 2010; Giovannelli et al., 2011). The genome sequence of these organisms showed that Epsilonproteobacteria encode the periplasmic nitrate reductase, NapA, and not for the membrane-bound enzyme, NarG (in contrast to, for example, E. coli, which encodes both enzymes).
The napA gene represents the core entity of the periplasmic nitrate reductase gene cluster. The napDAB genes encode the molybdopterin-containing reductase (NapA), the diheme component of the periplasmic electron transfer system (NapB, the redox partner of NapA) and a putative chaperone involved in the maturation of NapA (NapD). In Epsilonproteobacteria, the role of individual products of the nap gene cluster has been investigated in W. succinogenes (Kern et al., 2007). Consistent with the lack of the NapC (Simon et al., 2003), the electron transfer to NapAB in W. succinogenes is mediated by the NapGH membrane-bound iron–sulfur periplasmic quinone dehydrogenase complex (Kern and Simon, 2008). NapL is an iron–sulfur periplasmic protein present in all epsilonproteobacterial genomes. Although the function of NapL and NapF is currently unclear, recent data indicate that in W. succinogenes the NapGHF complex is involved in menaquinol oxidation, electron transfer to periplasmic NapA and maturation of the cytoplasmic NapA precursor (Kern et al., 2007; Kern and Simon, 2009b).
Although surveys of genes involved in nitrate reduction have been carried out in soils and estuarine environments (Bru et al., 2007; Smith et al., 2007; Dong et al., 2009), the distribution and diversity of the nitrate reductase in natural microbial communities from deep-sea geothermal environments are scarce. Recently, Perez-Rodriguez et al. (2013) identified Gamma- and Alphaproteobacteria-related, membrane-bound nitrate reductases, NarG, in vent microbial communities. However, there is no information on the distribution and diversity of the periplasmic nitrate reductase, NapA, in deep-sea environments. In this study we have reconstructed the structure of the nap gene cluster in Epsilonproteobacteria isolated from deep-sea hydrothermal vents whose genomes have been sequenced; we have established that reference strains of vent Epsilonproteobacteria encode and express the periplasmic nitrate reductase (NapA); and we have reconstructed the phylogeny of this enzyme. Furthermore, we have assessed the diversity, distribution and phylogeny of the NapA enzyme in natural microbial communities from different vent microhabitats. Our findings show that the periplasmic nitrate reductase is conserved and widespread in deep-sea vent microbial communities, and provide insight on alternative energy metabolism in vent microorganisms.
Materials and methods
Reference strains and genome-based reconstruction of the nitrate reduction pathway
The Epsilonproteobacteria used as reference strains in this study were cultured according to the conditions described in the original references (Supplementary Table S1 and Supplementary Information). The genomes of reference strains of Epsilonproteobacteria (Supplementary Table S2) were used to reconstruct the nap gene clusters. Both genes and predicted proteins were searched using BLAST against the public database. The Kyoto Encyclopedia of Genes and Genomes (http://www.genome.jp/kegg/) database, the Gene Orthology systems and BLAST algorithm were used to reconstruct the pathway of nitrate reduction in Caminibacter mediatlanticus.
RNA extraction and qualitative detection of napA transcripts
Total RNA was extracted from 20 ml of cultures of C. mediatlanticus and Caminibacter profundus grown in modified SME medium supplemented with either potassium nitrate or elemental sulfur (Voordeckers et al., 2005), using the UltraClean Microbial RNA Isolation Kit according to the protocol supplied with the kit (MoBio Laboratories, Solana Beach, CA, USA). Following DNase I treatment, total RNA was used as a template to detect napA transcripts by reverse transcriptase-PCR, using the SuperScript III One-Step RT-PCR System with primers NapV16F and NapV17R (Supplementary Table S4) according to the manufacturer's specifications (Invitrogen, Inc., Carlsbad, CA, USA). The identity of the transcripts was confirmed by sequencing.
Sample collection
Four types of samples were used in this study: (1) an active, high-temperature black smoker sulfide chimney was collected from the ‘Rainbow' vent field on the Mid-Atlantic Ridge (MAR); (2) hydrothermal fluids were collected in titanium samplers from two diffuse flow vents located at Alvinella Pillar (AP) and at East Wall (EW), respectively, on the East Pacific Rise (EPR) and were filtered shipboard on 0.2 μm Supor Gelman filters (Ann Arbor, MI, USA); (3) bacterial filaments were collected from an exclusion cage that had been deployed about 1 m above a diffuse flow vent located at Marker 89 on the EPR for 1 year. All samples were collected using the deep-submergence vehicle Alvin. Characteristics of samples and sampling locations are summarized in Supplementary Table S3.
DNA extraction
Individual replicate samples of the Rainbow chimney, the EW and AP hydrothermal fluids, and Mk 89 bacterial filaments (Supplementary Table S3) were collected and pooled together. Genomic DNA was extracted from pooled samples of each site and from cultures of each reference strain as previously described (Vetriani et al., 2003; Voordeckers et al., 2005, 2008).
DNA amplification by PCR
A fragment of about 1150 bp of the napA gene, coding for the periplasmic nitrate reductase, was amplified from the genomic DNA of reference strains (or was downloaded from GenBank, if available) and from the genomic DNA extracted from the four vent samples (Supplementary Table S3). PCR was carried out using primers NapV16F (5′-GCNCCNTGYMGNTTYTGYGG-3′) and NapV17R (5′-RTGYTGRTTRAANCCCATNGTCCA-3′), which were designed based on the napA sequence of Paracoccus pantotrophus, Ralstonia eutropha and E. coli (Flanagan et al., 1999), but which also target the epsiloproteobacterial napA gene (Supplementary Table S4). PCR conditions for amplification reactions were as follows: 30 cycles of denaturation at 94 °C for 1 min, annealing at 50 °C for 1 min and extension at 72 °C for 2 min, with a final extension time of 10 min during the last cycle. In order to reduce PCR bias, at least four independent PCR reactions per sample were carried out in parallel and pooled.
Library construction, restriction fragment length polymorphism screening and sequencing
The amplified napA gene fragments were cloned into the pCR4-TOPO plasmid vector and the ligation products were transformed into competent E. coli Oneshot cells (Invitrogen, Inc.). Four napA environmental libraries were constructed (Supplementary Table S3), and 189 napA clones were screened by restriction fragment length polymorphism analysis using the MnlI restriction endonuclease. Representative clones for each library showing unique restriction fragment length polymorphism patterns were selected and their sequences (about 1000 nucleotides) were determined for both strands.
Phylogenetic analyses
Translated amino acid sequences (http://www.ebi.ac.uk/emboss/transeq/) were aligned with ClustalX v 1.8 (Thompson et al., 1997) and manually adjusted using Seaview (Galtier et al., 1996). Phylogenetic distances were calculated using the Observed Divergence matrix and the neighbor-joining method was used to evaluate tree topologies. Phylo_win was used to plot tree topologies (Galtier et al., 1996) and their robustness was tested by bootstrap analysis with 1000 resamplings. 16S rRNA gene-based phylogenetic analyses were carried out as previously described (Perez-Rodriguez et al., 2013).
Cluster analysis of clone libraries
Cluster analysis of the four clone libraries was carried out using the R-software (R-Cran project, http://cran.r-project.org/). Briefly, the NapA sequences obtained from each library were sorted according to their phylogenetic grouping and fed to R for the determination of the distance matrix based on Jaccard dissimilarity. Cluster analysis was done using the average linkage method.
Nucleotide sequence accession numbers
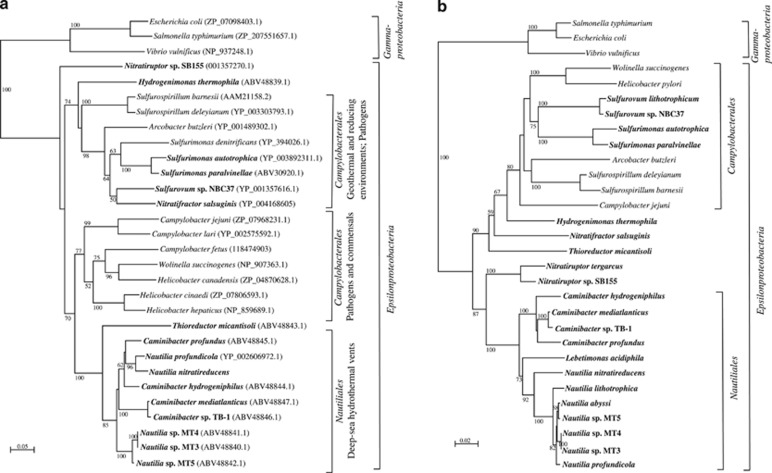
The sequences obtained in this study are available through GenBank under the following accession numbers: EF644686–EF644758, EF644772–EF644780, EF644815–EF644826 and EF683089–EF683090. The accession numbers of the NapA sequences of the reference strains are indicated in Figure 2a.
Figure 2.
(a) Neighbor-joining phylogenetic tree showing the position of the periplasmic nitrate reductase, NapA, of reference strains of Epsilonproteobacteria. Bootstrap values based on 1000 replications are shown at branch nodes. Bar, 5% estimated substitutions. (b) Neighbor-joining phylogenetic tree inferred from 16S rRNA gene sequences showing the position of reference strains of Epsilonproteobacteria. Bootstrap values based on 1000 replications are shown at branch nodes. Bar, 2% substitutions. Sequences from Epsilonproteobacteria isolated from deep-sea hydrothermal vents are in bold.
Results
Reconstruction of the nap gene cluster and of the nitrate reduction pathway in vent Epsilonproteobacteria
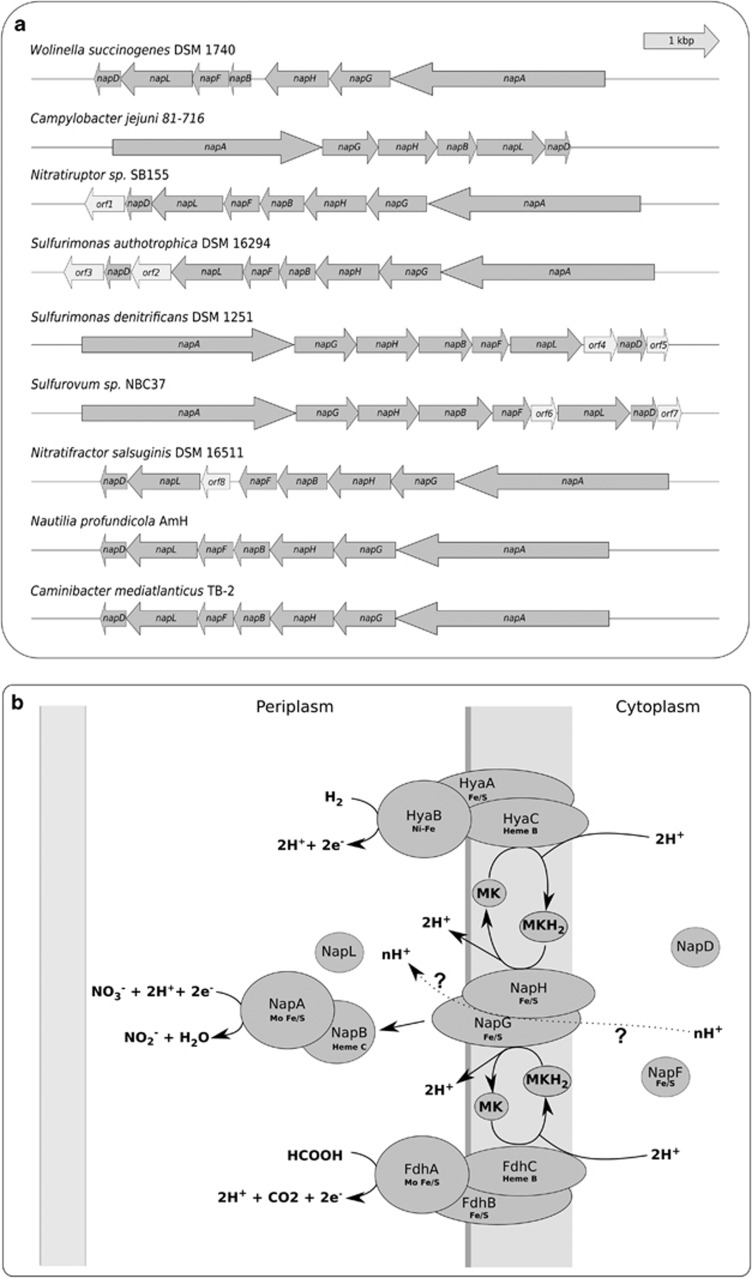
We reconstructed the nap gene clusters from the genomes of deep-sea vent Epsilonproteobacteria and compared them with those of the pathogen C. jejuni and the rumen commensal W. succinogenes (Figure 1a). The organization of the nap gene cluster, napAGHBFLD, was conserved in all vent Epsilonproteobacteria, whereas C. jejuni lacked the napF gene (Figure 1a). Furthermore, a number of open reading frames (ORFs) were found in the nap gene clusters of Sulfurimonas autotrophica, S. denitrificans, Sulfurovum sp. NBC37, Nitratiruptor sp. SB155 and Nitratiruptur salsuginis (Figure 1a).
Figure 1.
(a) Organization of the nap gene cluster in different Epsilonproteobacteria. Individual genes are drawn to scale. (b) Proposed model of the Nap respiratory nitrate reduction pathway in Caminibacter mediatlanticus. Question marks denote speculative processes and the dotted line indicates the proposed contribution of the Nap complex to the generation of the proton motive force. NapAB, periplasmic nitrate reductase complex; NapHG, membrane-bound iron–sulfur complex; NapD, putative chaperone; NapL, periplasmic iron–sulfur protein; NapF, putative iron–sulfur adaptor protein; HyaA, nickel–iron-dependent hydrogenase, large subunit; HyaB, nickel–iron-dependent hydrogenanse, small subunit; HyaC, nickel–iron-dependent hydrogenase, heme-b-containing subunit; FdhA, formate dehydrogenase, molibdenum-containing large subunit; FdhB, formate dehydrogenase, small subunit; FdhC, formate dehydrogenase, heme-b-containing subunit; MK, menaquinone.
The nitrate reduction pathway in C. mediatlanticus was reconstructed from its genome sequence and a model based on the gene functions assigned to W. succinogenes (Kern et al., 2007) was proposed (Figure 1b; see Discussion for further information).
Detection and phylogenetic analysis of the NapA periplasmic nitrate reductase in reference strains of vent Epsilonproteobacteria
Phylogenetic analysis of the amino acid sequence deduced from the napA genes placed the enzymes of all the Epsilonproteobacteria in a group distinct from the gammaproteobacterial NapA (Figure 2a). Three major clusters were identified: one of these clusters comprised sequences from the Nautiliales, which includes the genera Caminibacter and Nautilia, all isolated from deep-sea hydrothermal vents (Figure 2a). Within the Nautiliales, the enzymes from C. mediatlanticus and Caminibacter sp. strain TB-1 (98% sequence identity) formed a subgroup separated from those of Caminibacter hydrogeniphilus and C. profundus (93% sequence identity; Figure 2a). The sequence identity between the enzymes from the two subgroups ranged between 84 and 87%. The NapA from Nautilia profundicola and Nautilia nitratireducens formed a subgroup closely related to the enzymes from C. hydrogeniphilus and C. profundus (87–88% sequence identity), whereas the NapA from Nautilia spp. MT3, MT4 and MT5 formed a unique cluster (Figure 2a). In contrast with the NapA phylogeny, the 16S rRNA gene-based phylogenetic analysis placed all the Nautilia spp. in a unique group, closely related but separated from the Caminibacter spp. (compare Figures 2a and b). Taken together, these observations suggest an event of horizontal gene transfer of the napA gene.
The other two clusters contained NapA sequences from members of the Campylobacterales (Figure 2a). One of these two clusters included sequences from pathogenic and commensal Epsilonproteobacteria (Campylobacter spp., Helicobacter spp. and W. succinogenes; Figure 2a). The second cluster of NapA from the Campylobacterales included sequences from organisms isolated from geothermal and reducing environments (for example, Sulfurovum spp., Sulfurimonas spp. and Sulfurospirillum spp.) and from the pathogen A. butzleri (Figure 2a). The NapA from Nitratifractor salsuginis clustered with the enzyme of Sulfurovum sp. NBC37, whereas the 16S rRNA gene-based phylogeny placed this taxon in a unique lineage (compare Figures 2a and b).
The NapA from Thioreductor micantisoli was placed in an independent lineage related to the Nautiliales (77% identity to the NapA from C. mediatlanticus), whereas the sequence from Hydrogenimonas thermophila formed a discrete lineage related to one of the two clusters of Campylobacterales (68% identity to the NapA of Sulfurospirillum barnesii). Finally, the NapA from Nitratiruptor sp. SB155 formed a unique lineage outside of the three main clusters (Figure 2a).
Detection of napA transcripts in Caminibacter spp.
Total RNA was extracted from cultures of C. mediatlanticus and C. profundus, which were grown anaerobically at their optimal growth temperatures (Supplementary Table S1) under a CO2/H2 atmosphere (Supplementary Figure S1A, lanes 1 and 2). When the RNA extracted from cultures of the two species grown in the presence of nitrate as their sole terminal electron acceptor was subjected to reverse transcriptase-PCR with napA-specific primers, napA transcripts were detected in both cases (Supplementary Figure S1B, lanes 3 and 4). When the two cultures were grown in the presence of elemental sulfur as their sole terminal electron acceptor, faint signals for the napA transcript were also detected, indicating that the periplasmic nitrate reductase may be constitutively expressed at a basal level (data not shown).
Diversity and phylogenetic analysis of NapA nitrate reductase in vent microbial communities
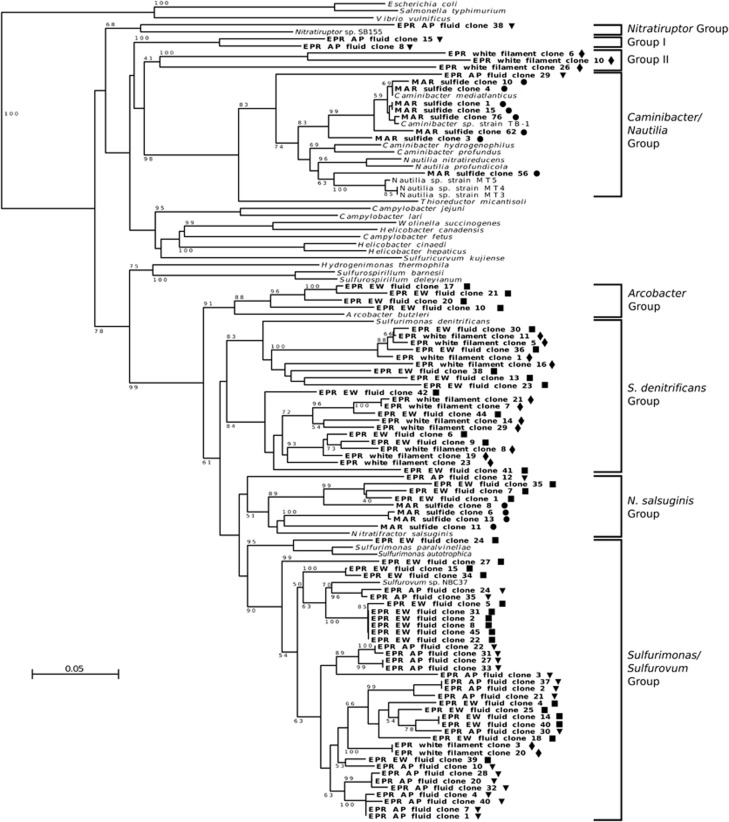
Genomic DNA was successfully extracted from four samples, representing vent microhabitats characterized by different temperature and redox regimes: an active, high-temperature sulfide chimney from the Rainbow vent site on the MAR sulfide (fluid emission at the orifice: 158 °C), hydrothermal fluids from two sites on the EPR AP fluid (40 °C) and EPR EW fluid (25 °C; maximum free sulfide concentration of 175 μM), and bacterial filaments attached to an artificial substrate (EPR white filaments; 2.5 °C; H2S concentration range: 0.2–32 μM; Supplementary Table S3). All the napA gene fragments retrieved from the vent natural microbial communities investigated in this study were phylogenetically related to the Epsilonproteobacteria, and a total of eight NapA groups were identified based on phylogenetic clustering (Figures 3 and 4). Out of these eight groups, six included sequences related to reference strains (Figure 3).
Figure 3.
Neighbor-joining phylogenetic tree of the amino acid sequence deduced from a fragment of the gene encoding for the periplasmic nitrate reductase (napA) from the Rainbow chimney (MAR sulfide; circles), the East Wall (EPR EW fluid; squares) and Alvinella Pillar (EPR AP fluid; triangles) diffuse flow vents, and from substrate-attached bacterial filaments (EPR white filament; diamonds). Representative sequences obtained in this study are in bold. Percentages >50% of 1000 bootstrap resampling that support each topological element are shown at the branch nodes. Bar, 5% estimated substitutions.
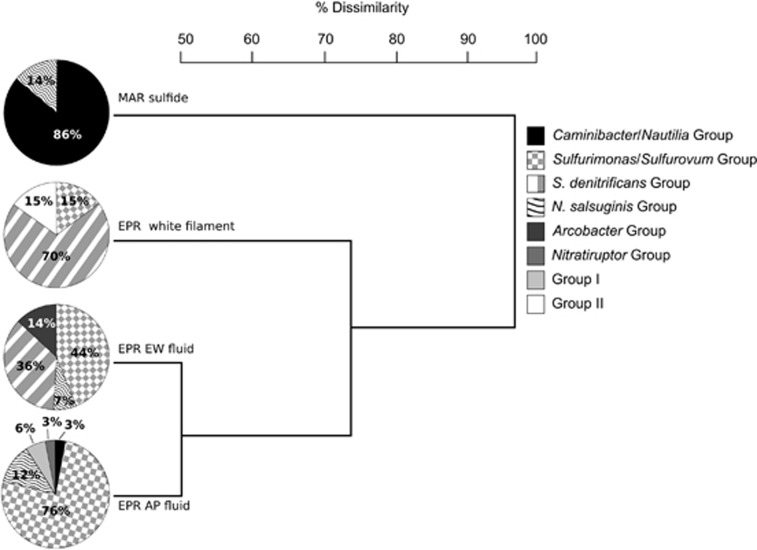
Figure 4.
Cluster analysis of the four vent microbial communities based on the phylogenetic affiliation of the NapA sequences. The frequency of the eight groups of sequences detected is shown (see text for details).
The MAR sulfide showed the lowest diversity of NapA sequences (Supplementary Figure S2), 86% of which were related to the enzymes of Caminibacter and Nautilia spp. (the amino acid sequence identity of these clones to the NapA of C. mediatlanticus ranged from 81 to 100%). The remaining 14% of the sequences from this site was related to the enzyme of N. salsuginis (83–88% identity to the NapA of N. salsuginis; Figures 3 and 4).
The EPR AP fluid showed the highest diversity of NapA sequences, which were grouped into five different clusters (Figures 3 and 4). The majority (76%) of the sequences retrieved from this sample were related to the NapA of Sulfurovum sp. NBC37 (84–90% identity), whereas 12% were related to N. salsuginis (82% identity) and 3% each to Nitratiruptur sp. SB155 (71% identity), and to Caminibacter and Nautilia spp. (81% and 79% identity to the enzymes of C. mediatlanticus and N. profundicola, respectively). Phylogenetic analysis placed the remaining 6% of the EPR AP fluid sequences in a unique cluster, designated as Group I, unrelated to the NapA of any cultured Epsilonproteobacteria (Figure 3). Within Group I, the amino acid sequence of the NapA fragment designated as EPR AP fluid clone 15 (Figure 3) was 69%, 70%, 71% and 72% identical to the enzymes of C. mediatlanticus, N. profundicola, W. succinogenes and Campylobacter lari, respectively.
The NapA sequences retrieved from the EPR EW fluid were grouped into four clusters (Figures 3 and 4). Forty-four percent of the sequences from this sample were related to Sulfurovum and Sulfurimonas spp., with 85–93% identity to the enzyme of Sulfurovum sp. NBC37 and 78–85% identity to the enzyme of S. autotrophica (Figure 3). The amino acid sequence of the NapA fragment designated as EPR EW fluid clone 24 was the most closely related to enzymes of Sulfurimonas spp. (87% and 85% identity to the enzymes of Sulfurimonas paralvinellae and S. autotrophica, respectively). The second major group (36%) of NapA sequences retrieved from the EPR EW fluid sample was related to S. denitrificans (∼80% identity). The remaining sequences from the EPR EW fluid clustered with A. butzleri (14% of the sequences; 80% identity) and with N. salsuginis (7% of the sequences; 83% identity).
The NapA sequences from the EPR white filament community were distributed into three groups: 70% of the sequences were related to S. denitrificans (80–81% identity), whereas 15% clustered with Sulfurovum and Sulfurimonas spp. (90% and 82% identity to the enzyme of Sulfurovum sp. NBC37 and S. autotrophica, respectively; Figures 3 and 4). The remaining 15% of the sequences from the EPR white filament community formed a unique cluster, designated as Group II, unrelated to the NapA of any cultured Epsilonproteobacteria (Figure 3). The amino acid sequence of EPR white filament clone 6, a NapA clone representative of Group II, was 66% or less identical to the enzymes of C. mediatlanticus, C. hydrogeniphilus, Nautila sp. MT3, W. succinogenes, Helicobacter cinaedi and Helicobacter canadensis.
Cluster analysis of the periplasmic nitrate reductase-encoding microbial communities indicated that EPR AP and EPR EW fluids were the most similar (51.1% Jaccard dissimilarity), whereas the MAR sulfide community shared the least number of napA genes with the other three communities (100%, 97.4% and 95.2% dissimilarity with the EPR white filaments, EPR EW fluid and EPR AP fluid, respectively; Figure 4). The EPR white filament community was more similar to the EPR EW fluid (56.4% dissimilarity) than to the EPR AP fluid communities (93.4% dissimilarity; Figure 4). This is consistent with the finding that Caminibacter and Nautilia-related periplasmic nitrate reductases were detected only in the MAR sulfide and in the EPR AP fluid libraries, whereas Sulfurovum-related enzymes were detected in the other three communities. S. denitrificans-related sequences were detected in both the EPR EW fluid and EPR white filament communities (Figure 4).
Discussion
The nitrate reduction pathway of vent Epsilonproteobacteria
Comparative analysis of the nap gene clusters showed that the napAGHBFLD structure is conserved in all vent Epsilonproteobacteria (Figure 1a). However, some differences were found. The nap gene clusters of Nitratiruptor sp. SB155, Sulfurovum sp. NBC31, N. salsuginis, S. denitrificans and S. autotrophica contained some predicted ORFs, which were not present in the gene clusters of the strict anaerobes, C. mediatlanticus and N. profundicola (Figure 1a). These ORFs are hypothetical proteins with no predicted conserved domain, except for orf1 (locus tag NIS_1801) in the operon of Nitratiruptor sp. SB155, orf2 (Saut_1245) in S. autotrophica and orf4 (Suden_1520) in S. denitrificans, which have a putative active site similar to the histidine kinase signal transduction sensor for oxygen. The identities of the deduced amino acid sequences of these ORFs are 66% between orf2 and orf4, 61% between orf1 and orf2, and 63% between orf1 and orf4, whereas all the other ORFs are unrelated. Although Nitratiruptor sp. SB155 and S. denitrificans are facultative microaerophiles, S. autotrophica is an obligate aerobe. Hence, in these bacteria the putative histidine kinase might mediate signal transduction in response to oxygen sensing.
It is worth noting that most of the mesophilic Epsilonproteobacteria isolated from deep-sea hydrothermal vents reduce nitrate to dinitrogen gas, whereas their thermophilic relatives produce ammonium as the end product of nitrate respiration (Supplementary Table S1 and references therein). This is consistent with data showing that ammonification is more prevalent than denitrification in nitrate-reducing thermophiles (Blochl et al., 1997; Vetriani et al., 2004; Perez-Rodriguez et al., 2012), and may be related to a possible decrease in nitrate concentrations as the hydrothermal fluid temperature increases. In line with this hypothesis, an increase in the significance of nitrate ammonification relative to denitrification as the concentration of nitrate decreased was observed in estuarine sediments (Dong et al., 2009). Phylogenetic analyses of the denitrification or ammonification pathways in these bacteria should provide further clues on their respective evolutionary histories.
The proposed model for the nitrate reduction pathway in C. mediatlanticus shows the NapAB reductase complex, containing the molybdenum-containing active site, which receives electrons from the membrane-bound NapGH heterodimer and carries out the reduction of nitrate to nitrite (Figure 1b). NapGH is a transmembrane Fe/S cluster that functions as a quinone oxidoreductase and mediates the transfer of electrons from the membrane quinone pool. In C. mediatlanticus and other thermophilic Epsilonproteobacteria, the reduction of nitrate is generally coupled with oxidation of hydrogen gas. The enzyme complex responsible for the oxidation of hydrogen (Hya complex, also known as Hyn, Hyd, Hup or Hox) includes three subunits that couple hydrogen oxidation with the transfer of electrons to the quinone pool and the generation of the proton gradient. In the proposed model, HyaAB (locus tags CMTB2_07241, CMTB2_07236) is responsible for the oxidation of hydrogen mediated by the Ni-Fe active site, whereas HyaC (CMTB2_07251) is a heme-b-containing protein responsible for the electron transfer to the quinone pool (Figure 1b). The Nap cluster in the genome of C. mediatlanticus is functionally linked to the formate dehydrogenase complex (genes fdhABC; CMTB2_03503, CMTB2_03508 and CMTB2_03513, respectively), which may provide reducing equivalents through a menaquinol–menaquinone transport chain (Figure 1b). However, at this point the role of the formate dehydrogenase in C. mediatlanticus is unclear, as this bacterium does not seem to use formate as an energy source (Voordeckers et al., 2005). Interestingly, the close relatives N. nitratireducens, Nautilia lithotrophica and N. profundicola use formate as an alternative energy source to hydrogen (Miroshnichenko et al., 2002; Smith et al., 2008; Perez-Rodriguez et al., 2010), and the fdhABC gene clusters of C. mediatlanticus and N. profundicola are highly conserved (86%, 83% and 65% amino acid identity for FdhA, B and C, respectively).
Community structure of nitrate-reducing Epsilonproteobacteria in deep-sea hydrothermal vents
The community structure of nitrate-reducing microorganisms was investigated by surveying the distribution of the periplasmic nitrate reductase in four vent microhabitats representing different temperature and redox regimes. Three independent lines of evidence gathered in this study strongly suggest that at deep-sea hydrothermal vents, chemolithoautotrophic Epsilonproteobacteria mediate NapA-catalyzed respiratory nitrate reduction to either dinitrogen gas or to ammonium: (1) nitrate-reducing Epsilonproteobacteria encode the periplasmic nitrate reductase, NapA (Figure 1a), and not for the membrane-bound enzyme, NarG; (2) the napA gene is expressed in vent Epsilonproteobacteria during growth with nitrate as the sole electron acceptor (Supplementary Figures S1 and S3); and (3) all the napA genes retrieved from the vent samples were related to epsilonproteobacterial enzymes (Figure 3). Furthermore, epsilonproteobacterial nap gene transcripts were recently detected in a chimney-associated microbial biofilm (Dahle et al., 2013). As Epsilonproteobacteria are widespread and, in some cases, abundant members in deep-sea hydrothermal vent microbial communities, nitrate reduction in these environments appears to be more relevant than previously recognized.
The observation that the NapA recovered from the MAR sulfide community were predominantly related to enzymes from Caminibacter and Nautila spp. (Figures 3 and 4) is in line with data obtained from 16S rRNA gene and ATP citrate lyase (an enzyme involved in CO2 fixation via the reductive tricarboxylic acid cycle) libraries constructed from the same chimney sample, which revealed a prevalence of Epsilonproteobacteria of the genus Caminibacter associated with this vent (Voordeckers et al., 2008). The isolation of C. mediatlanticus, Caminibacter sp. strain TB-1 (Voordeckers et al., 2005) and C. profundus (Miroshnichenko et al., 2004) from the Rainbow hydrothermal vent further confirmed the occurrence of these organisms at this site.
Analyses of the periplasmic nitrate reductase from the EPR EW and AP fluids suggest that the moderate temperatures (25–40 °C) and mildly reducing conditions (due to the advection of oxygenated seawater) usually associated with diffuse flow vents are conducive to the establishment of a diverse community of Epsilonproteobacteria. In general, a correlation exists between temperature and redox state of hydrothermal fluids in that higher-temperature fluids tend to be more reduced than lower-temperature ones (Luther et al., 2001; Le Bris et al., 2006). To some extent, this may explain the presence of clones related to the mesophilic, microaerobic, thiosulfate-oxidizing Sulfurovum and Sulfurimonas spp. in the EPR AP and EW fluid, and in the substrate-associated white filament communities (Figures 3 and 4). The distribution of napA genes related to mesophilic and thermophilic Epsilonproteobacteria appears to reflect the adaptation of these organisms to different temperature and redox regimes. For instance, the Caminibacter/Nautilia group dominated the high-temperature MAR sulfide (86%), which is in line with the anaerobic and thermophilic metabolism of these organisms (Figure 4).
Implications of napA expression in nitrate depleted environments
Although several microorganisms, including E. coli, encode for both the periplasmic and membrane-bound nitrate reductase, it is not clear why Epsilonproteobacteria encode the periplasmic type, NapA. In E. coli, the expression of the nap genes is induced at low concentrations of nitrate (μM range), whereas the cytoplasmic nitrate reductase (Nar) is expressed at nitrate concentrations in the mM range (Cole, 1996; Potter et al., 1999; Wang et al., 1999). Competition experiments, where mutant strains of E. coli lacking either Nar or Nap were carried out using continuous-culture techniques that maintained the concentration of nitrate in the μM range, showed that the Nap+ strain outcompeted the strain expressing only the Nar enzyme during nitrate-limited growth (Potter et al., 1999). The saturation constants (Ks) for nitrate for the two strains were estimated to be 15 and 50 μM for Nap and Nar, respectively (Potter et al., 1999). These observations, also supported by a study where NapA-mediated nitrate reduction in estuarine sediments remained important at low nitrate concentrations (Dong et al., 2009), imply that between the two enzymes NapA is better adapted to function in nitrate-depleted environments. Epsilonproteobacteria include very different organisms, such as deep-sea vent chemolithoautotrophs (for example, Caminibacter and Sulfurimonas spp.) and human pathogens (for example, Campylobacter and Helicobacter spp.). However, the Nap nitrate reductase system appears to be conserved within these different bacteria. Nitrate is depleted in hydrothermal fluids but it is present at μM concentrations in deep seawater (Millero, 1996), and vent microorganisms are exposed to it along redox gradients that form at the interface between fluids and seawater. As nitrate concentrations in the μM range are common to both deep seawater and fluids in the human body (Lentner, 1984; Millero, 1996), we hypothesize that the nitrate-deficient nature of these environments is a factor selecting for the high nitrate affinity NapA enzyme in both pathogenic and vent Epsilonproteobacteria. Future comparative analyses of the genes and enzymes comprising the nap cluster in pathogenic and vent Epsilonproteobacteria may provide insights into the evolution of the periplasmic nitrate reductase complex and nitrate scavenging strategies.
Acknowledgments
We thank the crew of R/V Atlantis and the crew and pilots of the deep-submergence vehicle Alvin, for their skilled operations at sea. We also thank Ileana Pérez-Rodríguez and Jessica Ricci for their work on N. nitratireducens. This research was supported by NSF grants MCB 04-56676, MCB 08-43678 and OCE 11-36451 to CV and OCE 03-27353 to CV and RAL, an NIH PhD Training Program in Biotechnology Fellowship (NIH NIGMS 5 T32 GM08339) to JV and an NSF Graduate Research Fellowship to MCM. This paper is C-DEBI Contribution 198.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Alain K, Zbinden M, Le Bris N, Lesongeur F, Querellou J, Gaill F, et al. Early steps in microbial colonization processes at deep-sea hydrothermal vents. Environ Microbiol. 2004;6:227–241. doi: 10.1111/j.1462-2920.2003.00557.x. [DOI] [PubMed] [Google Scholar]
- Blochl E, Rachel R, Burggraf S, Hafenbradl D, Jannasch HW, Stetter KO. Pyrolobus fumarii, gen. and sp. nov., represents a novel group of archaea, extending the upper temperature limit for life to 113 degrees C. Extremophiles. 1997;1:14–21. doi: 10.1007/s007920050010. [DOI] [PubMed] [Google Scholar]
- Bourbonnais A, Juniper SK, Butterfield DA, Devol AH, Kuypers MMM, Lavik G, et al. Activity and abundance of denitrifying bacteria in the subsurface biosphere of diffuse hydrothermal vents of the Juan de Fuca Ridge. Biogeosciences. 2012a;9:4661–4678. [Google Scholar]
- Bourbonnais A, Lehmann MF, Butterfield DA, Juniper SK. Subseafloor nitrogen transformations in diffuse hydrothermal vent fluids of the Juan de Fuca Ridge evidenced by the isotopic composition of nitrate and ammonium. Geochem Geophys Geosyst. 2012b;13:Q02T01. [Google Scholar]
- Bru D, Sarr A, Philippot L. Relative abundances of proteobacterial membrane-bound and periplasmic nitrate reductases in selected environments. Appl Environ Microbiol. 2007;73:5971–5974. doi: 10.1128/AEM.00643-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell BJ, Engel AS, Porter ML, Takai K. The versatile epsilon-proteobacteria: key players in sulphidic habitats. Nat Rev Microbiol. 2006;4:458–468. doi: 10.1038/nrmicro1414. [DOI] [PubMed] [Google Scholar]
- Campbell BJ, Smith JL, Hanson TE, Klotz MG, Stein LY, Lee CK, et al. Adaptations to submarine hydrothermal environments exemplified by the genome of Nautilia profundicola. PLoS Genet. 2009;5:e1000362. doi: 10.1371/journal.pgen.1000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole J. Nitrate reduction to ammonia by enteric bacteria: redundancy, or a strategy for survival during oxygen starvation. FEMS Microbiol Lett. 1996;136:1–11. doi: 10.1111/j.1574-6968.1996.tb08017.x. [DOI] [PubMed] [Google Scholar]
- Dahle H, Roalkvam I, Thorseth IH, Pedersen RB, Steen IH. The versatile in situ gene expression of an Epsilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ Microbiol Rep. 2013;5:282–290. doi: 10.1111/1758-2229.12016. [DOI] [PubMed] [Google Scholar]
- Dong LF, Smith CJ, Papaspyrou S, Stott A, Osborn AM, Nedwell DB. Changes in benthic denitrification, nitrate ammonification, and anammox process rates and nitrate and nitrite reductase gene abundances along an estuarine nutrient gradient (the Colne estuary, United Kingdom) Appl Environ Microbiol. 2009;75:3171–3179. doi: 10.1128/AEM.02511-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flanagan DA, Gregory LG, Carter JP, Karakas-Sen A, Richardson DJ, Spiro S. Detection of genes for periplasmic nitrate reductase in nitrate respiring bacteria and in community DNA. FEMS Microbiol Lett. 1999;177:263–270. doi: 10.1111/j.1574-6968.1999.tb13742.x. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M, Gautier C. SeaView and Phylo_win, two graphic tools for sequence alignment and molecular phylogeny. Comput Appl Biosci. 1996;12:543–548. doi: 10.1093/bioinformatics/12.6.543. [DOI] [PubMed] [Google Scholar]
- Giovannelli D, Ferriera S, Johnson J, Kravitz S, Perez-Rodriguez I, Ricci J, et al. Draft genome sequence of Caminibacter mediatlanticus strain TB-2(T), an epsilonproteobacterium isolated from a deep-sea hydrothermal vent. Stand Genomic Sci. 2011;5:135–143. doi: 10.4056/sigs.2094859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove J, Tanapongpipat S, Thomas G, Griffiths L, Crooke H, Cole J. Escherichia coli K-12 genes essential for the synthesis of c-type cytochromes and a third nitrate reductase located in the periplasm. Mol Microbiol. 1996;19:467–481. doi: 10.1046/j.1365-2958.1996.383914.x. [DOI] [PubMed] [Google Scholar]
- Inagaki F, Takai K, Kobayashi H, Nealson KH, Horikoshi K. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing epsilon-proteobacterium isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int J Syst Evol Microbiol. 2003;53:1801–1805. doi: 10.1099/ijs.0.02682-0. [DOI] [PubMed] [Google Scholar]
- Jannasch HW.1995Microbial interactions with hydrothermal fluidsIn: Humphris SE, Zierenberg RA, Mullineaux LS, Thomson RE (eds)Seafloor Hydrothermal Systems: Physical, Chemical, Biological and Geological Interactions American Geophysical Union: Washington, DC; 273–296. [Google Scholar]
- Karl DM.1995Ecology of free-living, hydrothermal vent microbial communitiesIn: Karl DM (ed)The Microbiology of Deep-Sea Hydrothermal Vents CRC Press, Inc.: Boca Raton, FL; 35–124. [Google Scholar]
- Kern M, Mager AM, Simon J. Role of individual nap gene cluster products in NapC-independent nitrate respiration of Wolinella succinogenes. Microbiology. 2007;153:3739–3747. doi: 10.1099/mic.0.2007/009928-0. [DOI] [PubMed] [Google Scholar]
- Kern M, Simon J. Characterization of the NapGH quinol dehydrogenase complex involved in Wolinella succinogenes nitrate respiration. Mol Microbiol. 2008;69:1137–1152. doi: 10.1111/j.1365-2958.2008.06361.x. [DOI] [PubMed] [Google Scholar]
- Kern M, Simon J. Electron transport chains and bioenergetics of respiratory nitrogen metabolism in Wolinella succinogenes and other Epsilonproteobacteria. Biochim Bioph Acta. 2009a;1787:646–656. doi: 10.1016/j.bbabio.2008.12.010. [DOI] [PubMed] [Google Scholar]
- Kern M, Simon J. Periplasmic nitrate reduction in Wolinella succinogenes: cytoplasmic NapF facilitates NapA maturation and requires the menaquinol dehydrogenase NapH for membrane attachment. Microbiology. 2009b;155:2784–2794. doi: 10.1099/mic.0.029983-0. [DOI] [PubMed] [Google Scholar]
- Le Bris N, Govenar B, Le Gall C, Fisher CR. Variability of physico-chemical conditions in 9 degrees 50′ N EPR diffuse flow vent habitats. Mar Chem. 2006;98:167–182. [Google Scholar]
- Lentner C. Geigy Scientific Tables. Ciba-Geigy Limited: Basel; 1984. Composition of blood; p. 81. [Google Scholar]
- Lin JT, Stewart V. Nitrate assimilation by bacteria. Adv Microb Physiol. 1998;39:379. doi: 10.1016/s0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- Lopez-Garcia P, Duperron S, Philippot P, Foriel J, Susini J, Moreira D. Bacterial diversity in hydrothermal sediment and epsilonproteobacterial dominance in experimental microcolonizers at the Mid-Atlantic Ridge. Environ Microbiol. 2003;5:961–976. doi: 10.1046/j.1462-2920.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- Luther GW, 3rd, Rozan TF, Taillefert M, Nuzzio DB, Di Meo C, Shank TM, et al. Chemical speciation drives hydrothermal vent ecology. Nature. 2001;410:813–816. doi: 10.1038/35071069. [DOI] [PubMed] [Google Scholar]
- Miller WG, Parker CT, Rubenfield M, Mendz GL, Wosten MM, Ussery DW, et al. The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One. 2007;2:e1358. doi: 10.1371/journal.pone.0001358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millero FJ.1996Micronutrients in the oceans Chemical Oceanography2nd ednCRC Press, Inc.: Boca Raton, FL; 281–305. [Google Scholar]
- Miroshnichenko ML, Kostrikina NA, L'Haridon S, Jeanthon C, Hippe H, Stackebrandt E, et al. Nautilia lithotrophica gen. nov., sp nov., a thermophilic sulfur-reducing epsilon-proteobacterium isolated from a deep- sea hydrothermal vent. Int J Syst Evol Microbiol. 2002;52:1299–1304. doi: 10.1099/00207713-52-4-1299. [DOI] [PubMed] [Google Scholar]
- Miroshnichenko ML, L'Haridon S, Schumann P, Spring S, Bonch-Osmolovskaya EA, Jeanthon C, et al. Caminibacter profundus sp. nov., a novel thermophile of Nautiliales ord. nov. within the class ‘Epsilonproteobacteria', isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2004;54:41–45. doi: 10.1099/ijs.0.02753-0. [DOI] [PubMed] [Google Scholar]
- Moreno-Vivian C, Cabello P, Martinez-Luque M, Blasco R, Castillo F. Prokaryotic nitrate reduction: molecular properties and functional distinction among bacterial nitrate reductases. J Bacteriol. 1999;181:6573–6584. doi: 10.1128/jb.181.21.6573-6584.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takai K, Inagaki F, Horikoshi K, Sako Y. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the epsilon-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int J Syst Evol Microbiol. 2005;55:925–933. doi: 10.1099/ijs.0.63480-0. [DOI] [PubMed] [Google Scholar]
- Nakagawa S, Takaki Y, Shimamura S, Reysenbach AL, Takai K, Horikoshi K. Deep-sea vent ε-proteobacterial genomes provide insights into emergence of pathogens. Proc Natl Acad Sci USA. 2007;104:12146–12150. doi: 10.1073/pnas.0700687104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, et al. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature. 2000;403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez I, Bohnert KA, Cuebas M, Keddis R, Vetriani C. Detection and phylogenetic analysis of the membrane-bound nitrate reductase (Nar) in pure cultures and microbial communities from deep-sea hydrothermal vents. FEMS Microbiol Ecol. 2013;86:256–267. doi: 10.1111/1574-6941.12158. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez I, Grosche A, Massenburg L, Starovoytov V, Lutz RA, Vetriani C. Phorcysia thermohydrogeniphila gen. nov., sp nov., a thermophilic, chemolithoautotrophic, nitrate-ammonifying bacterium from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2012;62:2388–2394. doi: 10.1099/ijs.0.035642-0. [DOI] [PubMed] [Google Scholar]
- Perez-Rodriguez I, Ricci J, Voordeckers JW, Starovoytov V, Vetriani C. Nautilia nitratireducens sp. nov., a thermophilic, anaerobic, chemosynthetic, nitrate-ammonifying bacterium isolated from a deep-sea hydrothermal vent. Int J Syst Evol Microbiol. 2010;60:1182–1186. doi: 10.1099/ijs.0.013904-0. [DOI] [PubMed] [Google Scholar]
- Potter L, Angove H, Richardson D, Cole J. Nitrate reduction in the periplasm of gram-negative bacteria. Adv Microb Physiol. 2001;45:51–112. doi: 10.1016/s0065-2911(01)45002-8. [DOI] [PubMed] [Google Scholar]
- Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. Competition between Escherichia coli strains expressing either a periplasmic or a membrane-bound nitrate reductase: does Nap confer a selective advantage during nitrate-limited growth. Biochem J. 1999;344Pt 1:77–84. [PMC free article] [PubMed] [Google Scholar]
- Reysenbach A-L, Gotz D, Yernool D. Biodiversity of Microbial Life. Wiley-Liss, Inc.: New York, NY; 2002. Microbial diversity of marine and terrestrial thermal springs; pp. 345–421. [Google Scholar]
- Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ. Functional, biochemical and genetic diversity of prokaryotic nitrate reductases. Cell Mol Life Sci. 2001;58:165–178. doi: 10.1007/PL00000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievert SM, Scott KM, Klotz MG, Chain PS, Hauser LJ, Hemp J, et al. Genome of the epsilonproteobacterial chemolithoautotroph Sulfurimonas denitrificans. Appl Environ Microbiol. 2008;74:1145–1156. doi: 10.1128/AEM.01844-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski J, Munk C, Lapidus A, Ngatchou Djao OD, Lucas S, Glavina Del Rio T, et al. Complete genome sequence of Sulfurimonas autotrophica type strain (OK10) Stand Genomic Sci. 2010;3:194–202. doi: 10.4056/sigs.1173118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon J, Sanger M, Schuster SC, Gross R. Electron transport to periplasmic nitrate reductase (NapA) of Wolinella succinogenes is independent of a NapC protein. Mol Microbiol. 2003;49:69–79. doi: 10.1046/j.1365-2958.2003.03544.x. [DOI] [PubMed] [Google Scholar]
- Smith CJ, Nedwell DB, Dong LF, Osborn AM. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl Environ Microbiol. 2007;73:3612–3622. doi: 10.1128/AEM.02894-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Campbell BJ, Hanson TE, Zhang CL, Cary SC. Nautilia profundicola sp. nov., a thermophilic, sulfur-reducing epsilonproteobacterium from deep-sea hydrothermal vents. Int J Syst Evol Microbiol. 2008;58:1598–1602. doi: 10.1099/ijs.0.65435-0. [DOI] [PubMed] [Google Scholar]
- Suerbaum S, Josenhans C, Sterzenbach T, Drescher B, Brandt P, Bell M, et al. The complete genome sequence of the carcinogenic bacterium Helicobacter hepaticus. Proc Natl Acad Sci USA. 2003;100:7901–7906. doi: 10.1073/pnas.1332093100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y, et al. Isolation and phylogenetic diversity of members of previously uncultivated ε-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol Lett. 2003;218:167–174. doi: 10.1111/j.1574-6968.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- Takai K, Nealson KH, Horikoshi K. Hydrogenimonas thermophila gen. nov., sp. nov., a novel thermophilic, hydrogen-oxidizing chemolithoautotroph within the epsilon-Proteobacteria, isolated from a black smoker in a Central Indian Ridge hydrothermal field. Int J Syst Evol Microbiol. 2004;54:25–32. doi: 10.1099/ijs.0.02787-0. [DOI] [PubMed] [Google Scholar]
- Takai K, Suzuki M, Nakagawa S, Miyazaki M, Suzuki Y, Inagaki F, et al. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sulfurimonas denitrificans comb. nov. and emended description of the genus Sulfurimonas. Int J Syst Evol Microbiol. 2006;56:1725–1733. doi: 10.1099/ijs.0.64255-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetriani C, Speck MD, Ellor SV, Lutz RA, Starovoytov V. Thermovibrio ammonificans sp. nov., a thermophilic, chemolithotrophic, nitrate ammonifying bacterium from deep-sea hydrothermal vents. Int J Syst Evol Microbiol. 2004;54:175–181. doi: 10.1099/ijs.0.02781-0. [DOI] [PubMed] [Google Scholar]
- Vetriani C, Tran HV, Kerkhof LJ. Fingerprinting microbial assemblages from the oxic/anoxic chemocline of the Black Sea. Appl Environ Microbiol. 2003;69:6481–6488. doi: 10.1128/AEM.69.11.6481-6488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voordeckers J, Starovoytov V, Vetriani C. Caminibacter mediatlanticus sp. nov., a thermophilic, chemolithoautotrophic, nitrate ammonifying bacterium isolated from a deep-sea hydrothermal vent on the Mid-Atlantic Ridge. Int J Syst Evol Microbiol. 2005;55:773–779. doi: 10.1099/ijs.0.63430-0. [DOI] [PubMed] [Google Scholar]
- Voordeckers JW, Do M, Hügler M, Ko V, Sievert SM, Vetriani C. Culture dependent and independent analyses of 16S rRNA and ATP citrate lyase genes: a comparison of microbial communities from different black smoker chimneys on the Mid-Atlantic Ridge. Extremophiles. 2008;12:627–640. doi: 10.1007/s00792-008-0167-5. [DOI] [PubMed] [Google Scholar]
- Wang H, Tseng CP, Gunsalus RP. The napF and narG nitrate reductase operons in Escherichia coli are differentially expressed in response to submicromolar concentrations of nitrate but not nitrite. J Bacteriol. 1999;181:5303–5308. doi: 10.1128/jb.181.17.5303-5308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.