Abstract
Most of the lineages of bacteria have remained unknown beyond environmental surveys using molecular markers. Until the recent characterisation of several strains, the phylum Armatimonadetes (formerly known as ‘candidate division OP10') was a dominant and globally-distributed lineage within this ‘uncultured majority'. Here we report the first Armatimonadetes genome from the thermophile Chthonomonas calidirosea T49T and its role as a saccharide scavenger in a geothermal steam-affected soil environment. Phylogenomic analysis indicates T49T to be related closely to the phylum Chloroflexi. The predicted genes encoding for carbohydrate transporters (27 carbohydrate ATP-binding cassette transporter-related genes) and carbohydrate-metabolising enzymes (including at least 55 putative enzymes with glycosyl hydrolase domains) within the 3.43 Mb genome help explain its ability to utilise a wide range of carbohydrates as well as its inability to break down extracellular cellulose. The presence of only a single class of branched amino acid transporter appears to be the causative step for the requirement of isoleucine for growth. The genome lacks many commonly conserved operons (for example, lac and trp). Potential causes for this, such as dispersion of functionally related genes via horizontal gene transfer from distant taxa or recent genome recombination, were rejected. Evidence suggests T49T relies on the relatively abundant σ-factors, instead of operonic organisation, as the primary means of transcriptional regulation. Examination of the genome with physiological data and environmental dynamics (including interspecific interactions) reveals ecological factors behind the apparent elusiveness of T49T to cultivation and, by extension, the remaining ‘uncultured majority' that have so far evaded conventional microbiological techniques.
Keywords: Armatimonadetes, Chthonomonas calidirosea, genome assembly, phylogenomics, soil, thermophiles
Introduction
Despite a growing scientific and industrial drive to cultivate and characterise the ‘uncultured majority', the bulk of known microbial diversity has evaded attempts at cultivation (Keller and Zengler, 2004; Fox, 2005). In fact, microbial strains from just five divisions of bacteria (Actinobacteria, Firmicutes, Proteobacteria, Bacteroidetes and Cyanobacteria) represent 95% of cultivated species (Keller and Zengler, 2004) and 88% of sequenced genomes (Liolios et al., 2010). Yet, cultured strains represent only a minor fraction of the global microbial and genetic diversity (Whitman et al., 1998; Wu et al., 2009a). This is a significant knowledge gap and fundamentally limits our ability to understand ecological and biogeochemical processes, and to discover novel proteins with biotechnological value (Wu et al., 2009a). The importance of investigating more representatives from undersampled phyla or first cultivars from phyla with no cultivated representatives (candidate divisions) can therefore not be overstated. Here we provide the first genomic analysis of Chthonomonas calidirosea T49T, the first cultivated strain of the newly described phylum Armatimonadetes (formerly candidate division OP10) and use these data to speculate on its ecological role.
Candidate division OP10 was a dominant bacterial candidate phylum first detected in a 16S rRNA gene survey of Obsidian Pool, Yellowstone National Park (Hugenholtz et al., 1998). Until recently, this division had no cultivated strains, yet contained just over 600 full-length phylotypes in the SILVA database version 114 (Pruesse et al., 2007). Phylotypes grouping within candidate division OP10 were detected in a wide range of environments, including soil, geothermal springs, freshwater sediments and bioreactors (Bond et al., 1995; Hugenholtz et al., 1998; Lehours et al., 2007; Stott et al., 2008; Dunfield et al., 2012), suggesting a broad range of metabolic capabilities and ecological roles. The first reported cultivated strains of candidate division OP10, strains T49T and P488, were isolated from geothermally heated soils in New Zealand (Stott et al., 2008). To date, three species from the Armatimonadetes have been formally described: C. calidirosea T49T (Lee et al., 2011), Fimbriimonas ginsengisoli GSoil348T (Im et al., 2012) and the phylum type species, Armatimonas rosea YO-36T (Tamaki et al., 2011). These strains are only distantly related, with 16S rRNA gene sequence similarities of <80% (Dunfield et al., 2012).
A phenotypic comparison of the three cultivated strains identifies a number of common traits; all strains are chemoheterotrophic with a carbohydrate-based metabolism, strictly aerobic, Gram-negative and pink-rose pigmented. However, in contrast to the mesophilic and neutrophilic A. rosea and F. ginsengisoli, T49T is a thermophile (Topt 68 °C, range: 50–73 °C) and a moderate acidophile (growth pH 5.3, range: pH 4.7–5.8). The three strains differ in mol% G+C content, cell morphology, fatty acid content, quinone and salt tolerance (Lee et al., 2011, Tamaki et al., 2011, Im et al., 2012; ). In addition, T49T appears to have a much broader carbohydrate utilisation range, including a partially cellulolytic phenotype; it can hydrolyse amorphous polysaccharides but not linear polysaccharides (Lee et al., 2011).
Here we present the genomic analysis of T49T and use these data along with experimental work to provide insight into the metabolism and ecology of this strain. These data allow us to infer its ecological role and speculate on possible reasons for the rarity in detection and cultivation of Armatimonadetes species.
Materials and methods
Genomic DNA extraction
The type strain T49T (DSM 23976T=ICMP 18418T) was grown for 7 days on solid medium (AOM1, pH 6.2) at 60 °C (Stott et al., 2008). Cell biomass was collected, washed seven times in sterile water to remove gellan gum and exopolysaccharides, and freeze dried. Total dry cell weight was ∼10 mg. Cells were resuspended in 500 μl TE buffer with 30 μl 10% SDS and 20 μl lysozyme at 37 °C and 700 r.p.m. for 1 h before proteinase K (100 μl) was added and incubation was continued overnight. Genomic DNA was obtained by phenol:chloroform extraction. RNA was removed with RNAse A. The product was re-extracted with phenol:choloform, precipitated with ethanol and acetate, and resuspended in 10 mM TE buffer.
DNA sequencing
A paired-end (8 kb) Titanium chemistry library of the T49T genome was constructed and then sequence through 454 GS-FLX system (Roche, Branford, CT, USA), which generated 171 649 reads with an average size of 400 bp. The reads assembled into three scaffolds (N50=3456 kb) and 60 contigs (N50=162 kb) using Newbler and resulted in 20 × sequencing coverage. A single draft scaffold was assembled from the contigs with the assistance of the paired-end data, and the remaining gaps were closed by primer walking and Sanger sequencing. The genomic data has been submitted to GenBank/EMBL/DDBJ databases under BioProject PRJEB1573 and accession number HF951689.
Genome annotation and analysis
Gene prediction, annotation and additional analysis were performed using the Integrated Microbial Genome-Expert Review (Markowitz et al., 2010) pipeline; anomalies in gene prediction were manually curated with the assistance of GenePRIMP (Pati et al., 2010). Carbohydrate-active enzymes were identified by hidden Markov model profiles through Integrated Microbial Genome-Expert Review and externally cross-referenced with MetaCyc (Caspi et al., 2012) and the CAZy database (Cantarel et al. 2009). Putative CRISPRs were identified via the CRISPRFinder webserver (Grissa et al., 2007) and σ-factors were identified and categorised based on the hidden Markov model profile hits of conserved σ-factor regions.
Putative genes from horizontal gene transfer (HGT) were identified using Alien_hunter (Vernikos and Parkhill, 2006) using a threshold of 12.015 and a window size of 5000 nt. BLASTX search of the output identified 357 open reading frames. Because of the large multigene windows used by Alien_hunter, not all candidate genes identified were HGT products. HGT was also assessed via IslandViewer (Langille and Brinkman, 2009). The package implements ‘IslandPath-DIMOB', a dinucleotide sequence composition bias and mobility genes analysis (Hsiao et al., 2005), and ‘SIGI-Hidden Markov Model', a codon usage hidden Markov model analysis (Waack et al., 2006). Finally, we utilised the deep phylogenetic roots of T49T and its lack of any known close neighbours to screen for candidate HGT genes by BLAST, selecting only genes with an E-value lower than 1e−100, as high similarity to genes in other phyla is unexpected for vertically transferred genes. In this way, both the compositional biases of the DNA (Alien_hunter) and sequence similarities (BLAST) were applied in order to identify the most likely putative HGT genes. A custom perl script was used to BLASTP search for top hits of all the predicted protein sequences against a ‘balanced' genome data set (Supplementary Table 1) to test the effect of sample size bias for Firmicutes in NCBI database.
Phylogenetic inference was conducted using PhyloPhlAN (Segata et al., 2013), which uses USEARCH (Edgar, 2010) and MUSCLE (Edgar, 2004) to identify and align the conserved proteins in a new genome against its built-in database, and FastTree (Price et al., 2010) to generate an approximate maximum-likelihood tree with local support values using Shimodaira–Hasegawa test (Shimodaira and Hasegawa, 1999). Pairwise genome comparisons were conducted by assigning predicted open reading frames from T49T to orthologous groups with the highest BLAST identity within the eggNOG 2.0 (Muller et al., 2010) database with a threshold of 1e−10. The resulting 2080 open reading frames with orthologous group assignment were compared with a subset of the database. All bacteria in the ‘core' eggNOG database belonging to the phyla Actinobacteria, Cyanobacteria, Aquificae, Thermotogae, Deinococcus-Thermus, Fusobacteria, Chloroflexi and Firmicutes (as Bacilli and Clostridia) were selected for comparison. A matrix of orthologous group occurrences was created for each genome and pairwise genome comparisons made using three similarity indices: presence–absence (Jacaard and Sørensen indices) or abundance (Bray–Curtis index; Legendre and Legendre, 1998).
Amino acid assimilation
The requirement of amino acid supplementation for growth was tested by growing T49T on a modified mineral salt medium FS1V (Stott et al., 2008) containing (g l−1): NH4Cl, 4.0; KH2PO4, 0.5; MgSO4.7H2O, 0.2; CaCl2, 0.1; and mannose, 3.0; and previously reported trace metal solutions. The medium was adjusted to pH 5.7 before sterilisation (121 °C, 15 psi, 15 min). Single amino acids (0.1 g l−1) were added aseptically via 0.22-μm filters after steam sterilisation. The liquid cultures were incubated in sealed bottles with 5:1 air to medium headspace at 60 °C and 180 r.p.m.
Carbon catabolite repression
Carbon catabolite repression was tested using the aforementioned modified FS1V without individual amino acid addition. Casamino acids (0.2 g l−1) and carbohydrate source(s) (3.0 g l−1) were added aseptically following steam sterilisation. Growth medium was supplemented with either a single or two-carbohydrate mixture. Single carbohydrate media (glucose, mannose, xylose, lactose or galactose) were used as controls for the baseline utilisation of the respective carbohydrates. Two-carbohydrate media utilised galactose, mannose, xylose or lactose with glucose in 1:1 mixtures (w/w; sum total 3.0 g l−1). Biomass generation was determined via optical density (600 nm) and carbohydrate utilisation via high-performance liquid chromatography.
Results and discussion
General genome characteristics
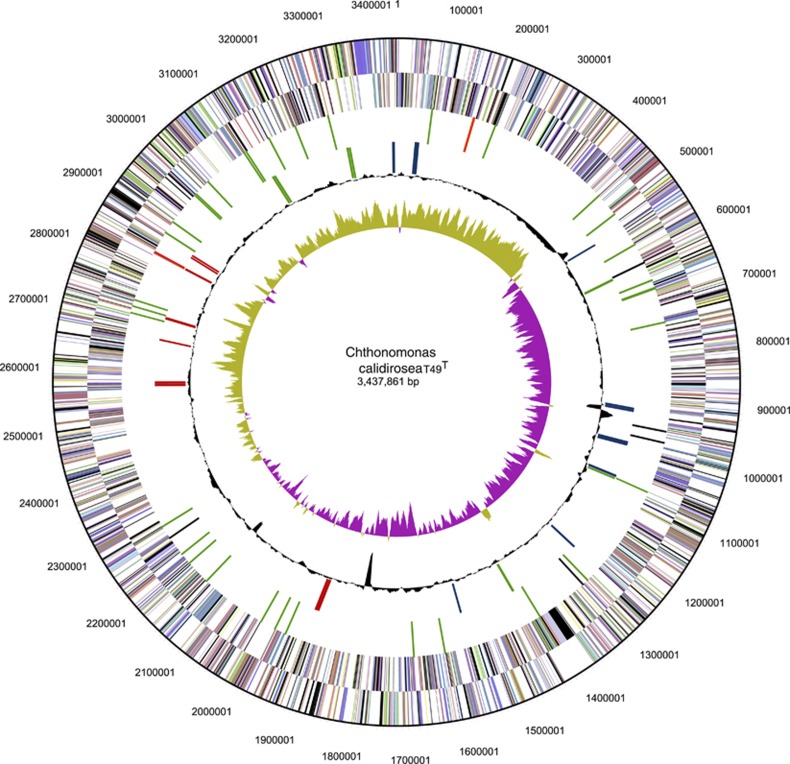
The T49T genome is a single circular chromosome of 3.43 Mb (Figure 1) with an average mol% G+C content of 54.4 and the total proportion of coding bases at 90.7%. The G+C content of the coding regions (90.1% of bases) was higher (55.4%) than that of the non-coding regions (49.1%). The genome contains 2877 predicted protein-coding genes; a functional prediction could be assigned for 2248 genes (∼78%), whereas 629 genes had no functional prediction of any kind. The putative origin of replication (OriC) was identified via DNA-Box-Complex using OriFinder (Gao and Zhang, 2008) and was in the vicinity of the GC skew minimum and adjacent to recF (CCALI_ 01940). The genome contained one complete rRNA operon (16S, 5S and 23S) and one additional copy of the 16S rRNA gene; both 16S rRNA genes (1416 bp) had identical nucleotide sequences. Forty-six transfer RNAs representing 43 anticodons were encoded in the genome (Supplementary Table 2).
Figure 1.
Circular representation of the C. calidirosea T49T genome. From outside to the centre: (1) genes on forward strand (colour by Clusters of Orthologous Groups categories); (2) genes on reverse strand (colour by Clusters of Orthologous Groups categories); (3) RNA genes (tRNAs green, rRNAs red, other RNAs black); (4) genes involved in: histidine biosynthesis (red), tryptophan biosynthesis (light green), purine biosynthesis (blue); (5) GC content; (6) GC skew.
A putative CRISPR element that consisted of a single spacer region (99 bp) and a pair of flanking direct repeats (46 bp each) was identified in the region 2 334 212–2 334 402. In addition, a cluster of CRISPR-associated proteins were found at a distant locus from the putative CRISPR. Interestingly, all the genes within the cluster (cmr1; CCALI_02398, cmr2; CCALI_02399 and CCALI_02400, and cmr3; CCALI_02401) appear to be pseudogenes due to frameshifts and premature stop codons.
Phylogenetic analysis
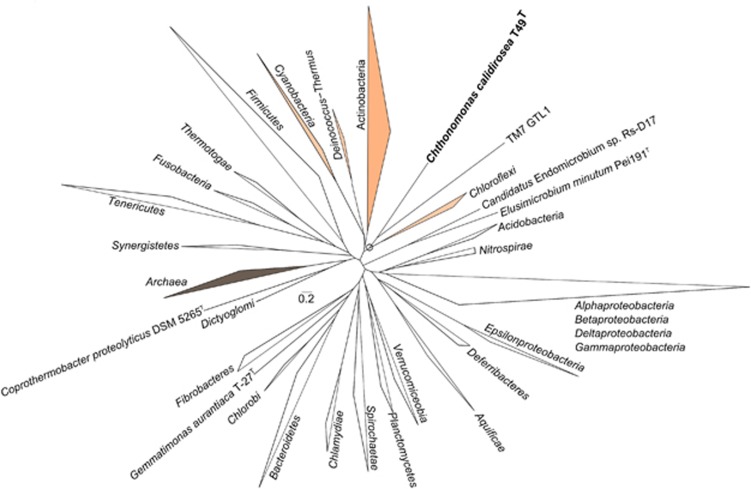
The placement of T49T within the phylum Armatimonadetes was previously investigated through 16S rRNA-based analysis (Lee et al., 2011). However, the 16S rRNA gene alone does not reliably resolve the relationships among deeply branching phyla (Delsuc et al., 2005). Here we utilised the recently reported phylogenetic pipeline ‘PhyloPhlAn' (Segata et al., 2013) to ensure a robust phylogenetic placement of T49T within the entire bacterial domain. The PhyloPhlAn pipeline presents an expansion in the scope of analysis, with the inclusion of both a large number (∼400) of highly conserved proteins and (currently 3737) microbial genomes. The analysis (Figure 2 and Supplementary Figure 1) confirmed previous findings (Dunfield et al., 2012) of the deeply branching nature of T49T/Armatimonadetes lineage and the strong association, and probably the common ancestry with the phylum Chloroflexi. In addition, the analysis confirmed our previously inferred cladal relationships with the other phyla, including Actinobacteria, Deinococcus-Thermus and Cyanobacteria. Interestingly, the closest phylogenetic neighbour of T49T was the partial genome sequence of an uncultivated representative of TM7 (Podar et al., 2007), a broadly distributed candidate division that has been detected in activated sludge, human oral cavities, soils and plant rhizospheres (Hugenholtz et al., 2001). Similarly to the three formally described Armatimonadetes, the recently sequenced TM7 metagenome had a saccharolytic-based metabolism (Albertsen et al., 2013). Finally, orthologous group-based functional similarity comparison with genomes from the most closely related phyla (see Materials and Methods) showed that the highest gene content similarities were with thermophiles of diverse phylogenetic backgrounds (Supplementary Table 3), indicating that T49T lacks high similarity to any other single described phylum, and thus represents a novel and distinct taxon.
Figure 2.
Unrooted tree representing the phylogenetic position of C. calidirosea T49T with major lineages (phyla) within the bacterial domain. The tree was constructed using PhyloPhlAn (Segata et al., 2013) with concatenated amino acid sequences of ∼400 conserved proteins among 3737 genomes. The circled node indicates the bifurcation between C. calidirosea T49T and TM7 GTL1 genomes with Chloroflexi, the closest other formal phylum (with SH-like local support value 85%). The scale bar represents 0.2 changes per amino acid position. The full phylogenetic tree is shown in Supplementary Figure 1.
Genome organisation
Many genes commonly found in conserved gene clusters such as the histidine, tryptophan and purine biosynthesis operons (Supplementary Figure 2) were scattered throughout the T49T genome (Figure 1). This scattering was not universal for all operon-like clusters, as genes encoding for flagella biosynthesis (CCALI_01237-44, 02077-85), coenzyme PQQ biosynthesis (CCALI_00346-48) and some ribosomal proteins (CCALI_02857-95) were present in conserved gene clusters. We examined the possibility that widespread HGT could explain the lack of commonly conserved operons in the genome. It has previously been reported (Davids and Zhang, 2008) that compared with strain- and species-specific genes, HGT genes are the least likely to be found in operons. However, very little HGT was detected in the T49T genome. Only 74 putative HGT genes (Supplementary Table 4) in small islands consisting of up to 7 genes were identified via DNA compositional bias using Alien_hunter, and IslandViewer detected no genomic islands. According to their Clusters of Orthologous Groups categories (Tatusov et al., 2003), most of the identified genes encode proteins involved in DNA replication, recombination and repair, and carbohydrate transport and metabolism, although there were also a number of hypothetical proteins. On the basis of protein sequence similarities, the putative sources of these genes appear to be a variety of bacterial groups. With the exception of Actinobacteria, the neighbouring phyla of Armatimonadetes (Chloroflexi, Deinococcus-Thermus and Cyanobacteria) appeared to be well represented. However, the lack of other Armatimonadetes genomes hinders the confirmation of candidate HGT genes, as genuine HGT events can be difficult to distinguish from highly conserved genes. Indeed, on further examination, most of the putative HGT genes identified via sequence similarity and/or DNA compositional bias analysis, including those most similar to homologues from distant phyla such as Aquificae (CCALI_00187) and Proteobacteria (CCALI_00606), appeared to be a result of conserved vertical inheritances instead of recent HGT events. These genes showed equally high similarity with homologues from a wide range of bacterial phyla. There were two notable exceptions: a type I restriction-modification system methyltransferase subunit (CCALI_00805) and an adenine-specific DNA methylase (CCALI_00449), which were both most similar to homologues to the firmicute Candidatus ‘Desulforudis audaxviator' (Chivian et al., 2008). These two putative HGT genes shared exceptionally high similarity scores (64% and 71% identity) with their best hits (accession numbers: YP_001716579 and YP_001716684) and significantly lower similarity scores (32% and 45% identity) in subsequent hits, suggesting a close phylogenetic relationship between the homologues rather than interphyla gene conservation.
Some bacterial phyla can be influenced by large-scale HGT from another phylum. This was demonstrated for the Thermotogae, which have apparently received many genes from Firmicutes (Zhaxybayeva et al., 2009). However, such a trend was not evident for T49T. When its inferred proteome was searched by BLASTP against the NCBI non-redundant protein database, most hits were to Firmicutes rather than to the phylogenetically closer Chloroflexi (Supplementary Table 1); however, this probably reflects a large bias in the database rather than extensive HGT with Firmicutes. Integrated Microbial Genome-Expert Review presently (August 2013) lists 1606 sequenced Firmicutes genomes and only 22 Chloroflexi genomes. BLASTP searches against a more balanced custom data set showed that most genes matched more closely to orthologues in Chloroflexi than Firmicutes, as expected based on the phylogenetic analyses (Supplementary Table 1). In addition, the eggNOG gene content analysis indicated that the mean gene content similarity of T49T to members of any other phylum was well below the mean similarity among members of that phylum (Supplementary Table 5). This suggests that T49T has a unique assemblage of genes and is not greatly influenced by HGT from another single phylum. In contrast, the Thermotogae show a much higher gene content similarity to Firmicutes than to other groups (Supplementary Table 5), in line with the theory that Thermotogae have been greatly affected by HGT from Firmicutes (Zhaxybayeva et al., 2009). These analyses support the view that the Armatimonadetes is a unique phylum with low affiliation to any other phylum. On the basis of all of these analyses, putative HGT events seem to be rare and are unlikely to explain the lack of gene clusters and conserved bacterial operons in the genome of T49T. The dispersal of genes is extensive, distributed through the genome, and does not correlate with the putative HGT sites.
Sigma factors
The genome of T49T contains a total of 30 σ-70-like proteins, including 22 extracytoplasmic function σ-factors (Supplementary Table 6), and has a high σ-factor to genome size (σ/Mb) ratio (Table 1). Diverse σ-factors have an important role in global transcriptional regulation by coordinating metabolic response genes, such as polysaccharide-degrading glycosyl hydrolases (GHs) and exopolysaccharide biosynthesis in Bacteroides thetaiotaomicron (Xu et al., 2004). Organisms lacking in commonly conserved gene clusters, such as the marine bacterium Pirelulla sp. strain 1 (Glöckner et al., 2003) and T49T, may rely more heavily on σ-factors to coordinate critical metabolic processes. Indeed, this hypothesis is supported by the identification of putative promoters and σ-70 family transcription factor binding sites of the dispersed histidine, tryptophan and purine biosynthesis genes (Supplementary Figure 3).
Table 1. A comparison of the number of σ-factors verses genome size for selected bacteria.
| Species | Genome size (MB) | No. of σ-factors | Ratio of no. of σ-factors to genome size |
|---|---|---|---|
| Escherichia coli (Blattner et al., 1997) | 4.7 | 18 | 3.8 |
| Pseudomonas aeruginosa (Potvin et al., 2008) | 6.3 | 24 | 3.8 |
| Solibacter usitatus (Ward et al., 2009) | 10 | 58 | 5.8 |
| Pirellula sp. (Glöckner et al., 2003) | 7.1 | 51 | 7.1 |
| Streptomyces coelicolor (Bentley et al., 2002) | 8.7 | 67 | 7.7 |
| Bacteroides thetaiotaomicron (Xu et al., 2003) | 6.3 | 54 | 8.6 |
| Chthonomonas calidirosea T49T | 3.4 | 30 | 8.8 |
| Nitrosomonas europaea (Chain et al., 2003) | 2.8 | 29 | 10.3 |
Bacterial strains isolated from soils (C. calidirosea T49T, S. usitatus and S. coelicolor) are included as a comparison with model organisms E. coli and P. aeruginosa.
Primary metabolism
T49T central metabolism and carbon fixation proceeds via routine glycolysis and the tricarboxylic acid cycle. Alpha- and β-D-glucose enter the glycolysis pathway via a glucose-6-phosphate isomerase and glucokinase. Acetyl CoA is synthesised from pyruvate via a complete pyruvate dehydrogenase complex, although the two copies of complex E3 (dihydrolipoamide dehydrogenase- CCALI_00365 and 00724) that convert dihydrolipoamide-E to lipoamide-E are located distantly from the other complex-encoding genes (CCALI_01202-04). Succinyl CoA is generated via isocitrate dehydrogenase and 2-oxoglutarate dehydrogenase complex.
T49T has previously been reported as exhibiting obligate chemoheterotrophic metabolism (Lee et al., 2011). In addition to these previous cultivation experiments, we predicted and experimentally confirmed its ability to utilise several less-common carbohydrates and associated derivatives, including sorbitol and galactan. Superoxide dismutase (CCALI_01521), catalase (CCALI_02206) and a complete oxidative phosphorylation pathway with a F-type ATPase were also identified, supporting the observed aerobic phenotype with no evidence of complete dissimilatory anaerobic respiration or fermentation pathways. No genes relating to photosynthesis were found in the genome, and genes related to major carbon fixation pathways were limited and fragmented. However, a complete pathway for anaplerotic CO2 fixation appears to be in place, proceeding via the reaction of phosphoenolpyruvate with CO2 by phosphoenolpyruvate carboxylase to form oxalacetate in a manner similar to the Wood–Werkman reaction. We have previously observed that growth of C. calidirosea strain P488 was improved when CO2 was supplemented into the aerobic headspace during growth (Stott et al., 2008). Although phosphoenolpyruvate carboxylase is a critical enzyme for the first step of carbon fixation in C4 and crassulacean acid metabolism plants (O'Leary and Diaz, 1982), the role of phosphoenolpyruvate carboxylase here appears to be limited to an anaplerotic role in maintaining the balance of intermediary molecules within the tricarboxylic acid cycle (Dunn, 2011); this is logical as the fixation of carbon dioxide is energetically unfavourable compared with the organic carbon substrates from which C. calidirosea T49T is capable of deriving energy.
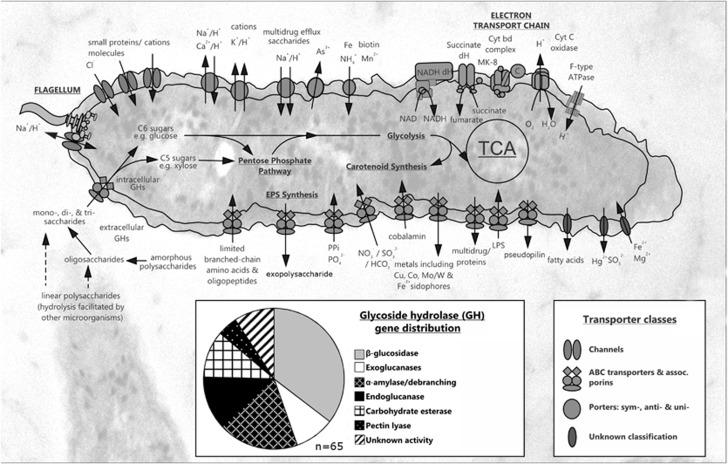
Nitrogen uptake by T49T was experimentally determined to be via ammonia assimilation and/or amino acid uptake (Lee et al., 2011). It appears that ammonia can be assimilated via glutamate or glycine and modified by a complete tetrahydrofolate one-carbon pathway. T49T possesses two ammonia permeases (CCALI_00383 and 01636). Interestingly, genes encoding both assimilatory nitrite reductase components (nirA and nirD) were identified, but no other genes encoding for nitrate dissimilation or assimilation, nor aerobic or anaerobic ammonia oxidation were found. We experimentally tested the assimilation of nitrite and nitrate, but no positive growth was detected, suggesting that the putative nitrite reductase genes may instead function as a means for sulphite/sulphate uptake. Previous growth experiments also showed that T49T obligately requires supplementary amino acid addition in the form of either yeast extract or casamino acids. No conclusive pathway deficiencies were identified. However, the requirement for supplementary amino acids may be a result of regulation difficulties or loss-of-function mutations rather than defunct pathways. The genome of T49T encodes only two amino acid transporters (Figure 3): a branched amino acid ATP-binding cassette (ABC) transporter complex (CCALI_00418-20) and a second oligopeptide transporter complex (CCALI_01041-45) proximal to an isoleucine synthetase. We tested various amino acid combinations in basal nutrient medium, including the branched amino acids valine, leucine and isoleucine, and determined that isoleucine alone can meet the amino acid requirement of T49T for growth. As a side note, we believe that these observations highlight the limitations of non-cultivation-based genome sequencing and associated genome prediction that has become prevalent since the advent of high-throughput sequencing technologies. An in silico-only-based analysis of the genome would have had difficulty identifying this metabolic deficiency and any resulting cultivation attempts based on a non-cultivation-based genome assembly would have been unsuccessful based on the lack of supplementary amino acids in the enrichment medium.
Figure 3.
The metabolic pathways of C. calidirosea T49T illustrating predicted channels, ABC transporters and associated proteins, symporters, antiporters and uniporters, and membrane transport proteins of unknown classification. A wide range of putative intracellular and extracellular GHs have been identified. The large number of carbohydrate ABC transporters (n⩾27) highlight scavenging capability C. calidirosea T49T for soluble carbohydrates, assisted by GH hydrolysis. The component sugars lead to the hexose and pentose pathways, glycolysis, tricarboxylic acid cycle and the electron transport chain. The high copy number of prepilin genes (n=57) may reflect the exportation of exopolysaccharide and formation of biofilm by C. calidirosea T49T.
Secondary metabolism features
An interesting feature of T49T is its growth within a narrow pH range, from 4.7 to 5.8 (Lee et al., 2011). This may result from a limited capacity to buffer cytoplasmic pH against external variation. A K+ transporting ATPase complex (CCALI_01552-54) may serve to reduce the electrical component (ΔΨ) of the proton motive force and facilitate the acidophilic phenotype. However, as expected from its narrow pH tolerance range, nearly all of the well-known inducible pH homeostasis mechanisms (Jain and Sinha, 2009; Krulwich et al., 2011) were missing from the genome. There was no evidence of glutamate, arginine or lysine decarboxylases for dealing with acid stress, nor of a urease system, agmatine deiminase or malolactic fermentation. The genome lacks any clear hydrogenases for proton removal with the possible exception of CCALI_00215, a protein of unknown function that contains several putative hydrogenase-like homologue domains. T49T cannot grow on acetate or lactate and lacks acetyl-CoA synthetase; hence, small organic acids might easily uncouple the membrane potential. There is no tryptophanase to compensate for alkali stress. However, T49T does have several other amino acid deaminases and two NhaP-type Na+/H+ (or K+/H+) antiporters (CCALI_00262 and 02512) that may respond to alkali stress. In general, the genome reflects a bacterium adapted to a pH-stable environment, with limited ability to respond to pH changes.
Another feature of T49T is the pink/orange pigmentation of cells associated with pellicle formation in aqueous medium (Lee et al., 2011). No pigmentation or pellicle formation has been noted in lag and exponential phase growth (including growth on solid medium). However, in stationary or decline phase, cells become pigmented and aggregate rapidly. This change can be brought about by low oxygen saturation, or more commonly by medium acidification due to carbohydrate hydrolysis, with a concomitant increase in carotenoid production. We have identified multiple genes related to carotenoid biosynthesis, including two phytoene desaturases (CCALI_00263 and 00231), a phytoene/squalene synthetase (CCALI_01286), a ζ-carotene desaturase (CCALI_01316) and a putative chlorobactene glucosyltransferase (CCALI_02059). Similar to many other metabolic features of the genome, these carotenoid biosynthesis genes were not arranged in operons as seen in other bacterial species. The spectrophotometric profile of the T49T acetone carotenoid extract is similar to that of the primary Thermomicrobium roseum carotenoid, oscillaxanthin (Jackson et al., 1973; Wu et al., 2009b). However, no homologues were identified for the 1,1′ hydroxylase-acyl transferase fusion gene of T. roseum, nor for cruF and cruD, the non-fusion equivalents in Salinibacter ruber, which have a key role in the modification of lycopene to oscillaxanthin. In addition, a wide array of genes present in the genome appears to be related to the production of exopolysaccharides, consistent with the observed pellicle formation. An operon-like gene cluster starting with exopolysaccharide synthesis protein (CCALI_01270) was identified. The cluster contains two polysaccharide export-related proteins (CCALI_01265 and 01269), a glycosyltransferase (CCALI_01268), an EpsI family protein (CCALI_01266) and a transmembrane exosortase (CCALI_01267). Together in a putative operon, these components bear some functional resemblance to known exopolysaccharide operons such as the ‘eps' genes in Bacillus subtilis (Nagorska et al., 2010). Additional exopolysaccharide-related genes were also identified outside this cluster, including exopolysaccharide biosynthesis polyprenyl glycosylphosphotransferase (CCALI_02642) and two putative alginate export channels (CCALI_00999 and 02397).
Regulation of carbohydrate metabolism
Carbohydrate utilisation experiments were conducted to test carbon catabolite repression in T49T. Growth was tested in various combinations of two-carbohydrate media (glucose+mannose, glucose+galactose, glucose+lactose and glucose+xylose). No diauxic shifts were observed and carbohydrates were simultaneously utilised in all of the carbohydrate combinations tested (Supplementary Figure 4), indicating a lack of carbon catabolite repression and non-diauxic growth. Non-diauxic growth has been observed previously in Sulfolobus acidocaldarius and Thermoanaerobacter thermohydrosulfuricus, and was attributed to the lack of a carbohydrate phosphotransferase system (PTS; Cook et al., 1993, 1994; Joshua et al., 2011). The T49T genome also lacks any transporter genes associated with PTS. Many other microbial species (including most Archaea) lack the PTS system (Koning et al., 2002; Silva et al., 2005), but the prevalence of diauxic versus non-diauxic growth as a physiological trait within the bacterial and archaeal domains is currently not well described. Archaeal and bacterial species without PTS transporters employ alternative carbohydrate transporter systems, including ABC transporters, and secondary transporters such as the major facilitator superfamily and major intrinsic protein (Koning et al., 2002; Silva et al., 2005; Joshua et al., 2011). These alternative transport systems are thought to be less involved in the regulation of metabolism and transcription than PTS (Saier, 2001). Eleven major facilitator superfamily and 99 ABC transporter-related genes have been identified in the T49T genome (Supplementary Table 7). Of the 99 ABC transporter genes identified, at least one quarter (27 genes) are putatively involved in carbohydrate transport. The presence of these alternative carbohydrate transporters may explain the concurrent importation of carbohydrates in the absence of PTS transporters, and thus the lack of diauxic growth in the two-carbohydrate conditions tested. Major facilitator superfamily transporters were less abundant than ABC transporters, with only nine genes related to carbohydrate transport. No major intrinsic protein transporters were identified.
Carbohydrate-active enzymes
The genome contains at least 65 genes encoding carbohydrate-active enzymes, including GHs, cellulose esterases and a pectin lyase (Supplementary Table 8), but excluding glycosyl transferases. The GHs belong to 26 described GH families; a further 16 putative carbohydrate-active enzymes were not associated with currently described families from the CAZy database (Cantarel et al., 2009).
Previous metabolic characterisation of T49T demonstrated that it can grow on a broad array of simple hemicellulosic (C5) and cellulosic (C6) carbohydrates, including monosaccharides, oligosaccharides and amorphous polysaccharides (for example, starch, xylan, glucomannan, pectin and carboxymethyl cellulose; Lee et al., 2011). In contrast, cells were unable to hydrolyse linear polysaccharides such as cotton, Avicel or lignocellulosic pulp preparations. The hydrolysis of linear or crystalline cellulose typically requires the synergistic action of endoglucanases, exoglucanases and β-glucosidases in order to obtain complete degradation (Lynd et al., 2002; Wang et al., 2011). In particular, endoglucanases from GH families 9 and/or 48 appear to be obligately required for linear cellulose hydrolysis (Tolonen et al., 2009; Olson et al., 2010), although GH families 5, 6, 12, 44, 45 and 74 have also been reported to be active against polysaccharides (Kaoutari et al., 2013). In agreement with T49T's observed carbohydrate hydrolysis activity, few putative endoglucanase GHs were detected. Of these, seven endoglucanases/mannanases (GH5, 6 and 44) and three β-1,4-xylanases (GH10) were detected and confirmed the utilisation capability of glucomannan and components of xylan for growth. In addition, three pectin lyases and two families of cellulose esterases were detected and these most likely act on plant cell wall polysaccharides. However, consistent with observations that T49T frequently lacks operon-based genome organisation, we were unable to identify polysaccharide utilisation loci previously reported (Martens et al., 2011) as essential for the degradation of plant pectins. The presence of endoglucanase-acting mannanases, xylanases and pectin lyases may reflect an adaption to the elevated mannan and hemicellulose contents of New Zealand native plants and exotic pines, which are abundant in the geothermal locations from which other Chthonomonas strains were isolated (Stott et al., 2008).
Nearly two-thirds of the annotated T49T GHs were related to β-glucosidases, exoglucanases and/or de-branching/starch-hydrolysing activities (Supplementary Table 8). These enzymes include β-galactosidases (GH families 2 and 42; 10 copies), α-arabinofuranosidases (GH family 51; 4 copies), and α- and β-amylases (GH families 13, 14 and 57; 6 copies). The high number of genes encoding these enzymes and the lack of crystalline cellulose-degrading endoglucanases suggests that C. calidirosea T49T may rely on the cellulolytic activity of other microorganisms to supply carbohydrate oligomers. A similar scenario may also apply during the degradation of the pectin and xylan components of complex plant wall polysaccharides, as we were unable to identify the full complement of genes reportedly required for complete hydrolysis. Thus, it is reasonable to infer that T49T does not act as a primary biomass degrader in its host ecosystem, but rather forms consortia with other soil-based cellulolytic bacteria to allow for complete hydrolysis.
Inferred ecology
In combination with the results of previous community and physiological studies (Stott et al., 2008; Lee et al., 2011; Dunfield et al., 2012), the genomic analysis presented here has allowed us to more accurately define the ecological role of T49T and provided some insight into the ecology of the phylum Armatimonadetes. Physiological data, characteristics of carbohydrate-active enzymes and a large array of carbohydrate transporters, all indicate that T49T has a chemoheterotrophic carbohydrate-based primary metabolism that targets soluble/amorphous carbohydrates.
The inability of T49T to directly utilise the complex and predominantly plant-based polysaccharides that are the primary carbohydrate source in its host environment suggests that it occupies its ecosystem niche as a scavenger, relying on the diffusion of various hydrolysis products generated by cellulose digesters, and possibly even acting as an oxygen barrier for cellulolytic anaerobes such as Clostridium thermocellum. In addition, T49T may contribute to a mutualistic relationship by removing C5 sugars (components of hemicelluloses, which many cellulolytic species cannot utilise) from the environment, thus facilitating further cellulose degradation. The regulation of carbohydrate utilisation in T49T does not appear to function via the standard well-described operon-based models of Escherichia coli and B. subtilis, but may reflect an ecological role as a scavenger in an environment in which the spectrum of available carbohydrate species is highly heterogeneous, in low concentration and in flux. Such environments would be detrimental to metabolisms adapted to maximise the utilisation of a single carbon source over others. Despite the relative lack of operons, the high abundance of σ-factors suggests that T49T can coordinate functionally related but dispersed genes to rapidly respond to ecosystem fluxes. In particular, the numerous extracytoplasmic function σ-factors may have a significant role in environment sensing (Butcher et al., 2008) and facilitation of biofilm formation (Bordi and de Bentzmann, 2011). Similarly to many other thermophilic aerobic bacteria, T49T produces carotenoids, likely to be regulated through stress response σ-factors in response to oxidative stress caused by the hostile environment. Surprisingly, despite the propensity for HGT within soil and biofilm environments, T49T showed few obvious signs of HGT. The genome topology based on GC skew gave no evidence of recent genomic rearrangement, despite T49T having a genome frequently lacking in typically conserved operon structures.
The physiology of T49T suggests a tight coupling with its environment due to its narrow pH range and specific nutrient (carbohydrate and branched amino acids) requirements. In addition, geothermal environments within the suitable pH range (∼4–5) for T49T are uncommon, as typical geochemical interactions cause a bimodal distribution of pH in geothermal features, with peaks at pH 2 and 7 (Brock, 1971). The combination of the fastidiousness of T49T and the rarity of suitable environments may explain why the species evaded isolation for almost a decade following the identification of the first phylotype within Armatimonadetes/candidate division OP10 (Hugenholtz et al., 1998).
Conclusion
Our analysis of the T49T genome has revealed adaptations of a thermophilic bacterium in a complex geothermal soil–biofilm environment, as well as the first glimpse into the genomic landscape of the novel phylum Armatimonadetes. Currently, T49T is the only complete Armatimonadetes genome that is publically available, and it has undergone extensive automated and manual improvement to remove gaps and resolve problematic regions in order to conform to the definition of ‘improved high-quality draft' (Chain et al., 2009). The prospect of additional Armatimonadetes genomic data (in particular the two known characterised species, A. rosea YO-36T (Tamaki et al., 2011) and F. ginsengisoli GSoil348T (Im et al., 2012)), provides an exciting opportunity to further understand common genomic features and evolution of this newly described phylum.
Acknowledgments
KCL was supported by the Sarah Beanland Scholarship and MS by the FRST grant C05 × 0801. PD and IT were supported by an NSERC Discovery Grant (355790, PD). We acknowledge Jean F Power for her technical work on gap resolving through primer walking.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–538. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Blattner FR, Plunkett G, Bloch CA, Perna NT, Burland V, Riley M, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- Bond PL, Hugenholtz P, Keller J, Blackall LL. Bacterial community structures of phosphate-removing and non-phosphate-removing activated sludges from sequencing batch reactors. Appl Environ Microbiol. 1995;61:1910–1916. doi: 10.1128/aem.61.5.1910-1916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordi C, de Bentzmann S. Hacking into bacterial biofilms: a new therapeutic challenge. Ann Intens Care. 2011;1:19. doi: 10.1186/2110-5820-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock TD. Bimodal distribution of pH values of thermal springs of the world. Geol Soc Am Bull. 1971;82:1393. [Google Scholar]
- Butcher BG, Mascher T, Helmann JD.2008Environmental sensing and the role of extracytoplasmic function sigma factorsIn: El-Sharoud WM, (ed)Bacterial Physiology Springer-Verlag: Berlin/Heidelberg, Germany; 233–261. [Google Scholar]
- Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 2009;37:D233–D238. doi: 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspi R, Altman T, Dreher K, Fulcher CA, Subhraveti P, Keseler IM, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2012;40:D742–D753. doi: 10.1093/nar/gkr1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain P, Grafham DV, Fulton RS, Fitzgerald MG, Hostetler J, Muzny D, et al. Genome project standards in a new era of sequencing. Science. 2009;326:236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chain P, Lamerdin J, Larimer F, Regala W, Lao V, Land M, et al. Complete genome sequence of the ammonia-oxidizing bacterium and obligate chemolithoautotroph Nitrosomonas europaea. J Bacteriol. 2003;185:2759–2773. doi: 10.1128/JB.185.9.2759-2773.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D, Brodie EL, Alm EJ, Culley DE, Dehal PS, DeSantis TZ, et al. Environmental genomics reveals a single-species ecosystem deep within Earth. Science. 2008;322:275–278. doi: 10.1126/science.1155495. [DOI] [PubMed] [Google Scholar]
- Cook GM, Janssen PH, Morgan HW. Uncoupler-resistant glucose uptake by the thermophilic glycolytic anaerobe Thermoanaerobacter thermosulfuricus (Clostridium thermohydrosulfuricum) Appl Environ Microbiol. 1993;59:2984–2990. doi: 10.1128/aem.59.9.2984-2990.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GM, Janssen PH, Russell JB, Morgan W. Dual mechanisms of xylose uptake in the thermophilic bacterium Thermoanaerobacter thermohydrosulfuricus. FEMS Microbiol Lett. 1994;116:257–262. [Google Scholar]
- Davids W, Zhang Z. The impact of horizontal gene transfer in shaping operons and protein interaction networks—direct evidence of preferential attachment. BMC Evol Biol. 2008;8:23. doi: 10.1186/1471-2148-8-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Dunfield PF, Tamas I, Lee KC, Morgan XC, McDonald IR, Stott MB. Electing a candidate: a speculative history of the bacterial phylum OP10. Environ Microbiol. 2012;14:3069–3080. doi: 10.1111/j.1462-2920.2012.02742.x. [DOI] [PubMed] [Google Scholar]
- Dunn MF. Anaplerotic function of phosphoenolpyruvate carboxylase in Bradyrhizobium japonicum USDA110. Curr Microbiol. 2011;62:1782–1788. doi: 10.1007/s00284-011-9928-y. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fox JL. Current topics: ribosomal gene milestone met, already left in dust. ASM News. 2005;71:6–7. [Google Scholar]
- Gao F, Zhang C-T. Ori-Finder: a web-based system for finding oriCs in unannotated bacterial genomes. BMC Bioinformatics. 2008;9:79. doi: 10.1186/1471-2105-9-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, et al. Complete genome sequence of the marine planctomycete Pirellula sp. strain 1. Proc Natl Acad Sci USA. 2003;100:8298–8303. doi: 10.1073/pnas.1431443100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007;35:W52–W57. doi: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao WWL, Ung K, Aeschliman D, Bryan J, Finlay BB, Brinkman FSL. Evidence of a large novel gene pool associated with prokaryotic genomic islands. PLoS Genet. 2005;1:e62. doi: 10.1371/journal.pgen.0010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugenholtz P, Tyson GW, Webb RI, Wagner AM, Blackall LL. Investigation of candidate division TM7, a recently recognized major lineage of the domain Bacteria with no known pure-culture representatives. Appl Environ Microbiol. 2001;67:411–419. doi: 10.1128/AEM.67.1.411-419.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im W-T, Hu Z-Y, Kim K-H, Rhee S-K, Meng H, Lee S-T, et al. Description of Fimbriimonas ginsengisoli gen. nov., sp. nov. within the Fimbriimonadia class nov., of the phylum Armatimonadetes. Antonie van Leeuwenhoek. 2012;102:307–317. doi: 10.1007/s10482-012-9739-6. [DOI] [PubMed] [Google Scholar]
- Jackson TJ, Ramaley RF, Meinschein WG. Thermomicrobium, a new genus of extremely thermophilic bacteria. Int J Syst Bacteriol. 1973;23:28–36. [Google Scholar]
- Jain P, Sinha S. Neutrophiles: acid challenge and comparison with acidophiles. Internet J Microbiol. 2009;7:1. [Google Scholar]
- Joshua CJ, Dahl R, Benke PI, Keasling JD. Absence of diauxie during simultaneous utilization of glucose and xylose by Sulfolobus acidocaldarius. J Bacteriol. 2011;193:1293–1301. doi: 10.1128/JB.01219-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaoutari AE, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Keller M, Zengler K. Tapping into microbial diversity. Nat Rev Microbiol. 2004;2:141–150. doi: 10.1038/nrmicro819. [DOI] [PubMed] [Google Scholar]
- Koning SM, Albers S-V, Konings WN, Driessen AJM. Sugar transport in (hyper)thermophilic archaea. Res Microbiol. 2002;153:61–67. doi: 10.1016/s0923-2508(01)01289-x. [DOI] [PubMed] [Google Scholar]
- Krulwich TA, Sachs G, Padan E. Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol. 2011;9:330–343. doi: 10.1038/nrmicro2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langille MGI, Brinkman FSL. IslandViewer: an integrated interface for computational identification and visualization of genomic islands. Bioinformatics. 2009;25:664–665. doi: 10.1093/bioinformatics/btp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KCY, Dunfield PF, Morgan XC, Crowe MA, Houghton KM, Vyssotski M, et al. Chthonomonas calidirosea gen. nov., sp. nov., an aerobic, pigmented, thermophilic micro-organism of a novel bacterial class, Chthonomonadetes classis nov., of the newly described phylum Armatimonadetes originally designated candidate division OP10. Int J Syst Evol Microbiol. 2011;61:2482–2490. doi: 10.1099/ijs.0.027235-0. [DOI] [PubMed] [Google Scholar]
- Legendre P, Legendre LFJ.1998Numerical Ecology(2nd edn).Elsevier: Amsterdam, The Netherlands [Google Scholar]
- Lehours AC, Evans P, Bardot C, Joblin K, Gerard F. Phylogenetic diversity of Archaea and Bacteria in the anoxic zone of a meromictic lake (Lake Pavin, France) Appl Environ Microbiol. 2007;73:2016–2019. doi: 10.1128/AEM.01490-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liolios K, Chen IM, Mavromatis K, Tavernarakis N, Hugenholtz P, Markowitz VM, et al. The Genomes On Line Database (GOLD) in 2009: status of genomic and metagenomic projects and their associated metadata. Nucleic Acids Res. 2010;38:D346–D354. doi: 10.1093/nar/gkp848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markowitz VM, Chen I-MA, Palaniappan K, Chu K, Szeto E, Grechkin Y, et al. The integrated microbial genomes system: an expanding comparative analysis resource. Nucleic Acids Res. 2010;38:D382–D390. doi: 10.1093/nar/gkp887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, et al. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9:e1001221. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagorska K, Ostrowski a, Hinc K, Holland IB, Obuchowski M. Importance of eps genes from Bacillus subtilis in biofilm formation and swarming. J Appl Genet. 2010;51:369–381. doi: 10.1007/BF03208867. [DOI] [PubMed] [Google Scholar]
- O'Leary MH, Diaz E. Phosphoenol-3-bromopyruvate. A mechanism-based inhibitor of phosphoenolpyruvate carboxylase from maize. J Biol Chem. 1982;257:14603–14605. [PubMed] [Google Scholar]
- Olson DG, Tripathi SA, Giannone RJ, Lo J, Caiazza NC, Hogsett DA, et al. Deletion of the Cel48S cellulase from Clostridium thermocellum. Proc Natl Acad Sci USA. 2010;107:17727–17732. doi: 10.1073/pnas.1003584107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pati A, Ivanova NN, Mikhailova N, Ovchinnikova G, Hooper SD, Lykidis A, et al. GenePRIMP: a gene prediction improvement pipeline for prokaryotic genomes. Nat Methods. 2010;7:455–457. doi: 10.1038/nmeth.1457. [DOI] [PubMed] [Google Scholar]
- Podar M, Abulencia CB, Walcher M, Hutchison D, Zengler K, Garcia JA, et al. Targeted access to the genomes of low-abundance organisms in complex microbial communities. Appl Environ Microbiol. 2007;73:3205–3214. doi: 10.1128/AEM.02985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potvin E, Sanschagrin F, Levesque RC. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol Rev. 2008;32:38–55. doi: 10.1111/j.1574-6976.2007.00092.x. [DOI] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, et al. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saier MH. The bacterial phosphotransferase system: structure, function, regulation and evolution. J Mol Microbiol Biotechno. 2001;3:325–327. [PubMed] [Google Scholar]
- Segata N, Börnigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- Silva Z, Sampaio M, Henne A, Böhm A, Gutzat R, Boos W, et al. The high-affinity maltose/trehalose ABC transporter in the extremely thermophilic bacterium Thermus thermophilus HB27 also recognizes sucrose and palatinose. J Bacteriol. 2005;187:1210–1218. doi: 10.1128/JB.187.4.1210-1218.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stott MB, Crowe MA, Mountain BW, Smirnova AV, Hou S, Alam M, et al. Isolation of novel bacteria, including a candidate division, from geothermal soils in New Zealand. Environ Microbiol. 2008;10:2030–2041. doi: 10.1111/j.1462-2920.2008.01621.x. [DOI] [PubMed] [Google Scholar]
- Tamaki H, Tanaka Y, Matsuzawa H, Muramatsu M, Meng X-Y, Hanada S, et al. Armatimonas rosea gen. nov., sp. nov., of a novel bacterial phylum, Armatimonadetes phyl. nov., formally called the candidate phylum OP10. Int J Syst Evol Microbiol. 2011;61:1442–1447. doi: 10.1099/ijs.0.025643-0. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolonen AC, Chilaka AC, Church GM. Targeted gene inactivation in Clostridium phytofermentans shows that cellulose degradation requires the family 9 hydrolase Cphy3367. Mol Microbiol. 2009;74:1300–1313. doi: 10.1111/j.1365-2958.2009.06890.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernikos GS, Parkhill J. Interpolated variable order motifs for identification of horizontally acquired DNA: revisiting the Salmonella pathogenicity islands. Bioinformatics. 2006;22:2196–2203. doi: 10.1093/bioinformatics/btl369. [DOI] [PubMed] [Google Scholar]
- Waack S, Keller O, Asper R, Brodag T, Damm C, Fricke WF, et al. Score-based prediction of genomic islands in prokaryotic genomes using hidden Markov models. BMC Bioinformatics. 2006;7:142. doi: 10.1186/1471-2105-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Qi M, Barboza P, Leigh MB, Ungerfeld E, Selinger LB, et al. Isolation of high-quality total RNA from rumen anaerobic bacteria and fungi, and subsequent detection of glycoside hydrolases. Can. J. Microbiol. 2011;57:590–598. doi: 10.1139/w11-048. [DOI] [PubMed] [Google Scholar]
- Ward NL, Challacombe JF, Janssen PH, Henrissat B, Coutinho PM, Wu M, et al. Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils. Appl Environ Microbiol. 2009;75:2046–2056. doi: 10.1128/AEM.02294-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ. Prokaryotes: the unseen majority. Proc Natl Acad Sci USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D, Raymond J, Wu M, Chatterji S, Ren Q, Graham JE, et al. Complete genome sequence of the aerobic CO-oxidizing thermophile Thermomicrobium roseum. PLoS One. 2009;4:e4207. doi: 10.1371/journal.pone.0004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, et al. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- Xu J, Chiang HC, Bjursell MK, Gordon JI. Message from a human gut symbiont: sensitivity is a prerequisite for sharing. Trends Microbiol. 2004;12:21–28. doi: 10.1016/j.tim.2003.11.007. [DOI] [PubMed] [Google Scholar]
- Zhaxybayeva O, Swithers KS, Lapierre P, Fournier GP, Bickhart DM, DeBoy RT, et al. On the chimeric nature, thermophilic origin, and phylogenetic placement of the Thermotogales. Proc Natl Acad Sci USA. 2009;106:5865–5870. doi: 10.1073/pnas.0901260106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.