Abstract
Background & objectives:
Malassezia species implicated with dandruff vary at different geographical locations. The present study was conducted to determine the spectrum and distribution of Malassezia species in dandruff patients and healthy individuals.
Methods:
Patients with dandruff from northern (Chandigarh) and southern (Manipal, Karnataka) parts of India (50 each) and healthy individuals (20) were included in the study. Dandruff severity was graded as mild, moderate and severe. Malassezia spp. isolated were quantified and identified by phenotypic characters and molecular methods including PCR-RFLP and DNA sequencing.
Results:
Number of Malassezia spp. retrieved was significantly higher (P<0.001) in dandruff cases (84%) as compared to healthy individuals (30%). Isolation of Malassezia spp. was significantly higher (P<0.01) in patients from southern India. In moderately severe cases M. restricta was single most predominant (37.8%) isolate from patients of northern part of India and M. furfur (46.4%) from patients of southern part of India. Malassezia density was significantly associated with the severity of dandruff (P<0.001).
Interpretation & conclusions:
Our results on a limited number of individuals show that Malassezia spp. associated with dandruff varies in different regions of the country and the density of yeasts increases with severity of disease.
Keywords: Dandruff, epidemiology, Malassezia, molecular identification
Dandruff is a common persistent, relapsing inflammatory condition affecting the areas rich in sebaceous glands1. The condition is characterized by the flaky white to yellowish scales seen on the scalp and less frequently on the nasolabial folds, behind the ears, eyebrows, and intertriginous areas. The prevalence of dandruff in population varies between 30-95 per cent2. Besides the discomfort, this disorder is also socially embarrassing and affects the self esteem of the patient3. Malassezia species play a role in pathogenesis of this condition along with stress, fatigue, weather extremes, oily nature of skin, use of shampoos, immunosuppressed status (AIDS), and neurological disorders. M. restricta and M. globosa are generally considered to be the causative agents of dandruff4; the species implicated vary depending on geographical location of the host. M. furfur, M. sympodialis, M. obtusa, and M. slooffiae are the other species associated with this condition5. Though cases of dandruff are widely prevalent in India3, the prevalence of this disease, associated factors including Malassezia species are not studied in Indian population. The present study was conducted to determine the spectrum of Malassezia spp. causing dandruff and existing as commensal on the scalp of healthy individuals at two geographical locations (northern and southern parts of India). In addition, features associated with dandruff were also studied.
Material & Methods
This study was conducted in the departments of Medical Microbiology and Dermatology, Venerology & Leprosy, Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh in collaboration of department of Microbiology, Kasturba Medical College, Manipal, Karnataka, India.
One hundred patients (50 each from northern and southern part of India) and 20 healthy individuals (10 each from northern and southern part of India) without features of dandruff (controls) were enrolled in this study. The study was conducted during September 2011 to August 2012. Based on the presence or absence of visible flakes over the scalp, the study population was categorized as dandruff patients or healthy individuals, respectively. The dandruff patients were defined as individuals having inflammation of the scalp in the form of red, scaly, itchy and flaking rash2. The study protocol was approved by the Institute's Ethics Committee of PGIMER, Chandigarh. The samples were collected after obtaining written informed consent of the patients. Study area was elected on the basis of convenience to perform field study. The patients from northern part of India belonged to two villages viz., Dhuhar (total population ~2500) and Nial (total population ~2400) of Punjab State, whereas patients from southern part of India belonged to two villages viz., Karki (total population ~2800) and Manki (total population 3000) of coastal Karnataka. The distance between the two study areas is approximately 2000 km. The climate of chosen area in southern part of India is hot with high humidity almost all through the year (temperature ~22-35°C, humidity ~65-90%), northern part of India is dry and hot (temperature ~35-45°C, humidity ~40-45%) in summer and cold (temperature ~5-15 °C, humidity ~70-80%) in winter. Sampling was carried out at convenience by random screening for the dandruff cases in the selected villages. Both children and adults (3-50 yr) were included in the study. On the day of sampling, the participants were advised not to wash their scalp. Patients and controls applying topical antifungals and/or steroid to the scalp preceding a month of sampling were not enrolled. The scalp hair were categorized as either oily or dry on visual observation. Hair fall of the patients was recorded as mild, moderate and excess on the basis of self assessment of the patients. In addition, use of cleansing agents (bath soap or shampoo), applying/not applying oil to the scalp, type of oil applied and its frequency were recorded. Dandruff severity was graded as mild, moderate and severe on the basis of size, colour and surface of the flakes or scales. The severity score was defined as mild (score up to 24), moderate (25-60) and severe (>60) dandruff6,7.
Samples were collected from relatively severely affected areas of the patients. Flakes or scales were collected by partitioning the hair with a sterile comb and scrapping approximately one inch area using blunt scalpel. In healthy individuals, the scrapings were collected from vertex and temporal regions. The samples were inoculated over the surface of modified Dixon's agar slants8, transferred to the laboratory on the same day and incubated at 30°C in a humid chamber up to four weeks. Colonies appearing like Malassezia were counted depending on the growth rate. Malassezia species isolated from southern region (Kasturba Medical College, Manipal) were transferred to PGIMER, Chandigarh for identification. At PGIMER all the isolates were identified on the basis of macroscopic and microscopic features (Fig. 1), biochemical, and physiological tests8,9.
Fig. 1.
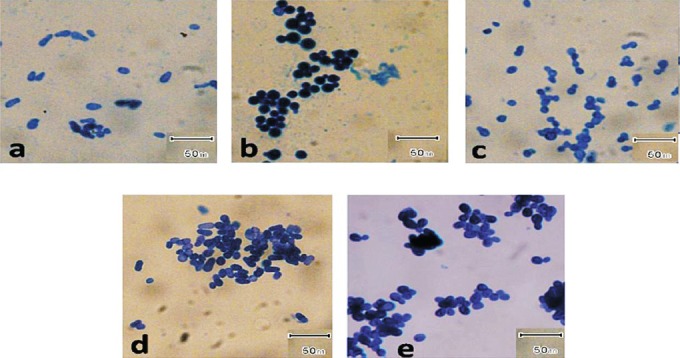
Photomicrographs of different Malassezia species stained by methylene blue. (a) M. furfur; (b) M. globosa; (c) M. restricta; (d) M. slooffiae; and (e) M. sympodialis. Magnification 1000X.
The identity of the isolates were further confirmed by PCR-RFLP (restriction fragment length polymorphisms) of ITS2 (internal transcribed spacer 2) region rDNA gene and DNA sequencing of 26s region of rDNA gene10,11. Standard strains of M. furfur (Microbial Type Culture Collection, Institute of Microbial Technology, Chandigarh, India, MTCC-1374), M. restricta (The Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands, CBS-7877), M. obtusa (CBS–7876), M. globosa (CBS-7966), M. pachydermatis (CBS–1879), M. slooffiae (CBS-7956), M. sympodialis (CBS-7222), M. japonica (CBS-9348), M. yamatoensis (CBS-9725) were used as controls. Genomic DNA isolation was performed by microwave irradiation technique12. Briefly, a colony of Malassezia yeast was transferred to a PCR tube and warmed three times for one minute each in a microwave. The supernatant was used as template for PCR analysis. For PCR- RFLP, amplification was done using the primers ITS3 (5’ GCATCGATGAAGAACGCAGC 3’) and ITS4 (5’ TCCTCCGCTTATTGATATGC 3’) (Sigma, Banglore). Enzymatic digestion was carried out using three restriction enzymes viz. Alu1, Ban1 and MspA1 (New England Biolabs, Ipswich, USA). The amplicons and enzyme mixture was incubated at 37°C for three hours. For each digestion reaction 12.5 μl of PCR product, 1.5 μl buffers and 1μl of corresponding enzyme were added. For each species, three separate reaction mixtures were made using three different restriction enzymes. The restriction patterns of clinical isolates were compared with band pattern of the standard strains of Malassezia (Table I and Fig. 2). Sequencing of the 26S rDNA was done by amplifying this region with universal primers NL-1 (5’-GCATATCAATAAGCGGAGGAAAAG) and NL-4 (5’-GGTCCGTGTTTCAAGACGG)11. Purification of amplified gene product was performed by gel extraction kit (QIAquick, Quiagen, Bangalore). The elute was used as the purified gene product for sequencing PCR. Sequencing PCR was performed for both the strands using above mentioned primers and Big Dye Terminator Cycle Sequencing Kit, Version 3.1 (Applied Biosystems, USA). All the sequencing reactions were purified and analyzed on ABI 3130 Genetic Analyzer (Applied Biosystems, USA). For each isolate, the consensus sequences were prepared from the sequence obtained by forward and reverse primers using Bionumerics software (version 6.5, Applied Maths, Ghent, Belgium). The consensus sequences were compared with the sequences of the GenBank DNA database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) and CBS-KNAW fungal biodiversity centre database (http://www.cbs.knaw.nl/).
Table I.
Amplified product size (bp) and expected length of restriction fragments (bp) obtained after digesting with different restriction enzymes
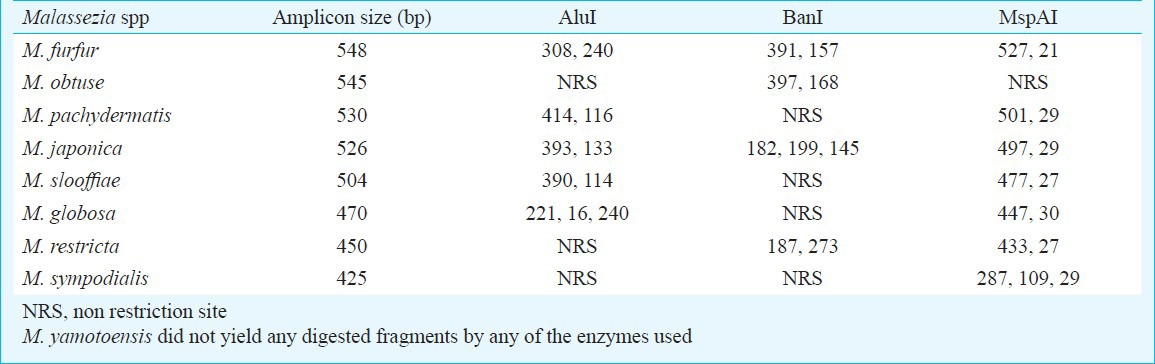
Fig. 2.
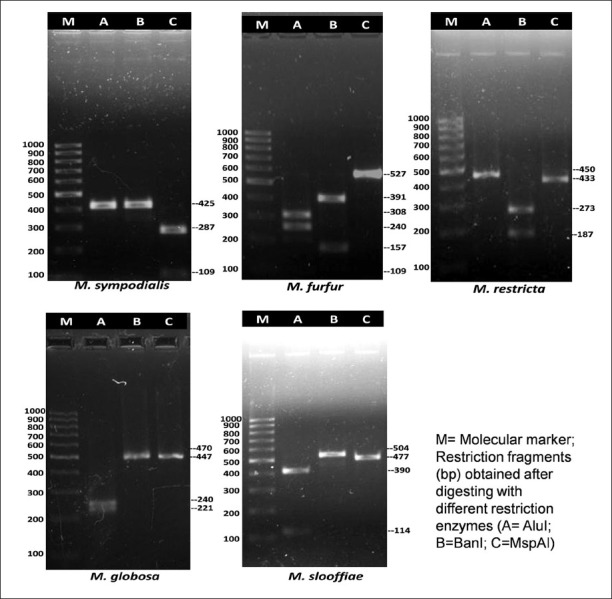
Agrose gel picture of PCR-RFLP profiles of few Malassezia species after digestion with restriction enzymes- AluI, BanI, MspAI.
Statistic analysis: Comparison of Malassezia spp. distribution based on severity, hair type and hair fall in different groups (mild, moderate and severe) of dandruff patients was done by Chi-squared test (Software- Epi Info™ 7.1.0.6 and OpenEpi version 2.3, Rolin School of Public Health, Emory University, Antlanta, Georgia, www.Openepi.com).
Results
Age of the patients varied between 3 and 50 years. Forty per cent of the patients were in the age range of 10 and 19 years and 34 per cent were between 20 and 29 years. Females were 61 per cent and males 39 per cent. Malassezia culture positivity rate was significantly higher in patients from southern India, compared to northern India (47 cases vs. 37 cases; P<0.01). M. furfur and M. restricta were isolated in significantly higher number (P<0.05) from patients of southern part of India as compared to patients from northern part of India (Table II). Of the 20 healthy individuals, Malassezia species could be isolated from 6 (30%) individuals from southern India, of whom 5 were M. furfur and one was M. slooffiae. None of the samples collected from the healthy individuals of northern India yielded Malassezia. The average number of colonies isolated from mild, moderate and severe dandruff cases from northern part of India was 4.5, 10.6 and 14.4 cfu, respectively, whereas from healthy individuals, mild and moderate dandruff cases of southern India was 6, 24 (samples from 5 patients had confluent growth and the colonies could not be counted) and 24 cfu (samples from 12 patients showed confluent growth), respectively. Isolation of Malassezia spp. among dandruff patients and healthy individuals using different cleansing agent and application of oil is given in Table III.
Table II.
Distribution of Malassezia spp. among dandruff patients according to geographical location, hair fall and severity
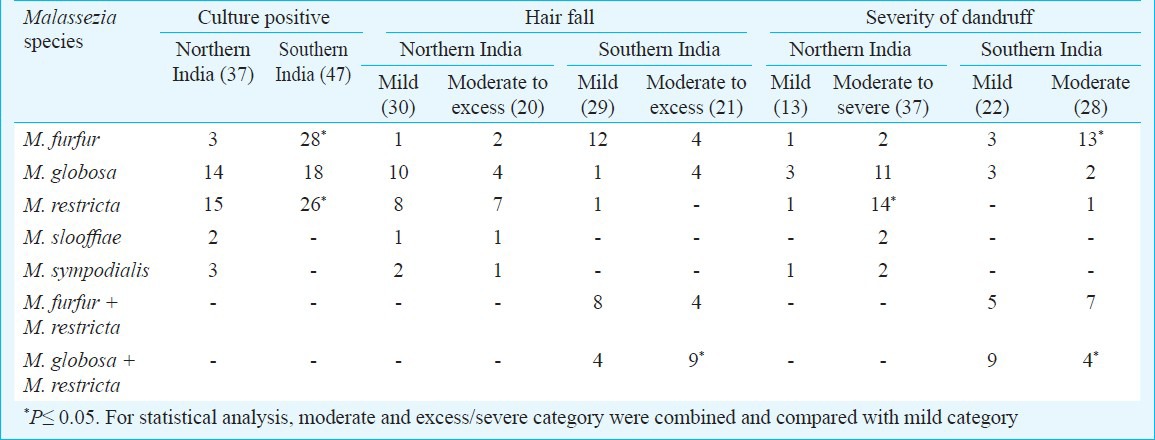
Table III.
Isolation of Malassezia spp. among dandruff patients and healthy individuals
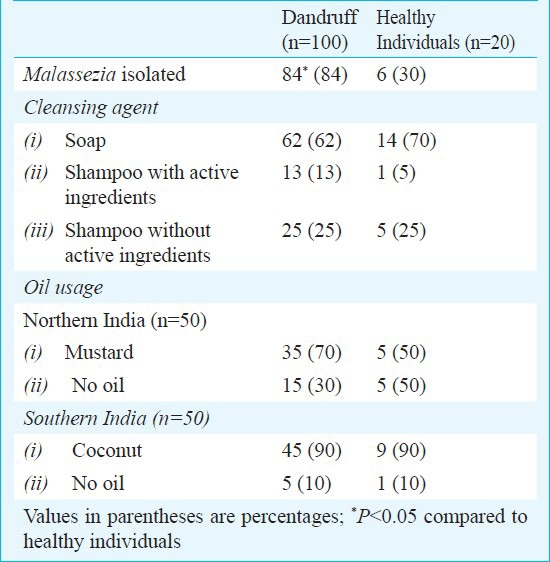
In northern India, M. globosa and M. restricta were isolated from all the age groups studied. In patients of southern India, M. furfur, M. globosa and M. restricta were almost equally distributed among all the age groups. Although statistically insignificant, in population of northern India, M. restricta was associated more in males (48%) and M. globosa in females (43%). In patients from southern India, M. furfur was predominantly isolated from male patients (60%) and mixture of M. globosa and M. restricta from females (32%). M. restricta was significantly isolated from majority (44.5%) of the patients having oily hair. The details of distribution of Malassezia spp. according to geographical location, hair fall and severity are given in Table II.
All the isolates were identified by the band pattern obtained after PCR-RFLP of ITS2 region and 37 of them were subjected to DNA sequencing. Representative sequences of the isolates are deposited in the GenBank database with the accession numbers JN651930 to JN651965 and JN642245. The isolates have been deposited at the National Culture Collection of Pathogenic Fungi, PGIMER, Chandigarh, India.
Discussion
In the present study, two geographically different locations were studied. Geographically, northern and southern regions of India are distinct in terms of environmental conditions. M. restricta and M. globosa were the two most prevalent species isolated from dandruff patients of northern India having moderate to severe lesions while M. furfur, M. restricta and M. globosa were either isolated singly or in mixture from patients from southern India. In Japan and USA, M. globosa is most frequently isolated from dandruff cases followed by M. restricta13. Whereas M. furfur and M. slooffiae are isolated from scalp of healthy individuals and are considered as commensal species. In the present study, the sample collection was carried out during the winter season (usually dry) from northern part of India and in summer from coastal region of southern India, which is humid
(~ 65-90%) all through the year. According to Pierard-Franchimont et al14 the severity of dandruff may fluctuate with seasons and often worsens in winter. High humidity may be the reason for occurrence of less severe cases from coastal part of southern India as compared to dry weather during winter in northern part of India. In addition, application of coconut oil in majority of the southern India patients (90%) might have prevented severe cases.
The isolation rate of Malassezia spp. is known to be highest in the age group of around twenty years when sebaceous glands activity is maximum15. Similar to the findings of Lee et al16, in the present study no significant age dependent variation of Malassezia species was noticed. This could be due to the relatively small number of the participants in our study.
Recovery of Malassezia species in culture usually depends on the culture medium and techniques used for its isolation. In a culture dependant study from Canada, Malassezia recovery rate was 82 per cent17. Similar studies from Sweden and Japan showed far less recovery rate. M.restricta was not isolated from any of these studies18,19. Tajima et al20 by using culture independent molecular technique showed that M. globosa and M. restricta were predominantly associated both with lesional and non-lesional area of Japanese SD/D patients. In the present study, isolation rate of Malassezia spp. was 74 per cent in northern India and 94 per cent from southern India patients.
Though association of dandruff is related to the species of Malassezia implicated, presence of this yeast in high number is also related in causation of dandruff. In a culture dependant study, Gupta et al17 showed higher number of Malassezia colony at non-lesional site than on the lesional site. But, in DNA based study Malassezia found at lesional site was higher in density than on non-lesional site20. In a few reports the quantity of yeasts was not found to be correlate positively with the severity of the disease4,21. In the present study, the higher colony count positively associated with the severity of dandruff. It is also important to note that accurate quantification of Malassezia on the scalp is difficult to perform as the Malassezia resides along with sebum at infandibulum of the hair follicle22,23. Hence possibly culture based technique is better than DNA based non-culture technique, as in the later only the surface of the skin is sampled without including hair infundibulum. Another disadvantage of DNA based non-culture technique is that the DNA from both viable and nonviable cells are amplified.
In our study, M. restricta was recovered from individuals having any type of hair (oily and dry). Severe hair loss was associated more with M. restricta and M. globosa than other Malassezia species, but the difference was not significant. This finding was in agreement with the study of Nematian et al24 where increased hair loss was shown to be associated with Pityrosporum ovale (M. globosa). Malassezia may cause hair loss by utilizing the lipids present in different strata of epidermis and dermis leading to weakening of hair root and hair fall25,26.
The major difficulty while conducting epidemiological studies on Malassezia associated disease is its isolation due to slow growing nature of this agent. A few species like M. globosa, M. restricta and M. obtusa take longer than a month to grow in sufficient amount to extract the DNA by phenol-chloroform extraction technique. In the present study, we used the rapid method of template preparation from the Malassezia colonies using microwave irradiation12. This technique is rapid, reliable, cost-effective, and simple. This technique could also be applied to amplify the DNA from a single isolated colony during the co-existence of multiple Malassezia species in a given culture.
In conclusion, M. globosa and M. restricta seemed to be the predominant species causing dandruff in Indian population and high density of these species on the scalp was related with the severity of dandruff. Small sample size was a major limitation of the present study. Such studies need to be done at various geographic locations in India with large carefully drawn sample.
Acknowledgment
Authors acknowledge the Indian Council of Medical Research, New Delhi, India for financial assistance. Authors also acknowledge Dr P.V.M. Lakshmi, Department of Community Medicine, PGIMER, Chandigarh and Shri Kapil Mukesh for their help in analysis and sample collection respectively.
References
- 1.Hay RJ, Graham-Brown RA. Dandruff and seborrhoeic dermatitis: causes and management. Clin Exp Dermatol. 1997;22:3–6. doi: 10.1046/j.1365-2230.1997.d01-231.x. [DOI] [PubMed] [Google Scholar]
- 2.Xu J, Saunders CW, Hu P, Grant RA, Boekhout T, Kuramae EE, et al. Dandruff-associated Malassezia genomes reveal convergent and divergent virulence traits shared with plant and human fungal pathogens. Proc Natl Acad Sci USA. 2007;104:18730–5. doi: 10.1073/pnas.0706756104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manuel F, Ranganathan S. A new postulate on two stages of dandruff: a clinical perspective. Int J Trichol. 2011;3:3–6. doi: 10.4103/0974-7753.82117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zisova LG. Malassezia species and seborrheic dermatitis. Folia Med (Plovdiv) 2009;51:23–33. [PubMed] [Google Scholar]
- 5.Gupta AK, Batra R, Bluhm R, Boekhout T, Dawson TL., Jr Skin diseases associated with Malassezia species. J Am Acad Dermatol. 2004;51:785–98. doi: 10.1016/j.jaad.2003.12.034. [DOI] [PubMed] [Google Scholar]
- 6.Loden M, Wessman C. The antidandruff efficacy of a shampoo containing piroctone olamine and salicylic acid in comparison to that of a zinc pyrithione shampoo. Int J Cosmet Sci. 2000;22:285–9. doi: 10.1046/j.1467-2494.2000.00024.x. [DOI] [PubMed] [Google Scholar]
- 7.Flutterer E. Evaluation of efficacy of antidandruff agents. J Soc Cosmet Chem. 1981;32:327–38. [Google Scholar]
- 8.Gueho E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996;69:337–55. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 9.Gueho E, Boekhout T, Ashbee HR, Guillot J, Van Belkum A, Faergemann J. The role of Malassezia species in the ecology of human skin and as pathogens. Med Mycol. 1998;36(Suppl 1):220–9. [PubMed] [Google Scholar]
- 10.Gaitanis G, Robert V, Velegraki A. Verifiable single nucleotide polymorphisms of the internal transcribed spacer 2 region for the identification of 11 Malassezia species. J Dermatol Sci. 2006;43:214–7. doi: 10.1016/j.jdermsci.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 11.Kurtzman CP, Robnett CJ. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5’ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol. 1997;35:1216–23. doi: 10.1128/jcm.35.5.1216-1223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim SM, Lim SH, Jung BR, Lee YW, Choe YB, Ahn KJ. The application of colony PCR in the molecular biological analysis of Malassezia yeasts. Korean J Med Mycol. 2007;12:180–8. [Google Scholar]
- 13.Sugita T, Velegraki A. Epidemiology of Malassezia-related skin diseases. In: Boekhout T, Gueho E, Mayser P, Velegraki A, editors. Malassezia and the skin. Berlin, Heidelberg: Springer-Verlag; 2010. pp. 65–120. [Google Scholar]
- 14.Pierard-Franchimont C, Pierard GE, Kligman A. Seasonal modulation of sebum excretion. Dermatologica. 1990;181:21–2. doi: 10.1159/000247853. [DOI] [PubMed] [Google Scholar]
- 15.Sugita T, Suzuki M, Goto S, Nishikawa A, Hiruma M, Yamazaki T, et al. Quantitative analysis of the cutaneous Malassezia microbiota in 770 healthy Japanese by age and gender using a real-time PCR assay. Med Mycol. 2010;48:229–33. doi: 10.1080/13693780902977976. [DOI] [PubMed] [Google Scholar]
- 16.Lee YW, Byun HJ, Kim BJ, Kim DH, Lim YY, Lee JW, et al. Distribution of Malassezia species on the scalp in korean seborrheic dermatitis patients. Ann Dermatol. 2011;23:156–61. doi: 10.5021/ad.2011.23.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta AK, Kohli Y, Summerbell RC, Faergemann J. Quantitative culture of Malassezia species from different body sites of individuals with or without dermatoses. Med Mycol. 2001;39:243–51. doi: 10.1080/mmy.39.3.243.251. [DOI] [PubMed] [Google Scholar]
- 18.Nakabayashi A, Sei Y, Guillot J. Identification of Malassezia species isolated from patients with seborrhoeic dermatitis, atopic dermatitis, pityriasis versicolor and normal subjects. Med Mycol. 2000;38:337–41. doi: 10.1080/mmy.38.5.337.341. [DOI] [PubMed] [Google Scholar]
- 19.Sandstrom Falk MH, Tengvall Linder M, Johansson C, Bartosik J, Back O, Sarnhult T, et al. The prevalence of Malassezia yeasts in patients with atopic dermatitis, seborrhoeic dermatitis and healthy controls. Acta Derm Venereol. 2005;85:17–23. doi: 10.1080/00015550410022276. [DOI] [PubMed] [Google Scholar]
- 20.Tajima M, Sugita T, Nishikawa A, Tsuboi R. Molecular analysis of Malassezia microflora in seborrheic dermatitis patients: comparison with other diseases and healthy subjects. J Invest Dermatol. 2008;128:345–51. doi: 10.1038/sj.jid.5701017. [DOI] [PubMed] [Google Scholar]
- 21.Clift DC, Dodd HJ, Kirby JD, Midgley G, Noble WC. Seborrheic dermatitis and malignancy. An investigation of the skin flora. Acta Derm Venereol. 1988;68:48–52. [PubMed] [Google Scholar]
- 22.Meyer LE, Otberg N, Tietz HJ, Sterry W, Lademann J. In vivo imaging of Malassezia yeasts on human skin using confocal laser scanning microscopy. Laser Phys Lett. 2005;2:148–52. [Google Scholar]
- 23.Schwartz JR, Shah R, Krigbaum H, Sacha J, Vogt A, Blume-Peytavi U. New insights on dandruff/seborrhoeic dermatitis: the role of the scalp follicular infundibulum in effective treatment strategies. Br J Dermatol. 2011;165(Suppl 2):18–23. doi: 10.1111/j.1365-2133.2011.10573.x. [DOI] [PubMed] [Google Scholar]
- 24.Nematian J, Ravaghi M, Gholamrezanezhad A, Nematian E. Increased hair shedding may be associated with the presence of Pityrosporum ovale. Am J Clin Dermatol. 2006;7:263–6. doi: 10.2165/00128071-200607040-00008. [DOI] [PubMed] [Google Scholar]
- 25.Pierard-Franchimont C, Hermanns JF, Degreef H, Pierard GE. From axioms to new insights into dandruff. Dermatology. 2000;200:93–8. doi: 10.1159/000018337. [DOI] [PubMed] [Google Scholar]
- 26.Pierard-Franchimont C, Xhauflaire-Uhoda E, Pierard GE. Revisiting dandruff. Int J Cosmet Sci. 2006;28:311–8. doi: 10.1111/j.1467-2494.2006.00326.x. [DOI] [PubMed] [Google Scholar]


