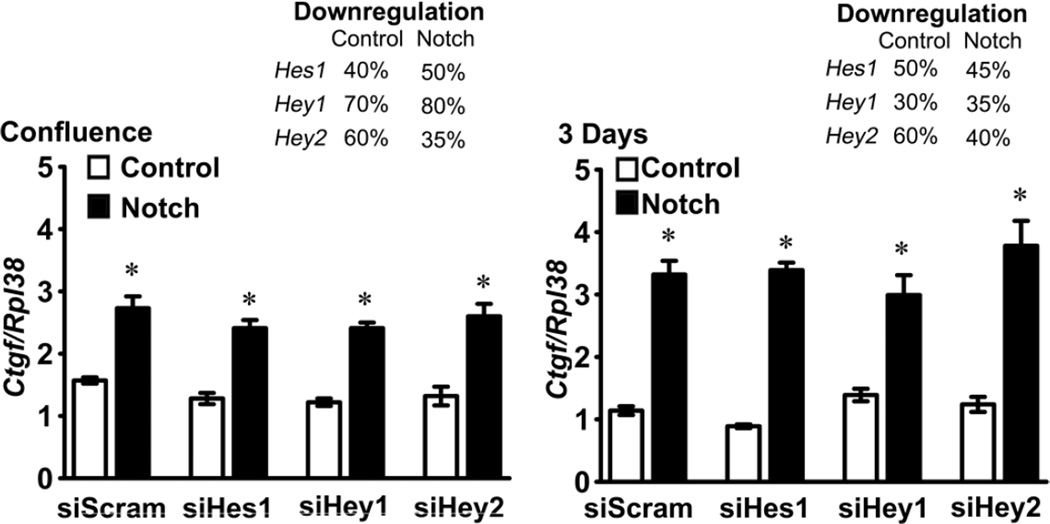
Figure 4.
Effect of Notch on Ctgf expression in the context of Hes1, Hey1 or Hey2 downregulation. Calvarial osteoblasts isolated from RosaNotch mice were transduced with Ad-CMV-Cre, to activate Notch (black bars), or with control Ad-CMV-GFP (white bars), transfected with Hes1, Hey1 or Hey2 small interfering RNA (si) or scrambled siRNA (siScram) and cultured to confluence or for 3 days after confluence. Total RNA was extracted, reverse transcribed and amplified by qRT-PCR. Data are expressed as copy number of Ctgf mRNA corrected for Rpl38. Downregulation of Hes1, Hey1 and Hey2 mRNA in control (Ad-CMV-GFP) and Notch activated (Ad-CMV-Cre) cells, expressed as the mean % of suppression relative to the mRNA expression in siScram cells is indicated in the right upper corners of both panels. Values are means ± SEM; n = 4. * Significantly different between siHes1, Hey1 or Hey2 and siScram, p < 0.05.