Abstract
Cervical cancer is the second malignancy in Mexico, little is known about the prognostic factors associated with this disease. Several cellular components are important in their transformation and progression. Alternative mRNA splice is an important mechanism for generating protein diversity, nevertheless, in cancer unknown mRNA diversity is expressed. Hyaluronan-mediated motility receptor (HMMR, RHAMM, CD168) is a family member of proteins, hyaluronan acid dependent, and has been associated with different malignant processes such as: angiogenesis, cell invasiveness, proliferation, metastasis and poor outcome in some tumors. In the present study we identified expression of HMMR in cervical cancer by means of RT-PCR and sequencing. Our results indicate co-expression of two HMMR variants in all samples, and one case expressed three alternative HMMR splice transcripts. These results showed the heterogeneity of mRNA transcripts of HMMR that could express in cancer and the expression of HMMR could be marker of malignancy in CC.
Keywords: Cervical cancer, alternative splicing, HMMR, expression
Introduction
Cervical cancer (CC) is one of the most common types of cancer worldwide, and in Mexico represents a leading cause of death in women over 25 years old, exhibiting an estimated incidence ~ 200,000 per year [1]. Human papillomavirus (HPV) infection is considered an etiological factor associated with CC, due to HPV oncoproteins that play a role in the dysregulation of the cell cycle, by interaction with pRB and TP53 suppressor proteins [2]. However, HPV infection is not an exclusive factor for cell transformation and progression. Several recent reports provide information of relevant contribution of other factors such as: cellular components, micro environmental, among others [3].
Alternative splicing (AS) is the most important mechanism involved in generation of protein diversity in eukaryotes. By bioinformatics methods, it has been estimated that over 90% of human genes will have two or more alternative mRNA variants [4]. However, there is little knowledge about alternative mRNA spliced expressed in diverse pathologies including cancer. There have been identified some mRNA spliced variants that have impact on cancer cell biology including control of cell proliferation, metabolism, angiogenesis, evasion of apoptosis, invasiveness, and metastasis [5,6]. Moreover, aberrant AS transcripts have been implicated in the clinical outcome of diverse cancer types such as: stomach, colon, breast, cervix, leukemia, and others [7,8].
Hyaluronan-mediated motility receptor (HMMR, RHAMM, CD168), is a family member of the hyaladherins proteins, and it is hyaluronan acid-dependent [9]. HMMR exhibits a variable cellular distribution, including the cytoskeleton [10], cell surface [11], mitochondria 12], and nucleus [10,13]. HMMR gene encodes four alternative proteins (725, 724, 709, and 638 aa), nevertheless, its roll in the cell is not clear. However HMMR expression has been associated with malignant processes including: angiogenesis [14,15], cell motility [16], poor outcome [17], proliferation and metastasis [16,18]; even though, most of them form part of the unknown functions in cancer. In this study we identified three mRNA alternative spliced of HMMR expressed in a case of cervical cancer.
Materials and methods
Biopsy sample, cell lines cultures and total RNA isolation
The cervical sample was obtained from the Oncology Department, General Hospital of Mexico SSA (Secretaría de Salud) from patients with diagnosis of cervical cancer. All samples were taken in accordance with the Helsinki Declaration and after informed consents were obtained. The samples were fractioned in two portions, then one was formalin-fixed and paraffin-embedded for posterior histopathology analysis; and the other was put in RNAlaterT solution (Qiagen, Valencia CA, USA) for total RNA isolation, and stored at -70°C. The diagnosis of carcinoma was confirmed with histological examination and the samples that presented > 80% of cervical squamous cells were recruited.
Furthermore, four cervical cell lines (CCL, as HeLa, SiHa, CasKi and CaLo) were included in the study. Cell cultures were grown in the Dulbecco’s Modified Eagle Medium (DMEM) with fetal bovine serum 10%, penicillin-streptomycin 100 U/mL Life technologyTM, in a 37°C incubator with a humidified atmosphere of 5% CO2 in air. For RNA isolation, tissue and cell lines were disrupted by means of the TissueLyser system (Qiagen, Valencia CA, USA) and total RNA was eluted using the RNeasy Mini kit (Qiagen, Valencia CA, USA). Total RNA was quantified using NanoDrop ND-1000 spectrophotometer and integrity was evaluated by means of agarose gel 1.5% (w/v) electrophoresis considering ribosomal subunits 28 s and 18 s.
HPV testing
Genomic DNA was isolated from tissue samples using Promega Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) in accordance with the manufacturer’s instructions. DNA quantity and purity was measured with a Nanodrop ND-1000 Spectrophotometer and the integrity was verified by 1% agarose gel (w/v) electrophoresis. DNA samples were subjected to HPV typing by PCR using E6F/E6R primers for HPV16 [19]. 100 ng of genomic DNA was taken and subjected to 40 amplification cycles as follows: 5 min at 94°C for pre-denaturation, 1 min at 94°C, 1 min at 44°C, 1 min at 72°C, and 5 min at 72°C as the final extension step. The PCR products were analyzed by 1% agarose gel and amplification size was 126 base pairs (bp).
cDNA synthesis and PCR
Total RNA was treated using DNase l, in brief, 2 μg total RNA was digested at 37°C for 30 min with 1U DNase l, in 1x DNase buffer. After 5 mM EDTA was added, it was incubated at 65°C for 10 min. Thereafter, the sample was placed in a master mix using the High Capacity cDNA Reverse Transcription Kit (Life Technologies, Carlsbad, CA, USA) according to the protocol. The cDNA was the endpoint in PCR amplification using housekeeping gene RPL4, according to previous reports [20], whereas HMMR was amplified with the following primers: 5′ ATCCGCATTCAGTTGTCGAGGAGT 3′ Fwd., 5′ AGTGCAGCATTTAGCCTTGCTTCC 3′ Rev., at 7.5 mM primer, 1 × Taq buffer, 2 mM MgCl2, 0.4 mM dNTP’s, 1.25 U Taq Pol, 1 μl cDNA. The Mixed reaction was incubated at 95°C for 10 min, and then was run for 40 cycles at 94°C 45 s, 60°C 45 s, 72°C 75 s; and finally at 72°C for 15 min.
Cloning sequences and sequencing
The PCR product was separated by electrophoresis at 3% of agarose gel, and purified using the gel extraction kit (Qiagen, Valencia CA, USA). The purified PCR products were ligated into pGem-T Easy VectorTM (Promega, Madinson, WI). Finally, the recombinant plasmid was purified using the Wizard Plus Miniprep DNA Purification SystemTM (Promega) and clones were sequenced with the M13 oligonucleotide and the BigDye Terminator 3.1 cycle sequencing kit (Applied Biosystems), and sequenced in an Applied Biosystems Abi Prism 3130 genetic analyzer automated sequencer according to previous reports [20]. The resulting sequences were assembled and compared to the reference transcript sequence reported in the NCBI nucleotide database.
Results
A total of four CT and four CCL were included in this study. Total RNA isolated was visualized in agarose gel at 1.5%, we did not observe any data that showed RNA degradation. The total RNA average concentration was 500 ng/μl. Thereafter, cDNA was subjected to PCR amplification; the housekeeping gene RPL4 showed height expression as expected (214 bp) Figure 1A, after HMMR was amplified. The amplification of HMMR showed two constant fragments in all samples. However, an additional fragment ~ 200 bp was observed Figure 1B.
Figure 1.
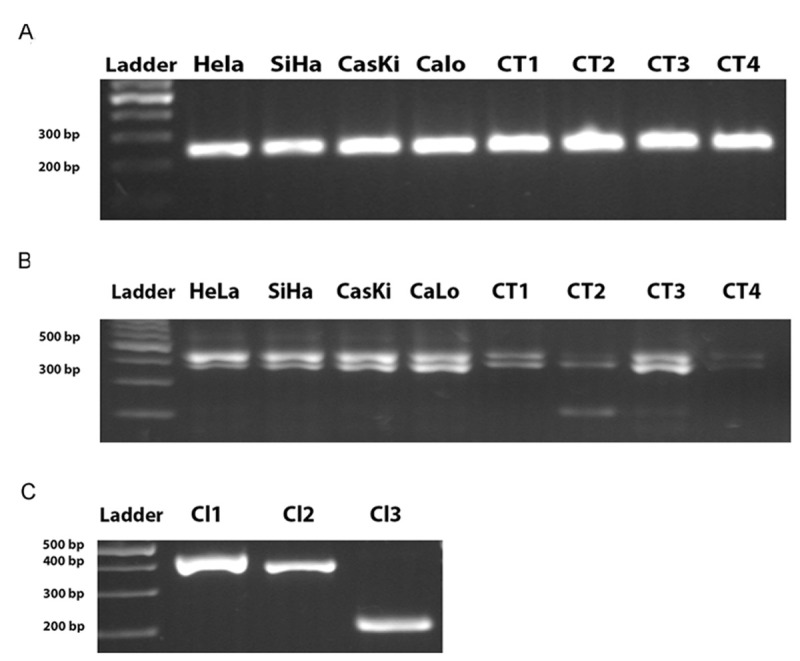
HMMR transcript is expressed in cervical cancer. A: Agarose gel showing raw expression of constitutive gene RPL4. B: RT-PCR products of HMMR were run in agarose gel at 2.5%. Cervical cell lines (HeLa, SiHa, CasKi, CaLo, one to four columns respectively) and Cervical tumors (CT, columns 5 to 8) showed two PCR products ~ 400 pb, CT2 sample showed expression of three amplicons in ~ 400 pb and ~ 200 pb. C: Three HMMR PCR Cloning (Cl1, Cl2, Cl3) were isolated and run in agarose gel.
Until this point, we identified two apparent isoforms of HMMR expressed in CCL and CT, although the CT2 sample showed the apparent expression of another transcript Figure 1B. In the rest of the study we focused in the characterization of the expression of three PCR products. CT2 sample belonged to a 28 year old woman. She had the clinical diagnosis of invasive squamous cervical cell carcinoma stage IB, and HPV infection type 16.
PCR products were isolated and cloned in pGem-T vector Figure 1C and sequenced Figure 2. All sequences matched with the HMMR transcript, however belong to different HMMR mRNA spliced. The top up PCR products showed sequences belong to HMMR variant one (NM_001142556.1) Figure 2A, the meddle belong to HMMR variant three (NM_012485.2) Figure 2B, the last product belong to HMMR variant four (NM_001142557.1) Figure 2C annotated in NCBI nucleotide database and HAVANA or ENSEMBL database. Thereafter, we aligned the sequence obtained of CT with the reference sequences of HMMR sequences reported in the NCBI database Figure 3. In the alignment, we could observe splice exons 2, 3 and 4, suggesting that HMMR could co-express several variants in cervical cancer. Finally, in the Figure 4 we are proposing a model of HMMR according to the results obtained, suggesting that HMMR could express variants in cervical cancer, nevertheless, not all tumors expressed three alternative splicing.
Figure 2.
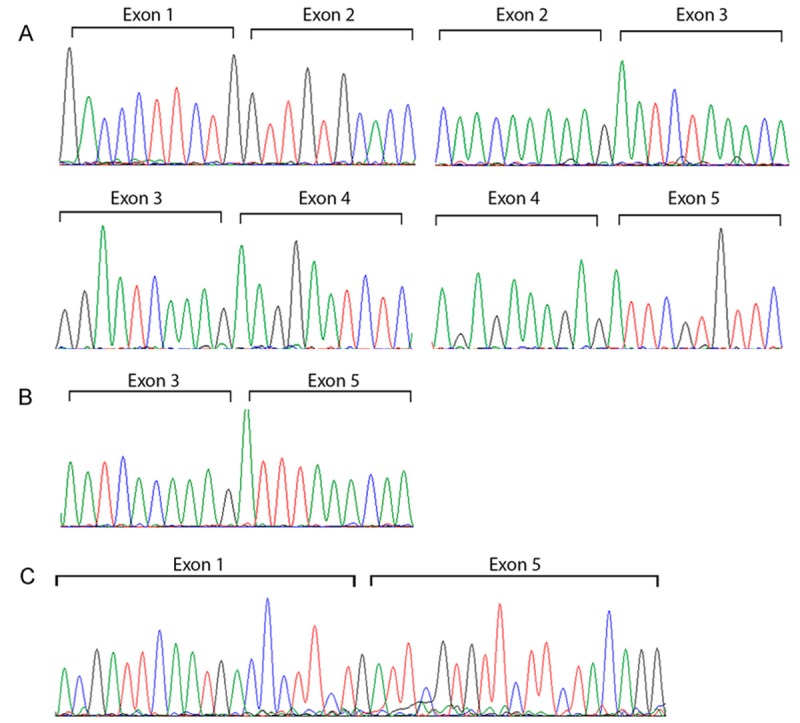
Cervical cancer expressed transcript variants of HMMR. A: Boundaries of HMMR variant one; HMMR variant ones is constituted of five exon junctions. B: Alternative splicing boundaries of exon 3 and 5. C: Alternative splicing of HMMR showed boundaries of exon 1 and 5 belonging to variant 4.
Figure 3.

Alignment HMMR sequences expressed in cervical cancer. HMMR expressed sequences in cervical cancer were aligned with the sequences reported in the NCBI nucleotide database.
Figure 4.
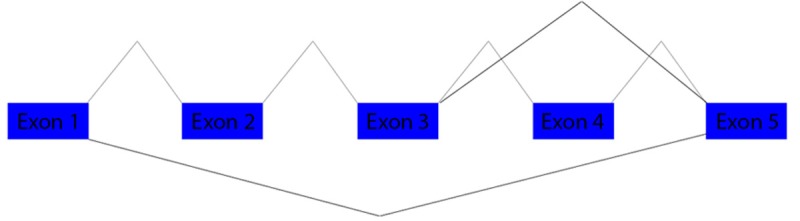
Alternative splicing model of HMMR gene. Primary splicing of exons 1-2-3-4-5 of HMMR gene containing the full sequence that corresponds to the largest amplicon. The junction of exons 3-5, which corresponds to the intermediate amplicon. The junction of exon 1-5, which are contained in the smaller amplicon.
Discussion
The characterization of malignant cellular components is a great challenge for understanding possible mechanisms involved in cancer process. Moreover, they give more opportunities for the identification of treatment targets in cancer. CC represents a major problem of health in developing countries such as Mexico. Actually, the HPV infection is the principal etiological factor for CC [21], however, cellular components play an important role in the generation of cellular conditions for cell survival, progression and transformation [22].
CC natural history is diverse, depending on its clinical features such as: tumor type, stage and treatment, overall given prognostic. However, in Mexico the clinical diagnosis generally takes place in advanced stages, worsening the prognosis. Most of the factors associated with tumor resistance, and relapses are unknown.
Alternative mRNA splicing is a biological process involved in mRNA editing and promotes an increase in RNA diversity, consequently more proteins are expressed. The expression of aberrant mRNA spliced is associated with cellular deregulation [23-25], malignant transformation [8,26], tumor development risk [7] and cellular transformation [27]. However, most of the mRNA spliced has an unknown cellular function.
Actually, there is little knowledge about the diversity of alternative mRNA splicing in health and disease. However, in humans > 90% of genes could be subject to AS [28], thereby estimating several millions of different expressed proteins [29]. AS could be differential and dependent cellular processes, specific tissue [30] even associated with malignancies [31,32]. CD antigens span over 200 members with many roles in the cell, including receptors, banding protein, immunoglobulin, among others. Hyaluronan-Mediated Motility Receptor (HMMR, RHAMM, CD168) that we found alternatively spliced in CCT and CCL belongs to a CD family of protein receptors. HMMR encodes four proteins: 725, 724, 709 and 638 aa are reported, moreover, HMMR proteins have several cell distribution [10-13], probably by means of expression of protein variants, however, little is known about the function of HMMR. HMMR proteins are involved in several cellular processes such as cell migration in development and [33,34], wound repair [35]. Moreover, it is associated with malignancy processes such as: angiogenesis [36,37], locomotion [38], tumor progression [39], cell proliferation [40], among others.
We found expression of two consistent mRNA variants (NM_001142556.1, NM_012485.2) of HMMR in CT and CCL; however, CT2 expressed three mRNA variants (NM_00114-2556.1, NM_012485.2 and NM_001142557.1) of HMMR. Interestingly, this case was of the youngest woman, suggesting that the four variants of HMMR could be associated with the aggressiveness. Another possibility is that the expression of the transcripts of HMMR is so low that we could not detect it. Mexican population is heterogeneous, and the tumor diversity could promote differential alternative mRNA splices. Interestingly, we found alternative mRNA splices of HMMR in CC. These results showed that HMMR encodes a variety of proteins expressed in CC, and its expression could be involved in malignant transformation and aggressiveness, among others. Moreover, HMMR expression is associated with different types of cancer including: breast, colon, leukemia, gastric and prostate [17,41-44], suggesting that HMMR expression in CC could be related to poor outcome. Nevertheless, there is little information about expression of HMMR variants in cancer. HMMR is distributed in multiple compartments such as: cytoskeleton [10], mitochondria [12], cell surface [11] and nucleus [10,13]. It is very likely that the distribution of HMMR could be by means of the AS transcript, consequently producing changes of the protein expressed.
The HPV infection was confirmed by PCR methods in all tumors, nevertheless, we did not found association between HPV infection and the expression of the three variants of HMMR. Probably, the HMMR expression could be a consequence of malignant cancer. Identification of alternative mRNA spliced in cancer generates more knowledge of the diversity of protein expression. We identified one patient that expressed three HMMR variants. Therefore, we consider that HMMR spliced variants one, three and four are potential oncogenes that play and important role in the malignancy of CC.
Acknowledgements
This work was partial supported by a Basic Science grant 61742 from SEP-CONACyT México and federal funds INP.
Disclosure of conflict of interest
None to disclose.
References
- 1.Ortega-Cervantes L, Rojas-Garcia AE, Robledo-Marenco Mde L, Barron-Vivanco BS, Giron-Perez MI, Vallejo-Ruiz V, Lopez-Flores JF, Carrillo-Cortez A, Cantu-De Leon D, Rodriguez-Trejo A, Medina-Diaz IM. Morbidity of breast cancer and cervico-uterine cancer in women from the occidental region of Mexico. Rev Invest Clin. 2013;65:221–227. [PubMed] [Google Scholar]
- 2.Moody CA, Laimins LA. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer. 2010;10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 3.Li H, Fan X, Houghton J. Tumor microenvironment: the role of the tumor stroma in cancer. J Cell Biochem. 2007;101:805–815. doi: 10.1002/jcb.21159. [DOI] [PubMed] [Google Scholar]
- 4.Modrek B, Resch A, Grasso C, Lee C. Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res. 2001;29:2850–2859. doi: 10.1093/nar/29.13.2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Akgul C, Moulding DA, Edwards SW. Alternative splicing of Bcl-2-related genes: functional consequences and potential therapeutic applications. Cell Mol Life Sci. 2004;61:2189–2199. doi: 10.1007/s00018-004-4001-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonomi S, Gallo S, Catillo M, Pignataro D, Biamonti G, Ghigna C. Oncogenic alternative splicing switches: role in cancer progression and prospects for therapy. Int J Cell Biol. 2013;2013:962038. doi: 10.1155/2013/962038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Adamia S, Reiman T, Crainie M, Mant MJ, Belch AR, Pilarski LM. Intronic splicing of hyaluronan synthase 1 (HAS1): a biologically relevant indicator of poor outcome in multiple myeloma. Blood. 2005;105:4836–4844. doi: 10.1182/blood-2004-10-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng C, Sharp PA. Regulation of CD44 alternative splicing by SRm160 and its potential role in tumor cell invasion. Mol Cell Biol. 2006;26:362–370. doi: 10.1128/MCB.26.1.362-370.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toole BP. Hyaluronan and its binding proteins, the hyaladherins. Curr Opin Cell Biol. 1990;2:839–844. doi: 10.1016/0955-0674(90)90081-o. [DOI] [PubMed] [Google Scholar]
- 10.Assmann V, Jenkinson D, Marshall JF, Hart IR. The intracellular hyaluronan receptor RHAMM/IHABP interacts with microtubules and actin filaments. J Cell Sci. 1999;112:3943–3954. doi: 10.1242/jcs.112.22.3943. [DOI] [PubMed] [Google Scholar]
- 11.Crainie M, Belch AR, Mant MJ, Pilarski LM. Overexpression of the receptor for hyaluronan-mediated motility (RHAMM) characterizes the malignant clone in multiple myeloma: identification of three distinct RHAMM variants. Blood. 1999;93:1684–1696. [PubMed] [Google Scholar]
- 12.Lynn BD, Turley EA, Nagy JI. Subcellular distribution, calmodulin interaction, and mitochondrial association of the hyaluronan-binding protein RHAMM in rat brain. J Neurosci Res. 2001;65:6–16. doi: 10.1002/jnr.1122. [DOI] [PubMed] [Google Scholar]
- 13.Entwistle J, Hall CL, Turley EA. HA receptors: regulators of signalling to the cytoskeleton. J Cell Biochem. 1996;61:569–577. doi: 10.1002/(sici)1097-4644(19960616)61:4<569::aid-jcb10>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 14.Golshani R, Lopez L, Estrella V, Kramer M, Iida N, Lokeshwar VB. Hyaluronic acid synthase-1 expression regulates bladder cancer growth, invasion, and angiogenesis through CD44. Cancer Res. 2008;68:483–491. doi: 10.1158/0008-5472.CAN-07-2140. [DOI] [PubMed] [Google Scholar]
- 15.Koyama H, Hibi T, Isogai Z, Yoneda M, Fujimori M, Amano J, Kawakubo M, Kannagi R, Kimata K, Taniguchi S, Itano N. Hyperproduction of hyaluronan in neu-induced mammary tumor accelerates angiogenesis through stromal cell recruitment: possible involvement of versican/PG-M. Am J Pathol. 2007;170:1086–1099. doi: 10.2353/ajpath.2007.060793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamilton SR, Fard SF, Paiwand FF, Tolg C, Veiseh M, Wang C, McCarthy JB, Bissell MJ, Koropatnick J, Turley EA. The hyaluronan receptors CD44 and Rhamm (CD168) form complexes with ERK1,2 that sustain high basal motility in breast cancer cells. J Biol Chem. 2007;282:16667–16680. doi: 10.1074/jbc.M702078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishigami S, Ueno S, Nishizono Y, Matsumoto M, Kurahara H, Arigami T, Uchikado Y, Setoyama T, Arima H, Yoshiaki K, Kijima Y, Kitazono M, Natsugoe S. Prognostic impact of CD168 expression in gastric cancer. BMC Cancer. 2011;11:106. doi: 10.1186/1471-2407-11-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamano Y, Uzawa K, Shinozuka K, Fushimi K, Ishigami T, Nomura H, Ogawara K, Shiiba M, Yokoe H, Tanzawa H. Hyaluronan-mediated motility: a target in oral squamous cell carcinoma. Int J Oncol. 2008;32:1001–1009. [PubMed] [Google Scholar]
- 19.Terris MK, Peehl DM. Human papillomavirus detection by polymerase chain reaction in benign and malignant prostate tissue is dependent on the primer set utilized. Urology. 1997;50:150–156. doi: 10.1016/S0090-4295(97)00126-X. [DOI] [PubMed] [Google Scholar]
- 20.Juarez-Mendez S, Zentella A, Villegas-Ruiz V, Perez-Gonzalez OA, Salcedo M, Lopez-Romero R, Roman-Basaure E, Lazos-Ochoa M, Montes de Oca-Fuentes VE, Vazquez-Ortiz G, Moreno J. Splice variants of zinc finger protein 695 mRNA associated to ovarian cancer. J Ovarian Res. 2013;6:61. doi: 10.1186/1757-2215-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 24.Garcia-Blanco MA, Baraniak AP, Lasda EL. Alternative splicing in disease and therapy. Nat Biotechnol. 2004;22:535–546. doi: 10.1038/nbt964. [DOI] [PubMed] [Google Scholar]
- 25.Blencowe BJ. Exonic splicing enhancers: mechanism of action, diversity and role in human genetic diseases. Trends Biochem Sci. 2000;25:106–110. doi: 10.1016/s0968-0004(00)01549-8. [DOI] [PubMed] [Google Scholar]
- 26.Zardi L, Carnemolla B, Siri A, Petersen TE, Paolella G, Sebastio G, Baralle FE. Transformed human cells produce a new fibronectin isoform by preferential alternative splicing of a previously unobserved exon. EMBO J. 1987;6:2337–2342. doi: 10.1002/j.1460-2075.1987.tb02509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A, Karnoub AE, Palmby TR, Lengyel E, Sondek J, Der CJ. Rac1b, a tumor associated, constitutively active Rac1 splice variant, promotes cellular transformation. Oncogene. 2004;23:9369–9380. doi: 10.1038/sj.onc.1208182. [DOI] [PubMed] [Google Scholar]
- 28.Carninci P. Constructing the landscape of the mammalian transcriptome. J Exp Biol. 2007;210:1497–1506. doi: 10.1242/jeb.000406. [DOI] [PubMed] [Google Scholar]
- 29.Missler M, Sudhof TC. Neurexins: three genes and 1001 products. Trends Genet. 1998;14:20–26. doi: 10.1016/S0168-9525(97)01324-3. [DOI] [PubMed] [Google Scholar]
- 30.Joseph R, Dou D, Tsang W. Neuronatin mRNA: alternatively spliced forms of a novel brain-specific mammalian developmental gene. Brain Res. 1995;690:92–98. doi: 10.1016/0006-8993(95)00621-v. [DOI] [PubMed] [Google Scholar]
- 31.Merdzhanova G, Gout S, Keramidas M, Edmond V, Coll JL, Brambilla C, Brambilla E, Gazzeri S, Eymin B. The transcription factor E2F1 and the SR protein SC35 control the ratio of pro-angiogenic versus antiangiogenic isoforms of vascular endothelial growth factor-A to inhibit neovascularization in vivo. Oncogene. 2010;29:5392–5403. doi: 10.1038/onc.2010.281. [DOI] [PubMed] [Google Scholar]
- 32.Guo M, Liu W, Serra S, Asa SL, Ezzat S. FGFR2 isoforms support epithelial-stromal interactions in thyroid cancer progression. Cancer Res. 2012;72:2017–2027. doi: 10.1158/0008-5472.CAN-11-3985. [DOI] [PubMed] [Google Scholar]
- 33.Toole BP, Munaim SI, Welles S, Knudson CB. Hyaluronate-cell interactions and growth factor regulation of hyaluronate synthesis during limb development. Ciba Found Symp. 1989;143:138–145. doi: 10.1002/9780470513774.ch9. discussion 145-139 281-135. [DOI] [PubMed] [Google Scholar]
- 34.Toole BP. Developmental role of hyaluronate. Connect Tissue Res. 1982;10:93–100. doi: 10.3109/03008208209034409. [DOI] [PubMed] [Google Scholar]
- 35.Goldberg RL, Seidman JD, Chi-Rosso G, Toole BP. Endogenous hyaluronate-cell surface interactions in 3T3 and simian virus-transformed 3T3 cells. J Biol Chem. 1984;259:9440–9446. [PubMed] [Google Scholar]
- 36.Slevin M, Krupinski J, Gaffney J, Matou S, West D, Delisser H, Savani RC, Kumar S. Hyaluronan-mediated angiogenesis in vascular disease: uncovering RHAMM and CD44 receptor signaling pathways. Matrix Biol. 2007;26:58–68. doi: 10.1016/j.matbio.2006.08.261. [DOI] [PubMed] [Google Scholar]
- 37.Park D, Kim Y, Kim H, Kim K, Lee YS, Choe J, Hahn JH, Lee H, Jeon J, Choi C, Kim YM, Jeoung D. Hyaluronic acid promotes angiogenesis by inducing RHAMM-TGFbeta receptor interaction via CD44-PKCdelta. Mol Cells. 2012;33:563–574. doi: 10.1007/s10059-012-2294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turley EA, Austen L, Vandeligt K, Clary C. Hyaluronan and a cell-associated hyaluronan binding protein regulate the locomotion of ras-transformed cells. J Cell Biol. 1991;112:1041–1047. doi: 10.1083/jcb.112.5.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bourguignon LY. Hyaluronan-mediated CD44 activation of RhoGTPase signaling and cytoskeleton function promotes tumor progression. Semin Cancer Biol. 2008;18:251–259. doi: 10.1016/j.semcancer.2008.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatano H, Shigeishi H, Kudo Y, Higashikawa K, Tobiume K, Takata T, Kamata N. Overexpression of receptor for hyaluronan-mediated motility (RHAMM) in MC3T3-E1 cells induces proliferation and differentiation through phosphorylation of ERK1/2. J Bone Miner Metab. 2012;30:293–303. doi: 10.1007/s00774-011-0318-0. [DOI] [PubMed] [Google Scholar]
- 41.Maxwell CA, Benítez J, Gómez-Baldó L, Osorio A, Bonifaci N, Fernández-Ramires R, Costes SV, Guinó E, Chen H, Evans GJ, Mohan P, Català I, Petit A, Aguilar H, Villanueva A, Aytes A, Serra-Musach J, Rennert G, Lejbkowicz F, Peterlongo P, Manoukian S, Peissel B, Ripamonti CB, Bonanni B, Viel A, Allavena A, Bernard L, Radice P, Friedman E, Kaufman B, Laitman Y, Dubrovsky M, Milgrom R, Jakubowska A, Cybulski C, Gorski B, Jaworska K, Durda K, Sukiennicki G, Lubiński J, Shugart YY, Domchek SM, Letrero R, Weber BL, Hogervorst FB, Rookus MA, Collee JM, Devilee P, Ligtenberg MJ, Luijt RB, Aalfs CM, Waisfisz Q, Wijnen J, Roozendaal CE HEBON; EMBRACE; Easton DF, Peock S, Cook M, Oliver C, Frost D, Harrington P, Evans DG, Lalloo F, Eeles R, Izatt L, Chu C, Eccles D, Douglas F, Brewer C, Nevanlinna H, Heikkinen T, Couch FJ, Lindor NM, Wang X, Godwin AK, Caligo MA, Lombardi G, Loman N, Karlsson P, Ehrencrona H, Wachenfeldt Av SWE-BRCA; Barkardottir RB, Hamann U, Rashid MU, Lasa A, Caldés T, Andrés R, Schmitt M, Assmann V, Stevens K, Offit K, Curado J, Tilgner H, Guigó R, Aiza G, Brunet J, Castellsagué J, Martrat G, Urruticoechea A, Blanco I, Tihomirova L, Goldgar DE, Buys S, John EM, Miron A, Southey M, Daly MB BCFR; Schmutzler RK, Wappenschmidt B, Meindl A, Arnold N, Deissler H, Varon-Mateeva R, Sutter C, Niederacher D, Imyamitov E, Sinilnikova OM, Stoppa-Lyonne D, Mazoyer S, Verny-Pierre C, Castera L, de Pauw A, Bignon YJ, Uhrhammer N, Peyrat JP, Vennin P, Fert Ferrer S, Collonge-Rame MA, Mortemousque I GEMO Study Collaborators; Spurdle AB, Beesley J, Chen X, Healey S kConFab. Barcellos-Hoff MH, Vidal M, Gruber SB, Lázaro C, Capellá G, McGuffog L, Nathanson KL, Antoniou AC, Chenevix-Trench G, Fleisch MC, Moreno V, Pujana MA. Interplay between BRCA1 and RHAMM regulates epithelial apicobasal polarization and may influence risk of breast cancer. PLoS Biol. 2011;9:e1001199. doi: 10.1371/journal.pbio.1001199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mohan P, Castellsague J, Jiang J, Allen K, Chen H, Nemirovsky O, Spyra M, Hu K, Kluwe L, Pujana MA, Villanueva A, Mautner VF, Keats JJ, Dunn SE, Lazaro C, Maxwell CA. Genomic imbalance of HMMR/RHAMM regulates the sensitivity and response of malignant peripheral nerve sheath tumour cells to aurora kinase inhibition. Oncotarget. 2013;4:80–93. doi: 10.18632/oncotarget.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tzankov A, Strasser U, Dirnhofer S, Menter T, Arber C, Jotterand M, Rovo A, Tichelli A, Stauder R, Gunthert U. In situ RHAMM protein expression in acute myeloid leukemia blasts suggests poor overall survival. Ann Hematol. 2011;90:901–909. doi: 10.1007/s00277-011-1159-6. [DOI] [PubMed] [Google Scholar]
- 44.Gust KM, Hofer MD, Perner SR, Kim R, Chinnaiyan AM, Varambally S, Moller P, Rinnab L, Rubin MA, Greiner J, Schmitt M, Kuefer R, Ringhoffer M. RHAMM (CD168) is overexpressed at the protein level and may constitute an immunogenic antigen in advanced prostate cancer disease. Neoplasia. 2009;11:956–963. doi: 10.1593/neo.09694. [DOI] [PMC free article] [PubMed] [Google Scholar]


