Abstract
The aim of the present study was to investigate the association of the expression of members in the miR-200 family with clinicopathological characteristics and their impacts on overall survival in patients with epithelial ovarian cancer (EOC). Expression levels of members in the miR-200 family, including miR-200a, miR-200b, miR-200c, miR-141, and miR-429, were detected by using miRNA qRT-PCR and in situ hybridization. Associations of their expression with clinicopathological factors and overall survival were statistically evaluated. Among five members in the miR-200 family, the expression levels of miR-200a, miR-200b and miR-200c were significantly higher in EOC tissues than those in normal surface ovarian epithelium tissues, in line with the findings ofin situ hybridization analysis. In addition, tumors with high miR-200a and miR-200 bexpressionwere both more likely to have advanced stage (both P=0.006) and higher grade (P=0.01 and 0.02), whilehighmiR-200 cexpression was onlysignificantly associated with advanced stage disease (P=0.01). Moreover, univariate analysis showed that the patients with high miR-200a, miR-200b and miR-200c expression all correlated with shorter overall survival in EOC patients (all P<0.001). Multivariate statistical analysis further identified miR-200a, miR-200b and miR-200c asindependent prognostic factorsfor EOC (all P=0.01). In conclusion, these findings suggest that miR-200a, miR-200b and miR-200c overexpression may promote the aggressive tumor progression and be recognized as reliable markers to predict the survival in patients with EOCs. The three miRNAs could be attractive therapeutic targets in patients with advanced-stage EOCs.
Keywords: miR-200 family, clinicopathology, epithelial ovarian cancer, prognosis
Introduction
Epithelial ovarian cancer (EOC) is the most lethal gynecological malignancies and the second leading cause of cancer-related death in women worldwide [1]. In spite of the development of diagnostic and therapeutic technologies, the clinical outcome of EOC patients remains poor, because the majorities of patients are diagnosed at an advanced stage of disease due to mild and diffuse symptoms, and develop recurrence [2]. The 5-year overall survival of patients with advanced-stage EOCs is only 40~45% [3]. Thus, it is of great clinical significance to identify and validate tumor-specific markers for early-stage diagnosis in order to improve the therapeutic levels and individualize therapeutic strategies.
MicroRNAs (miRNAs), originally identified in Caenorhabditis elegans, are a class of single-stranded, evolutionarily conserved, small (17~25 ribonucleotides) non-coding RNAs [4]. Subsequently, miRNAs was found in a large variety of Metazoens from Drosophila to humans [5]. More than 700 miRNAs have been discovered in the human genome (www.microRNA.org), regulating approximately 30% of all human transcripts [6]. miRNAs have demonstrated to regulate gene expression by binding to imperfect complementary sites in the 3’-untranslated region of their target messenger RNA transcripts [7]. Bioinformatics and microarray studies have revealed that a single miRNA can bind to as many as 200 mRNA targets which can be diverse in their function, include transcription factors, secreted factors, receptors, and transporters [8]. Accumulating evidence have shown that miRNAs are involved in regulating various biological processes, such as cellular differentiation, proliferation, angiogenesis, metabolism and cancer development [9]. In tumorigenesis, miRNAs can act either as oncogenes or tumor suppressors depending on the cellular context and the expression of the miRNA targets in the particular malignant tissues. miRNAs overexpressed in tumors may contribute to oncogenesis by down-regulating tumor suppressors, whereas miRNAs lost by tumors generally contribute to oncogene overexpression [10]. An increasing number of investigators have carried out miRNAs expression profiling studies in cell lines, tissue or serum samples in order to identify new biomarkers of EOC. For example, miRNAs with oncogenic functions, such as miR-20a, miR-106b, miR-143, miR-145, miR-125b and the let-7 family of miRNAs, have been well-known onco-miRNAs [11-15]; Several miRNAs such as miR-30, miR-124 and miR-199 have shown downregulation in EOC tissues and have tumor inhibitory effects [16-18]. Although dozens of miRNAs are identified to be differentially expressed and can be either up- or down-regulated, depending on their target downstream genes, only a small fraction of them may actually be of clinical utility as diagnostic/prognostic biomarkers or therapeutic targets.
The miR-200 family, containing miR-200a, miR-200b, miR-200c, miR-141 and miR-429, is organized into two groups based on a single nucleotide difference in their seed sequence (group A: miR-141 and -200a are located on chromosome 2; group B: miR-200b, -200c and -429 are located on chromosome 1), allowing each group to have overlapping and non-overlapping target genes [19]. These miRNAs have been reported as powerful markers and determining factors of the epithelial phenotype of cancer cells. More interestingly, a large number of investigators have found the associations of the aberrant expression of the miR-200 family with tumor prognosis in patients with ovarian cancer [20-23]. However, some studies reported contradictory findings because of the use of different normal controls or the heterogeneity of ovarian cancer. For example, Hu et al. [20] using 55 advanced tumor samples showed that the overexpression of miR-200a, miR-200b, and miR-429 were associated with improved survival in patients with ovarian cancer, while Nam et al. [21] using 20 serous carcinomas found that high expression of the miR-200 family members significantly correlated with poor prognosis; Chen et al. [22] combined systematic review profiling studies and experimental validation to reveal that miR-200a, miR-200b, miR-200c, and miR-141 were highly expressed in EOC tissues; Prislei et al. [23] indicated that the prognostic value of miR-200c in ovarian cancer may be depend on the cellular localization of HuR: when HuR is nuclear, high expression of miR-200c may result in a good prognosis, whereas when HuR occurs in cytoplasm, the same miRNA produced a poor outcome. According to these findings, there are two problems should be addressed. At first, no previous studies revealed the associations between miR-200 expression and the clinicopathological characteristics of EOC. Secondly, the prognostic value of the miR-200 family should be validated using a large cohort of EOC patients. Therefore, the aim of the current study was to investigate the association of members in the miR-200 family with clinicopathological characteristics and their impacts on overall survival in patients with EOCs.
Materials and methods
Patients and tissue samples
The study was approved by the Research Ethics Committee of the 180th Hospital, China. Written informed consent was obtainedfrom all of the patients. All specimens were handled and made anonymous according to the ethical and legal standards.
A total of 100 EOC tissues were collected from the Department of Obstetrics and Gynecology, the 180th Hospital, between December 2006 and January 2009. In addition, 50 normal ovarian epithelia tissues, used as controls, were obtained from 50 women who underwent surgery for benign or malignant gynecological diseases other than EOC. All fresh surgical specimens were snap-frozen in liquid nitrogen immediately after resection, and were stored at -80°C for miRNA qRT-PCR and in situ hybridization. All patients who had received chemotherapy and/or radiotherapy before surgery were excluded. After surgery, 100 EOC patients were received intravenous paclitaxel (175 mg/m2) or docetaxel (75 mg/m2) plus carboplatin (AUC 5) combination chemotherapy every 3 weeks for 6-8 cycles. The age of the EOC patients ranged from 26 to 88 years, with a median of 58 years. Tumor stage was in accordance with the International Federation of Gynecology and Obstetrics (FIGO) criteria, whereas tumor grade and histological type were determined following World Health Organization standards. The clinicopathological parameters are shown in Table 1.
Table 1.
Association between miR-200a, miR-200b, miR-200c expression and clinicopathological characteristics of epithelial ovarian cancer
| Factor | Cases no. | miR-200a expression | P | miR-200b expression | P | miR-200c expression | P |
|---|---|---|---|---|---|---|---|
|
|
|
|
|||||
| High (n, %) | High (n, %) | High (n, %) | |||||
| Age (year) | |||||||
| <58 | 58 | 31 (53.45) | NS | 30 (51.72) | NS | 31 (53.45) | |
| ≥58 | 42 | 22 (52.38) | 20 (47.62) | 21 (50.00) | |||
| FIGO stage | |||||||
| I~II | 56 | 17 (30.35) | 0.006 | 14 (25.00) | 0.006 | 20 (35.71) | 0.01 |
| III~IV | 44 | 36 (81.82) | 36 (81.82) | 32 (72.73) | |||
| Histologic type | |||||||
| Serous carcinoma | 60 | 36 (60.00) | NS | 36 (60.00) | NS | 36 (60.00) | NS |
| Mucinous carcinoma | 13 | 9 (69.23) | 6 (46.15) | 8 (61.54) | |||
| Endometrioid carcinoma | 15 | 5 (33.33) | 5 (33.33) | 5 (33.33) | |||
| Clear cell carcinoma | 10 | 2 (20.0) | 2 (20.0) | 2 (20.0) | |||
| Others | 2 | 1 (50.00) | 1 (50.00) | 1 (50.00) | |||
| Histologic grade | |||||||
| Grade 1 | 23 | 7 (30.43) | 0.01 | 7 (30.43) | 0.02 | 11 (47.83) | NS |
| Grade 2 | 29 | 13 (44.83) | 13 (44.83) | 11 (37.93) | |||
| Grade 3 | 48 | 33 (68.75) | 30 (62.50) | 30 (62.50) | |||
| Residual tumor size | |||||||
| <1 cm | 64 | 30 (46.88) | NS | 30 (46.88) | NS | 30 (46.88) | NS |
| ≥1 cm | 36 | 23 (63.89) | 20 (55.56) | 22 (61.11) | |||
| Chemoresistance | |||||||
| Yes | 37 | 24 (64.86) | NS | 21 (56.76) | NS | 23 (62.16) | NS |
| No | 63 | 29 (46.03) | 29 (46.03) | 29 (46.03) | |||
| Recurrence | |||||||
| Yes | 54 | 30 (55.56) | NS | 27 (50.00) | NS | 29 (53.70) | NS |
| No | 46 | 23 (50.00) | 23 (50.00) | 23 (50.00) |
All EOC patients were followed until death or the end of the follow-up period (January 3, 2014). The median follow-up period was 36.8 months, ranging from 6 to 56 months. The patients were evaluated monthly by clinical history, physical examination, ultrasound examination, and computed tomography when necessary. Study outcomes included overall survival, each measured from the time of definitive surgery. The duration of overall survival was the interval between definitive surgery and death.
RNA extraction
Small RNA from EOC and normal ovarian epithelia tissues was extracted using mirVana miRNA Isolation Kit (Ambion, Austin, TX, USA) according to the manufacture’s instruction. Only those RNA samples with OD A260/A280 ratio close to value of 2.0, suggesting that the RNA is pure, were subsequently analyzed. Real-time absolute quantification was utilized to insure the quality of samples.
miRNA qRT-PCR analysis
To detect the expression levels of five members in the miR-200 family (miR-200a, miR-200b, miR-200c, miR-141 and miR-429) in 100 EOC and 50 normal ovarian epithelia tissues, small RNA (10 ng) was synthesised with TaqMan MicroRNA RT Kit (Applied Biosystems, Foster City, CA, USA). QRT-PCR was performed on StepOne Plus Real-Time PCR Systems (Applied Biosystems) using EXPRESS qPCR Supermixes (Invitrogen, Carlsbad, CA, USA). U6B was used as endogenous controls. The following PCR primers were used in the following PCR reactions: miR-200a-Fwd, 5’-GCCGTCTAACACTGTCTGGTA-3’, and reverse, 5’-CCTACGCCACAATTAACAAGCC-3’; miR-200b-Fwd, 5’-GCG GCT AAT ACT GCC TGG TAA-3’, and reverse, 5’-GTG CAG GGT CCG AGG T-3’; miR-200c-Fwd, 5’-CCAACGTAATACTGCCGGGT-3’, and reverse, 5’-CTGCTGGCGAATTAGTAGACCA-3’; miR-141-Fwd, 5’-CCGCCTTAACACTGTCTGGTA-3’, and reverse, 5’-ATCGCCAGGATAAATTGACGCA-3’; miR-429-Fwd, 5’-GGAGTCGGCAATTCAGTTGAGACGGTTTT-3’, and reverse, 5’-ACACTCCAGCTGGGTAATACTGTCTGGTA-A-3’; U6-Fwd, 5’-CGC TTC GGC AGC ACA TAT ACT A-3’; and reverse, 5’-CGC TTC ACG AAT TTG CGT GTC A-3’. PCR cycles were as follows: 94°C for 5 min, followed by 40 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s.
Data analysis was performed by the comparative threshold cycle (Ct) method according to User Bulletin no.2 (Applied Biosystems). Each sample was examined in triplicate and the amounts of the PCR products produced were normalized to the internal controls.
In situ hybridization
In situ hybridization was performed to detect the expression level and subcellular localization of miR-200a, miR-200b, and miR-200c in 100 EOC and 50 normal ovarian epithelia tissues. Briefly, the tissue slides were hybridized with 200 nM of 5’-Digoxigenin (DIG) LNA-modified-miR-200a, LNA-modified-miR-200b and LNA-modified-miR-200c (Blossom Biotechnologies Inc, Blossom, USA) using IsHyb in Situ Hybridization kit (Biochain, Eureka Drive Newark, USA) according to manufacturer’s instructions. Sequences of the LNA/DNA oligonucleotides contained locked nucleic acids at eight consecutive centrally located bases (indicated by the underline) as shown: hsa-miR-200a 5’-ACA TCG TTA CCA GAC GAC AGT GTT A-3’; hsa-miR-200b 5’-TCA TCA TTA CCA GGC AGT ATT A-3’; and hsa-miR-200c 5’-TCC ATC ATT ACC CGG CAG TAT TA-3’.
In situ hybridization results were scored by two independent experienced pathologists, who were blinded to the clinicopathological data and clinical outcomes of the patients. The scores of the two pathologists were compared and any discrepant scores were re-examined by both pathologists to reach a consensus score. The number of positive-staining cells in ten representative microscopic fields was counted and the percentage of positive cells was calculated. The percentage scoring of positive tumor cells was as follows: 0 (0%), 1 (1-10%), 2 (11-50%) and 3 (>50%). The staining intensity was visually scored and stratified as follows: 0 (negative), 1 (weak), 2 (moderate) and 3 (strong). A final score was obtained for each case by multiplying the percentage and the intensity score. Therefore, tumors with a multiplied score exceeding median of total score for miR-200a, miR-200b, or miR-200c were deemed to be low expressions of miR-200a, miR-200b, or miR-200c; all other scores were considered to be high expressions of miR-200a, miR-200b, or miR-200c.
Statistical analysis
The software of SPSS version 13.0 for Windows (SPSS Inc, IL, USA) and SAS 9.1 (SAS Institute, Cary, NC) was used for statistical analysis. Continuous variables were expressed as x̅±s. Statistical analysis were performed with Fisher’s exact test for any 2×2 tables, Pearson X2 test for non- 2×2 tables, chi-square trend test for ordinal datum, Kaplan-Meier and Cox Regression methods for the question of survival analysis. Differences were considered statistically significant when p value was less than 0.05.
Results
Expression levels of the miR-200 family in EOC tissues
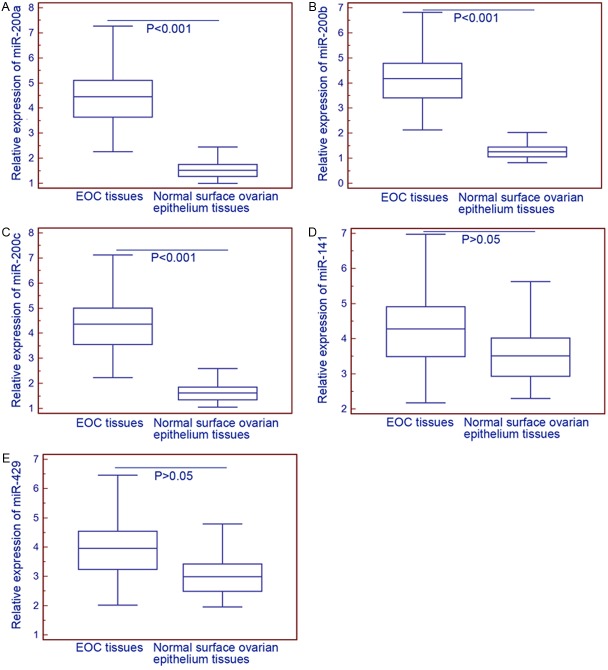
As shown in Figure 1, the expression levels of miR-200a (EOC vs. normal: 4.42±1.14 vs. 1.57±0.39, P<0.001), miR-200b (EOC vs. normal: 4.15±1.07 vs. 1.30±0.32, P<0.001) and miR-200c (EOC vs. normal: 4.33±1.12 vs. 1.67±0.41, P<0.001) were significantly higher in EOC tissues than those in normal surface ovarian epithelium tissues, while the expression levels of miR-141 (EOC vs. normal: 4.25±1.10 vs. 3.61±0.89, P>0.05) and miR-429 (EOC vs. normal: 3.94±1.02 vs. 3.08±0.76, P>0.05) had no significant differences between EOC and normal surface ovarian epithelium tissues.
Figure 1.
Relative expression levels of miR-200a (A), miR-200b (B), miR-200c (C), miR-141 (D) and miR-429 (E) in epithelial ovarian cancer (EOC) and normal ovarian surface epithelium tissues. The expression levels of miR-200a (EOC vs. normal: 4.42±1.14 vs. 1.57±0.39, P<0.001), miR-200b (EOC vs. normal: 4.15±1.07 vs. 1.30±0.32, P<0.001) and miR-200c (EOC vs. normal: 4.33±1.12 vs. 1.67±0.41, P<0.001) were significantly higher in EOC tissues than those in normal surface ovarian epithelium tissues, while the expression levels of miR-141 (EOC vs. normal: 4.25±1.10 vs. 3.61±0.89, P>0.05) and miR-429 (EOC vs. normal: 3.94±1.02 vs. 3.08±0.76, P>0.05) had no significant differences between EOC and normal surface ovarian epithelium tissues.
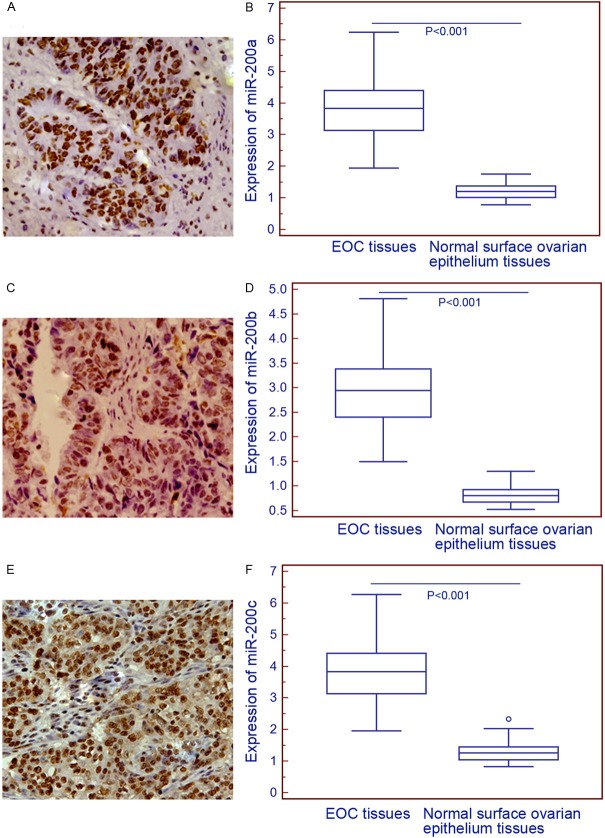
In order to validate the expression patterns and subcellular localizations of miR-200a, miR-200b and miR-200c in EOC and normal surface ovarian epithelium tissues, we performed in situ hybridization analysis. As shown in Figure 2, the three miRNAs all predominantly localized in nucleus of tumor cells in EOC tissues, while negatively stained in normal surface ovarian epithelium tissues. Statistical analysis shown the overexpression of miR-200a, miR-200b and miR-200c in EOC compared to normal surface ovarian epithelium tissues, which was in line with the findings of miRNA qRT-PCR analysis.
Figure 2.
Representative in situ hybridizationimages for miR-200a (A), miR-200b (C), and miR-200c (E) expression in epithelial ovarian cancer (EOC) tissues (Original magnification×200). Compared with normal ovarian surface epithelium tissues, the expression levels of miR-200a (EOC vs. normal: 3.80±0.98 vs. 1.24±0.31, P<0.001, (B) miR-200b (EOC vs. normal: 2.92±0.76 vs. 0.83±0.21, P<0.001, (D) and miR-200c (EOC vs. normal: 3.81±0.99 vs. 1.30±0.32, P<0.001, (F) were all significantly increased in EOC tissues.
Association ofmiR-200a, miR-200b and miR-200c overexpression with the clinicopathological characteristics of EOC
The median values of miR-200a (3.83), miR-200b (2.95) and miR-200c (3.84) expression levels detected by in situ hybridization analysis in all EOC tissues were used as cutoff points to classified 100 EOC patientsinto miR-200a-low (n=47), miR-200a-high (n=53), miR-200b-low (n=50), miR-200b-high (n=50), miR-200c-low (n=48), miR-200c-high (n=52) expression groups. Table 1 summarized the association of their expression with the clinicopathological characteristics of patients with EOCs. Statistical analysis showed that tumors with high miR-200a and miR-200b expression were both more likely to have advanced stage (both P=0.006, Table 1) and higher grade (P=0.01 and 0.02, Table 1), whilehighmiR-200c expression was onlysignificantly associated with advanced stage disease (P=0.01, Table 1). There was no significant association between miR-200a, miR-200b and miR-200c expression and other clinicopathologic characteristics of EOC, including age of patients, histologic type, residual tumor size, chemoresistance and tumor recurrence (all P>0.05).
Prognostic value of miR-200a, miR-200b and miR-200c overexpression in EOC
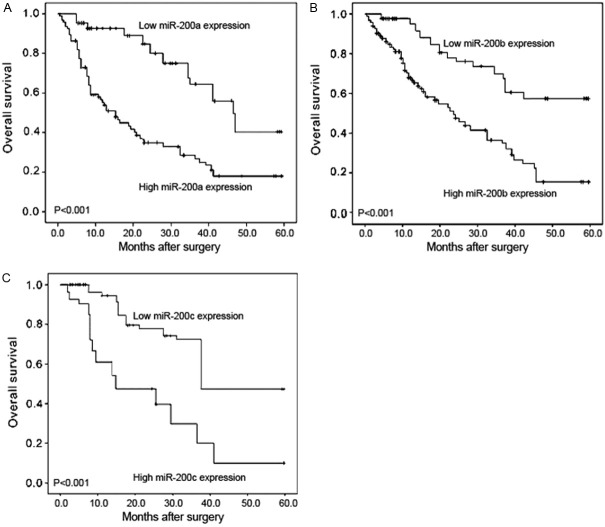
The univariate and multivariate survival analyses were performed to determine the correlation of miR-200a, miR-200b and miR-200c expression with overall survival of EOC patients. In Kaplan-Meier analysis, the overall survival of EOC patients with high miR-200a, miR-200b and miR-200c expression all correlated with shorter overall survival in EOC patients than the corresponding controls (all P<0.001, Figure 3). In addition, the univariate logistic regression analysis revealed that high miR-200a expression (HR 22.69, CI 1.32-50.53, P<0.001), high miR-200b expression (HR 20.28, CI 1.20-42.28, P<0.001), high miR-200c expression (HR 21.42, CI 1.26-48.33, P<0.001), advanced FIGO stage (HR 19.83, CI 1.18-40.62, P<0.001) and high tumor grade (HR 15.57, CI 1.0-33.16, P=0.008) were significantly associated with poor overall survival (Table 2). Moreover, the multivariate COX regression analysis adjusted for other prognostic factors, further identified miR-200a, miR-200b and miR-200c as independent prognostic factors for EOC (all P=0.01, Table 3).
Figure 3.
Kaplan-Meier survival curves for miR-200a (A), miR-200b (B) and miR-200c (C) expressionin epithelial ovarian cancer (EOC). In Kaplan-Meier analysis, the overall survival of EOC patients with high miR-200a, miR-200b and miR-200c expression all correlated with shorter overall survival in EOC patients than the corresponding controls (all P<0.001).
Table 2.
Univariate analysis: factors predicting overall surviva
| Characteristic | N | Overall survival | |
|---|---|---|---|
|
| |||
| P value | Hazard Ratio (95% CI) | ||
| FIGO stage | |||
| I~II | 56 | <0.001 | 19.83 (1.18-40.62) |
| III~IV | 44 | ||
| Histologic type | |||
| Serous carcinoma | 60 | NS | 5.21 (0.5-10.89) |
| Mucinous carcinoma | 13 | ||
| Endometrioid carcinoma | 15 | ||
| Clear cell carcinoma | 10 | ||
| Others | 2 | ||
| Histologic grade | |||
| Grade 1 | 23 | 0.008 | 15.57 (1.00-33.16) |
| Grade 2 | 29 | ||
| Grade 3 | 48 | ||
| Residual tumor size | |||
| <1 cm | 64 | NS | 4.28 (0.67-8.71) |
| ≥1 cm | 36 | ||
| Chemoresistance | NS | 7.09 (0.82-14.20) | |
| Yes | 37 | ||
| No | 63 | ||
| Recurrence | NS | 8.15 (0.89-17.36) | |
| Yes | 54 | ||
| No | 46 | ||
| miR-200a status | |||
| High | 53 | <0.001 | 22.69 (1.32-50.53) |
| Low | 47 | ||
| miR-200b status | |||
| High | 50 | <0.001 | 20.28 (1.20-42.28) |
| Low | 50 | ||
| miR-200c status | |||
| High | 52 | <0.001 | 21.42 (1.26-48.33) |
| Low | 48 | ||
Table 3.
Multivariate logistic regression analysis
| Characteristic | Hazard Ratio (95% CI) | P-value |
|---|---|---|
| FIGO stage | 15.08 (1.51-30.83) | 0.01 |
| I~II vs. III~IV | ||
| Histologic grade | 10.91 (1.08-21.02) | 0.03 |
| Grade 1~2 vs. Grade 3 | ||
| miR-200a status | 17.26 (1.36-36.98) | 0.01 |
| Low vs. High | ||
| miR-200b status | 15.41 (1.13-31.36) | 0.01 |
| Low vs. High | ||
| miR-200c status | 16.22 (1.27-33.81) | 0.01 |
| Low vs. High |
Discussion
No characteristic early symptoms or tumor markers lead to disappointing clinical outcome of EOC. The remission and relapse are frequently seen in patients undergoing therapy for EOC. Accumulating studies have demonstrated that the discovery and investigation of the genetic changes and molecular events may contribute to a better understanding of the molecular mechanisms on tumorigenesis of EOC. The miR-200 family is known as the main suppressor of the epithelial-to mesenchymal transition (EMT), which is a reversible embryonic program aberrantly activated in tumor progression and metastasis [24]. Although several studies have attempted to investigatethe prognostic importance of the miR-200 family in EOCs, no definitive conclusions could be drawn. In the current study, our data provide the convincing evidence that three members of the miR-200 family (miR-200a, miR-200b and miR-200c) may be all upregulated in human EOC tissues. Moreover, the aberrant expression of these three miRNAs may be associated with aggressive clinicopathologic characteristics and poor prognosis of patients with EOC.
According to the functions, the miR-200 family can be grouped into two subfamilies: miR-200b, miR-200c and miR-429 have the same seed region while those of miR-200a and miR-141 are different [19]. Recently, this miRNA family has been identified as a powerful marker and determining factor of the epithelial phenotype of cancer cells. Members in this family have been observed to be downregulated in several types of cancers and upregulated in others, suggesting that the miR-200 family can act as either an oncogene or as a tumor-suppressor gene [25-26]. Among them, miR-200a upregulation may underline genomic amplification. Especially in EOC, several studies have shown frequent chromosomal gains in that region [27]; miR-200b has been demonstrated to be deregulated in many human cancers and considered a negative regulator of EMT in tumor cell migration and invasion; miR-200b has also been identified as a novel target for enhancing chemosensitivity or inducing cell death in EOC [28]; miR-200c expression may be correlated with chemoresistance to paclitaxel and cisplatin in a panel of ovarian adenocarcinoma cell lines with inherent or acquired drug-resistance [23]. In addition, accumulating recent studies have observed the altered expression of the miR-200 family in EOC. However, the reported results from these studies are often inconsistent with each other. Some of these studies belong to miRNA profiling analysis and were performed with different microarray platforms, which may lead to part of the discrepancy. In contrast, the miRNA qRT-PCR and the in situ hybridization analyses were both performed in our study, which can validate the results each other. Moreover, some of these studies have differences in experimental design, which may affect result interpretation. For instance, the recent studies investigated the prognostic value of the miR-200 family using a small number (all n<100) of EOC samples. In contrast, a larger number of tumor samples (n=100) were analyzed in our study, which renders higher statistical power to determine the associations between the miR-200 family and the clinicopathologic characteristics and prognosis in patients with EOC.
In conclusion,these findings suggest that miR-200a, miR-200b and miR-200c overexpression may promote the aggressive tumor progression and be recognized as reliable markers to predict the survival in patients with EOCs. The three miRNAs could be attractive therapeutic targets in patients with advanced-stage EOCs.Further functional study and perspective clinical trail are needed in the future.
Disclosure of conflict of interest
None.
References
- 1.Konstantinopoulos PA, Matulonis UA. Current Status and Evolution of Preclinical Drug Development Models of Epithelial Ovarian Cancer. Front Oncol. 2013;3:296. doi: 10.3389/fonc.2013.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoyer T, Bekkers R, Gooszen H, Massuger L, Rovers M, Grutters JP. Cost-effectiveness of early-initiated treatment for advanced-stage epithelial ovarian cancer patients: a modeling study. Int J Gynecol Cancer. 2014;24:75–84. doi: 10.1097/IGC.0000000000000025. [DOI] [PubMed] [Google Scholar]
- 3.Nasu K, Kai K, Hirakawa T, Nishida M, Matsumoto H, Kawano Y, Narahara H. Retrospective analysis of outcomes of secondary debulking surgery for recurrent epithelial ovarian cancer with favorable prognostic factors. J Obstet Gynaecol Res. 2014;40:791–796. doi: 10.1111/jog.12222. [DOI] [PubMed] [Google Scholar]
- 4.Stahlhut C, Slack FJ. MicroRNAs and the cancer phenotype: profiling, signatures and clinical implications. Genome Med. 2013;5:111. doi: 10.1186/gm516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nalejska E, Mączyńska E, Lewandowska MA. Prognostic and Predictive Biomarkers: Tools in Personalized Oncology. Mol Diagn Ther. 2014 doi: 10.1007/s40291-013-0077-9. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.To KK. MicroRNA: a prognostic biomarker and a possible druggable target for circumventing multidrug resistance in cancer chemotherapy. J Biomed Sci. 2013;20:99. doi: 10.1186/1423-0127-20-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Song H, Liu Y, Pan J, Zhao ST. Expression profile analysis reveals putative prostate cancer-related microRNAs. Genet Mol Res. 2013;12:4934–4943. doi: 10.4238/2013.October.24.4. [DOI] [PubMed] [Google Scholar]
- 8.Stoyianni A, Pentheroudakis G, Benjamin H, Cervantes A, Ashkenazi K, Lazaridis G, Pavlidis N, Spector Y. Insights into the epithelial mesenchymal transition phenotype in cancer of unknown primary from a global microRNA profiling study. Clin Transl Oncol. 2013 doi: 10.1007/s12094-013-1139-5. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 9.Costa PM, Pedroso de Lima MC. MicroRNAs as Molecular Targets for Cancer Therapy: On the Modulation of MicroRNA Expression. Pharmaceuticals (Basel) 2013;6:1195–1220. doi: 10.3390/ph6101195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Avci CB, Baran Y. Use of MicroRNAs in Personalized Medicine. Methods Mol Biol. 2014;1107:311–325. doi: 10.1007/978-1-62703-748-8_19. [DOI] [PubMed] [Google Scholar]
- 11.Shapira I, Oswald M, Lovecchio J, Khalili H, Menzin A, Whyte J, Dos Santos L, Liang S, Bhuiya T, Keogh M, Mason C, Sultan K, Budman D, Gregersen PK, Lee AT. Circulating biomarkers for detection of ovarian cancer and predicting cancer outcomes. Br J Cancer. 2014;110:976–983. doi: 10.1038/bjc.2013.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, Lee KW. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Xiao Z, Wang K, Liu W, Hao Q. MiR-145 is downregulated in human ovarian cancer and modulates cell growth and invasion by targeting p70S6K1 and MUC1. Biochem Biophys Res Commun. 2013;441:693–700. doi: 10.1016/j.bbrc.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Wang Q, Yu M, Wu N, Wang H. MicroRNA-145 Function as a Cell Growth Repressor by Directly Targeting c-Myc in Human Ovarian Cancer. Technol Cancer Res Treat. 2014;13:161–168. doi: 10.7785/tcrt.2012.500367. [DOI] [PubMed] [Google Scholar]
- 15.Chung YW, Bae HS, Song JY, Lee JK, Lee NW, Kim T, Lee KW. Detection of microRNA as novel biomarkers of epithelial ovarian cancer from the serum of ovarian cancer patient. Int J Gynecol Cancer. 2013;23:673–679. doi: 10.1097/IGC.0b013e31828c166d. [DOI] [PubMed] [Google Scholar]
- 16.Zhang H, Wang Q, Zhao Q, Di W. MiR-124 inhibits the migration and invasion of ovarian cancer cells by targeting SphK1. J Ovarian Res. 2013;6:84. doi: 10.1186/1757-2215-6-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng H, Zhang L, Zhao Y, Yang D, Song F, Wen Y, Hao Q, Hu Z, Zhang W, Chen K. Plasma miRNAs as Diagnostic and Prognostic Biomarkers for Ovarian Cancer. PLoS One. 2013;8:e77853. doi: 10.1371/journal.pone.0077853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miles GD, Seiler M, Rodriguez L, Rajagopal G, Bhanot G. Identifying microRNA/mRNA dysregulations in ovarian cancer. BMC Res Notes. 2012;5:164. doi: 10.1186/1756-0500-5-164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pecot CV, Rupaimoole R, Yang D, Akbani R, Ivan C, Lu C, Wu S, Han HD, Shah MY, Rodriguez-Aguayo C, Bottsford-Miller J, Liu Y, Kim SB, Unruh A, Gonzalez-Villasana V, Huang L, Zand B, Moreno-Smith M, Mangala LS, Taylor M, Dalton HJ, Sehgal V, Wen Y, Kang Y, Baggerly KA, Lee JS, Ram PT, Ravoori MK, Kundra V, Zhang X, Ali-Fehmi R, Gonzalez-Angulo AM, Massion PP, Calin GA, Lopez-Berestein G, Zhang W, Sood AK. Tumour angiogenesis regulation by the miR-200 family. Nat Commun. 2013;4:2427. doi: 10.1038/ncomms3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim JH, Kim JW, Kim S. MicroRNA expression profiles in serous ovarian carcinoma. Clin Cancer Res. 2008;14:2690–2695. doi: 10.1158/1078-0432.CCR-07-1731. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y, Zhang L, Hao Q. Candidate microRNA biomarkers in human epithelial ovarian cancer: systematic review profiling studies and experimental validation. Cancer Cell Int. 2013;13:86. doi: 10.1186/1475-2867-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prislei S, Martinelli E, Mariani M, Raspaglio G, Sieber S, Ferrandina G, Shahabi S, Scambia G, Ferlini C. MiR-200c and HuR in ovarian cancer. BMC Cancer. 2013;13:72. doi: 10.1186/1471-2407-13-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, Wellner U, Dimmler A, Faller G, Schubert J, Brabletz T. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–782. doi: 10.1038/emboj.2010.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Engelsvold DH, Utheim TP, Olstad OK, Gonzalez P, Eidet JR, Lyberg T, Trøseid AM, Dartt DA, Raeder S. miRNA and mRNA expression profiling identifies members of the miR-200 family as potential regulators of epithelial-mesenchymal transition in pterygium. Exp Eye Res. 2013;115:189–198. doi: 10.1016/j.exer.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manavalan TT, Teng Y, Litchfield LM, Muluhngwi P, Al-Rayyan N, Klinge CM. Reduced expression of miR-200 family members contributes to antiestrogen resistance in LY2 human breast cancer cells. PLoS One. 2013;8:e62334. doi: 10.1371/journal.pone.0062334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu S, Xu P, Wu W, Ou Y, Xu J, Zhang G, Li J, Xu G. The biphasic expression pattern of miR-200a and E-cadherin in epithelial ovarian cancer and its correlation with clinicopathological features. Curr Pharm Des. 2014;20:1888–1895. doi: 10.2174/13816128113199990523. [DOI] [PubMed] [Google Scholar]
- 28.Knouf EC, Garg K, Arroyo JD, Correa Y, Sarkar D, Parkin RK, Wurz K, O’Briant KC, Godwin AK, Urban ND, Ruzzo WL, Gentleman R, Drescher CW, Swisher EM, Tewari M. Integrative genomic approach identifies p73 and p63 as activators of miR-200 microRNA family transcription. Nucleic Acids Res. 2012;40:499–510. doi: 10.1093/nar/gkr731. [DOI] [PMC free article] [PubMed] [Google Scholar]