Abstract
Objective: In a recent study we determined a strong differential expression of DCC in OA compared to normal chondrocytes and a strong impact of the DCC receptor on cellular mobility triggered by its ligand Netrin-1. Migration of chondrocytes or their progenitor cells may play a role in remodeling of cartilage and pathological conditions. The purpose of this study is to identify subsets of chondrocytes expressing DCC and to understand signaling pathways used by DCC in chondrocytes. Methods: Immunofluorescent histology of human cartilage was used to determine the expression pattern of CD166, DCC and p-CREB. Cell culture of chondrocytes and SW1353, transient transfection, siRNA transfection, EMSA, luciferase assay, quantitative RT-PCR, ELISA, and Western Blotting were used to study signaling down-stream of DCC. Results: DCC expressing chondrocytes are mainly located in the surface layers of OA cartilage. These also express CD166 indicating that DCC expressing chondrocytes are progenitor cells. Interestingly, expression of DCC reduces cAMP levels, CREB DNA-binding activity and CRE activity in chondrocytes, whereas down-regulation of DCC results in induction of CRE signaling. Conclusion: In summary, DCC is up-regulated in CD166-positive chondrogenic progenitor cells in OA and induces down-regulation of CREB. These findings indicate that migration of CD166 positive progenitor cells to sites of cartilage damage may be directed by regulation of DCC signaling.
Keywords: Repellent factors, DCC, migration, osteoarthritis, CRE activity, CREB
Introduction
Until recently, chondrocytes have been viewed as stationary cells imbedded in an impenetrable matrix. Today, several in vitro observations object this theory. Chondrocytes were shown to be able to migrate in vitro in response to chemoattractant factors, such as cytokines, growth factors and components of the extracellular matrix (for review see Morales [1]). Moreover, chondrocyte movements were observed in planar and 3D matrices [1], in vivo in the growth plate [2] and during development of the intervertebral disc [3,4]. Further, re-colonization of a cell-free auto-transplant of meniscal tissue by α-smooth muscle expressing chondrocytes was observed in a canine meniscus repair model indicating the possibility of migration of chondrocytes into the transplant [5]. Recently, CD105+/CD166+ chondrogenic progenitor cells were observed migrating to cartilage lesions along the joint surface in cartilage explants [6]. The latter findings indicate that chondrocytes or their progenitors may contribute to repair mechanisms in cartilage by actively moving to sites of damage. Therefore, the migratory activities have to be directed in some way.
A group of receptors and ligands has been shown to be involved in migratory activity of synovial fibroblasts in rheumatoid arthritis (RA) and OA [7]. These so called “repellent factors” have originally been identified in the developing nervous system [8,9] and were shown to regulate cell-cell interactions and cell-matrix interactions of migrating cells during embryonic development. These factors are Semaphorins and their receptors, Netrins and their receptors, Deleted in Colon Cancer (DCC) and UNCs, Slits and Roundabout receptors (Robo) as well as Ephrins and the Eph receptors [8,10,11]. Recently, the presence of Semaphorin, Netrin and Slit in cartilage was demonstrated [12-14]. Furthermore, the Netrin receptor DCC was revealed to be strongly expressed in human joint cartilage obtained from patients suffering from osteoarthritis (OA). We reported that Netrin-1 induced migration of chondrocytes expressing DCC in vitro [13].
In this study, we analyzed the expression patterns of DCC in human OA cartilage in vivo and signaling of DCC in chondrocytes obtained from OA patients and healthy donors.
Materials and methods
OA-primary human chondrocytes and human cartilage
Tissues were obtained from probands or from patients after informed consent following the standards of the Ethics Commission of the University of Regensburg. Normal cartilage with no significant softening or surface fibrillation was obtained at autopsy within 48 h after death. OA cartilage was removed under sterile conditions from femoral condyles of knee joints of patients aged 40-65 years undergoing total knee arthroplasty. Tissue fragments were cut into 1 mm slices and were washed four to five times with Krebs buffer containing 100 units/ml penicillin and 100 μg/ml streptomycin. After two further rinses with calcium-free Dulbecco’s modified Eagle’s medium 4.5 g/l glucose (DMEM, Sigma-Aldrich, Munich, Germany), the slices were further cut into small pieces and were incubated for 16-24 h with 1.5 mg/ml collagenase B (Roche, Mannheim, Germany) and 1 mM cysteine in DMEM. The cell suspension was filtered through nylon gauze (100 μm pore size) and was washed three times with calcium-free DMEM. For immunohistochemical studies normal human cartilage and OA cartilage was fixed in 5% buffered formalin.
Normal human chondrocytes were purchased (Provitro, Berlin, Germany) and cultivated as suggested by the manufacturer.
Cell lines and tissue culture
Primary chondrocytes and the chondrosarcoma cell line SW1353 were used in the experiments. Cells were grown at 37°C/8% CO2 in DMEM (Sigma-Aldrich) supplemented with penicillin (100 U/ml), streptomycin (10 μg/ml) (both PAA) and 10% fetal calf serum (PAN Biotech, Aidenbach, Germany) and split 1:4 at confluence. Cells were detached by incubation with 0.05% trypsin, 0.04% EDTA (PAA) in PBS for 5 minutes at 37°C.
Transient transfection
Primary chondrocytes were transfected using the Amaxa Nucleofector System (Lonza, Basel, Switzerland) according to the manufacturer’s instructions.
Recombinant human pDCC-CMV-S was a kindly gift of Patrick Mehlen (University of Lyon [15]).
Transfection of SW1353 cells was performed using Lipofectamine LTX (Life technologies, Darmstadt, Germany). Briefly, the cells were cultured in 6-well plates. For transient transfection with expression plasmids each cationic lipid/plasmid DNA suspension was prepared of 0.5 μg plasmid within the transfection solutions according to the manufacturer’s instructions. The cells were harvested 48 h later, and RNA was isolated.
siRNA transfection
The siRNAs against DCC (siDCC_1, siDCC_2) and the control siRNAs were synthesized by Qiagen (Hilden, Germany). Cells were grown to 70-80% confluence in culture dishes, and harvested in the proliferative growth phase. Cells were transfected with the HiPerFect Transfection Reagent (Qiagen), according to the manufacturer’s protocol. Cells were transfected in six-well culture plates, and RNA was isolated 48 h after transfection.
Reporter gene assays
Cells were cultured in six-well plates for 24 h and then treated with cationic lipid/plasmid DNA suspension: 0.5 μg of luciferase reporter plasmid (CREB, NFκB or an empty pGL2basic vector as a control (all Promega Corporation, Madison, WI, USA)), 0.1 μg of the internal control plasmid pRL-TK. 24 h after the transfection cells were harvested and the lysate was analyzed for luciferase activity with a luminometer using Promega dual-luciferase assay reagent (Promega).
Nuclear extract preparation and EMSA (electrophoretic mobility shift assay)
Nuclear extract preparation and EMSAs were performed as described before [16,17]. For EMSAs the following oligonucleotides were used: CREB (5’-AGA GAT TGC CTG ACG TCA GAG AGC TAG-3’), AP-1 (5’-CGC TTG ATG AGT CAG CCG GAA-3’) or NFκB (5’-AGT TGA GGG GAC TTT CCC AGG C-3’).
Rabbit anti-CREB antibody (antibodies-online GmbH, Aachen, Germany) was used for supershift experiments.
Protein analysis in vitro (WB)
Trypsinized cells were incubated in 200 μl of RIPA-buffer (Roche) for 15 min at 4°C. Insoluble fragments were sedimented at 11,000 g for 10 min and the supernatant lysate was stored at -20°C. RIPA-cell lysate was loaded and separated on 10% SDS-PAGE and subsequently blotted onto polyvinylidene fluoride (PVDF) membrane. After blocking for 1 h with 3% dried milk powder/TBST the membrane was incubated for 16 h with a primary antibody against p-CREB Ser133 (1:1000; Merk-Millipore, Darmstadt, Germany), DCC (1:500; BD biosciences, Heidelberg, Germany) or β-actin (1:5000; Sigma-Aldrich). Then the membrane was washed three times in TBST, incubated for 1 h with an alkaline phosphate-conjugated secondary anti-rabbit antibody (1:3000; Chemicon, Hampshire, UK) or anti-mouse antibody (1:4000; Chemicon) and then washed again. Finally immunoreactions were visualized by NBT/BCIP (Sigma-Aldrich) staining.
Isolation of RNA and generation of cDNA
Total cellular RNA was isolated from the respective cells using the Micro Elute Total RNA Kit (VWR, Darmstadt, Germany) and cDNA was generated by reverse transcriptase reaction as described before [18].
Analysis of mRNA expression by quantitative real time PCR
Quantitative real time PCR was performed on a LightCycler® 480 II technology (Roche), cDNA template (1 μl), 0,5 μl (20 mM) of primers (hDCC for: 5’-GAC TCC AAT CCC AGG TGA CT, hDCC rev: 5’-TGA CTT CCT CGC CTC GTA AC; β-actin for: 5’-CTA CGT CGC CCT GGA CTT CGA GC; β-actin rev: 5’-GAT GGA GCC GCC GAT CCA CAC GG) and 10 μl LightCycler® 480 SYBR Green I Master in a total of 20 μl were applied to the following PCR program: initial denaturation at 95°C for 10 minutes, followed by 45 cycles of amplification with 10 s at 95°C, 10 s at 60°C (annealing) and 20 s at 72°C (elongation).
The PCR reaction was evaluated by melting curve analysis and analyzing the PCR products on 1.8% agarose gels.
Cyclic AMP ELISA
Intracellular cAMP was quantified using a direct cAMP-ELISA kit (Enzo Life Sciences, Lorrach, Germany) according to the manufacturer’s protocol.
Immunofluorescence
Three cartilage specimens from healthy donors and eight specimens from OA patients were studied by immunofluorescence microscopy. Therefore, standard 3 μm sections of formalin-fixed and paraffin-embedded tissue blocks were used. The tissue sections were deparaffinized, rehydrated and subsequently heated in a water bath for 40 min (90°C) in 0.01 M sodium citrate buffer (pH 7.2) and placed on microscope slides. After permeabilization with 0.3% Triton-X (Sigma-Aldrich) and blocking with 20% BSA/PBS the cartilage sections were first incubated with primary antibody (mouse-α-DCC (BD biosciences, Heidelberg, Germany) 1:1000 in PBS, rabbit-α-CREB (Merk-Millipore) 1:1000 in PBS) or rabbit-α-CD166 (Santa Cruz Biotechnologies, Santa Cruz, CA, USA) 1:50 in PBS) and the with the according secondary antibody (α-mouse-Alexa Fluor 488 or α-rabbit-Alexa Fluor 594 (Life technologies)). Evaluation of the staining was performed by means of immunofluorescent microscopy using 20 and 40-fold magnification. (AxioVisionAxio Imager Z1, Zeiss, Oberkochen, Germany).
Statistical analysis
Results are expressed as mean ± SD (range) or percent. Comparison between groups was made using the Student’s unpaired t-test. Correlation between parameters was calculated with the Spearman test. A p value < 0.05 was considered statistically significant. All calculations were performed by using the GraphPad Prism Software (GraphPad Software, Inc., San Diego, USA).
Results
The repellent factor DCC (deleted in colon cancer) was previously shown to be strongly induced in chondrocytes obtained from OA patients [13]. Overexpression of DCC in normal chondrocytes resulted in induction of chondrocyte migration revealing marked effects of DCC. In this study, we aimed to understand the expression pattern of DCC in cartilage and the signaling pathways modulating DCC effects on chondrocytes.
First, we studied normal cartilage and OA cartilage by means of immunofluorescence. In normal cartilage no signal for DCC-staining was detectable. To identify chondrogenic progenitor cells within the cartilage, the sections were also stained using a CD166 antibody. CD166 positive cells were few and were restricted to superficial layers (Figure 1A). In OA cartilage numerous DCC-positive cells were found in superficial layers and in the zone bordering the synovium. Using double labeling technique most of the DCC positive cells were also positive for CD166 indicating that DCC expressing cells are chondrogenic progenitor cells (Figure 1B). These cells were frequently oriented parallel to the cartilage surface and displayed a flattened spindly shape. At the border to the synovium CD166 positive cells had a roundish shape.
Figure 1.
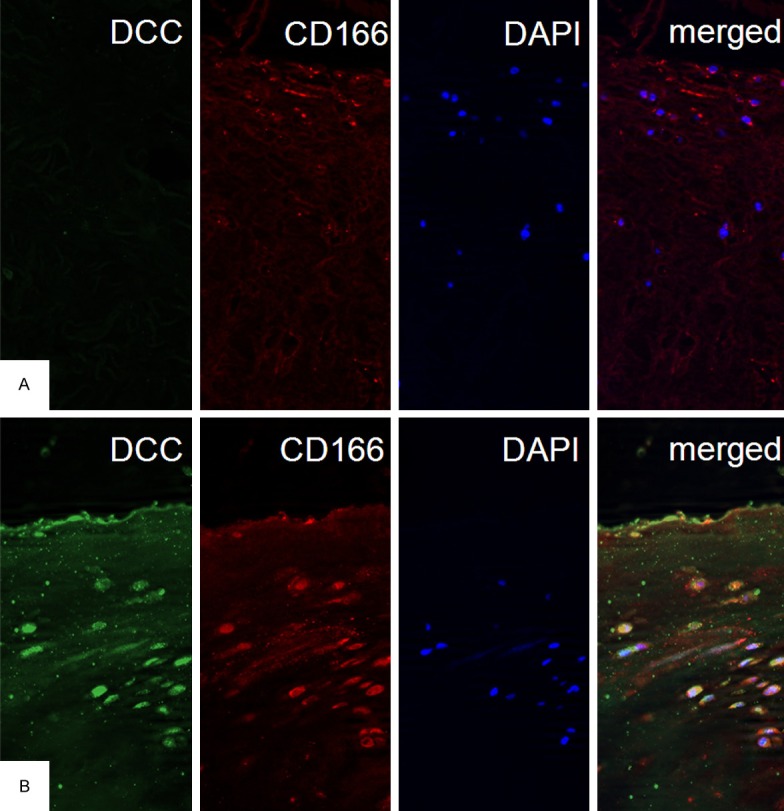
DCC is increased in CD166 positive progenitor cells in OA cartilage. A. In immunofluorescent histology of normal cartilage CD166 positive cells are situated in the upper layers and do not express DCC (magnification × 20). B. Immunofluorescence stainings for DCC verified induction of DCC in OA cartilage preferentially in CD166 positive cells (magnification × 20).
In the next step, we characterized the activity of NFκB, AP-1 and CREB signaling in normal chondrocytes after DCC or mock transfection. Interestingly, DCC expression had no detectable effects on NFκB and AP-1 signaling as observed by EMSA, however resulted in marked reduction of CREB DNA-binding activity (Figure 2A). The impact of DCC on CRE signaling was confirmed in chondrocytes of three additional donors (Figure 2B). Specificity of the EMSA was revealed by cold-competition of binding to the probes and by achieving a supershifted band using a specific anti-CREB antibody (Figure 2C). To confirm the results showing marked effects on CRE-signaling luciferase reporter gene assays were performed in DCC-transfected chondrocytes versus controls which showed a significant reduction of CRE activity after transfection of DCC (Figure 2D), whereas NFkB activity, as expected based on the EMSAs, stayed unchanged.
Figure 2.
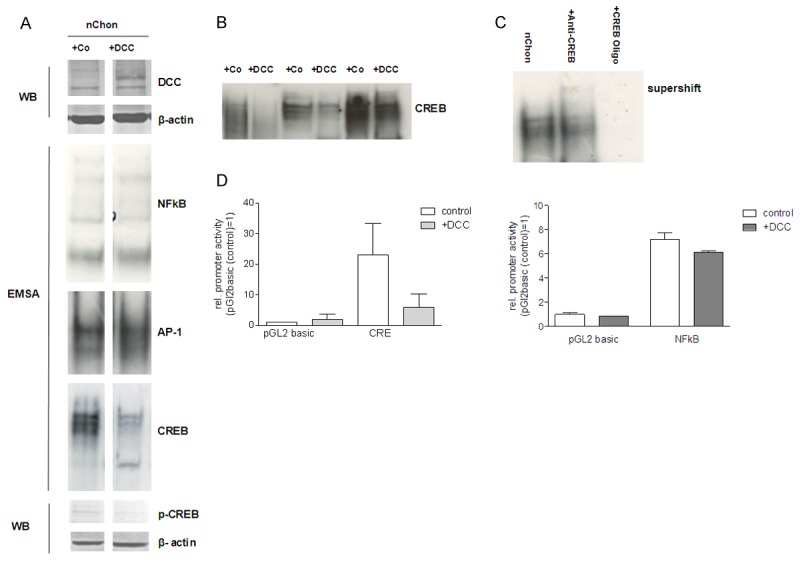
Activity of NFkB, AP-1 and CREB signaling in normal chondrocytes after DCC or mock transfection. (A) DCC expression had no detectable effects on NFkB and AP-1 signaling by EMSA, whereas marked reduction of CREB DNA-binding activity was observed in normal chondrocytes. (B) The impact of DCC on CREB signaling was confirmed in chondrocytes of three additional donors (C) Specificity of the EMSA was confirmed by cold-competition of binding and by achieving a supershifted band using a specific anti-CREB antibody. (D) A luciferase reporter gene assay was performed in DCC-transfected chondrocytes versus controls and determined significant reduction of CREB activity after transfection of DCC, whereas NFkB activity stayed unchanged.
CRE luciferase assays were further performed comparing normal chondrocytes to chondrocytes derived from OA patients (Figure 3). A significant difference in CRE activity was observed comparing normal and OA chondrocytes, however, NFkB signaling was unchanged.
Figure 3.
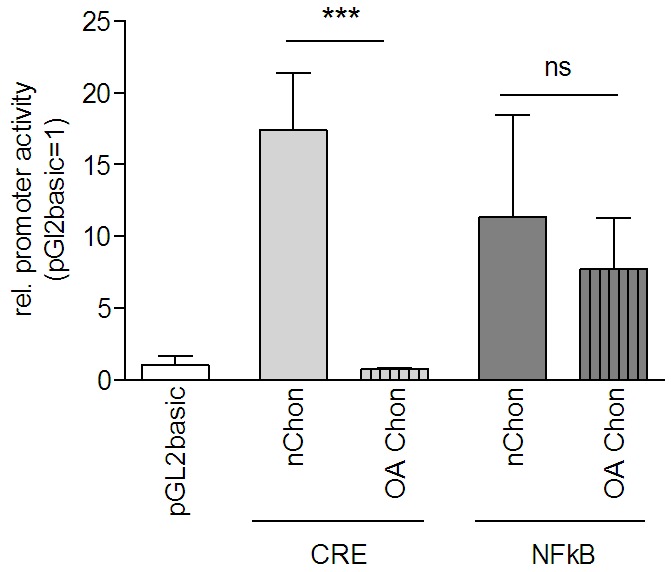
CREB activity is reduced in OA chondrocytes. CREB luciferase reporter gene assays were performed comparing normal chondrocytes to chondrocytes derived from OA patients. A significant difference in CREB activity was observed comparing normal and OA chondrocytes, whereas NFkB signaling was unchanged. (***: p < 0.001; ns: p > 0.05)
To confirm the influence of DCC on CREB signaling OA chondrocytes were transfected with siRNA against DCC or control and CREB activity was determined by EMSA and CRE luciferase reporter gene assays. Down-regulation of DCC expression was confirmed on mRNA level (OA chondrocytes, Figure 4A), Further, the SW1353 cell line was used as a model cell line in these experiments. As SW1353 do not express DCC, expression was achieved by transient transfection of a DCC-expression plasmid prior to siRNA transfection. DCC down-regulation was confirmed on mRNA level or protein level (Figure 4A and 4B). EMSAs and luciferase reporter gene assays showed that down-regulation of DCC expression in OA chondrocytes and SW1353 resulted in induction of CREB signaling (Figure 4C and 4D).
Figure 4.
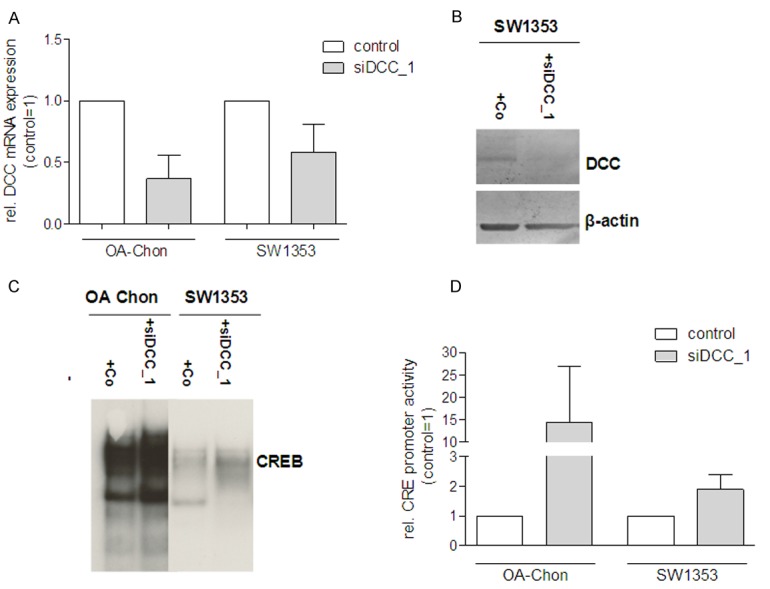
Down-regulation of DCC expression in OA chondrocytes and SW1353 chondrosarcoma cells results in induction of CRE signaling. OA chondrocytes were transfected with siRNA against DCC or control and CREB activity was determined by EMSA and CREB luciferase reporter gene assays. Down-regulation of DCC expression was confirmed on (A) mRNA level (OA chondrocytes and SW1353) and (B) protein level (SW1353). As SW1353 do not express DCC, expression was achieved by transient transfection of a DCC-expression plasmid prior to the experiment. DCC down-regulation resulted in higher CREB activity in (C) EMSAs and (D) luciferase reporter gene assays.
To analyze the role of cAMP in DCC signaling normal chondrocytes were transfected with DCC. After transfection a reduced cytoplasmic concentration of cAMP was measured by ELISA (Figure 5A). Expression of DCC after transfection was demonstrated by qRT-PCR (Figure 5B). Therefore, DCC expression led to lower cAMP levels in chondrocytes.
Figure 5.
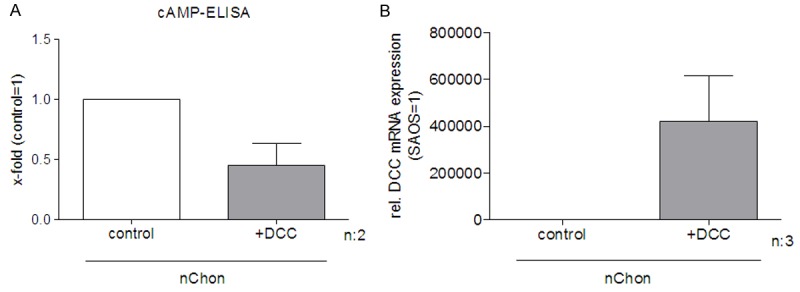
cAMP levels are down-regulated in chondrocytes after DCC transfection. After DCC transfection normal chondrocytes show a reduced cytoplasmic concentration of cAMP measured by ELISA (A). Expression of DCC after transfection was demonstrated by qRT-PCR (B).
In order to confirm the significance of our in vitro data, immunohistochemical double labeling reactions against DCC and p-CREB were performed in human cartilage obtained from healthy donors and OA patients. In normal cartilage strong nuclear staining of chondrocytes was observed for p-CREB accompanied by a barely visible cytoplasmic signal for DCC (Figure 6A). However, the majority of DCC positive superficial cells in OA cartilage showed a strong reduction of p-CREB in their nuclei (Figure 6B). Stationary chondrocytes in clusters showed faint DCC staining and strong nuclear staining for p-CREB (Figure 6C).
Figure 6.
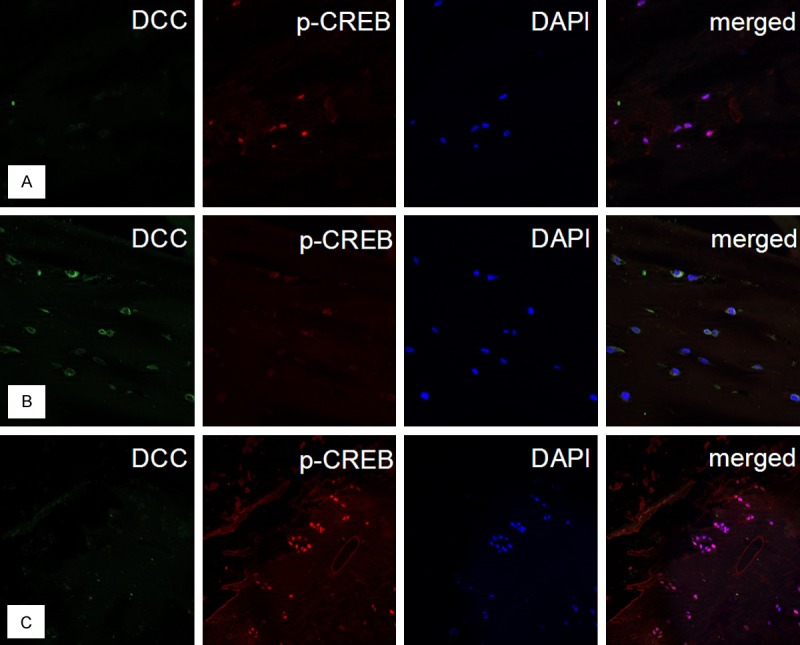
DCC is increased and nuclear p-CREB is decreased in CD166 positive progenitor cells in OA cartilage. (A) In healthy cartilage chondrocytes of upper layers display a strong nuclear signal for p-CREB and a faint cytoplasmic signal for DCC (magnification x 40). (B) No nuclear staining for p-CREB was found in strongly DCC positive superficial chondrocytes of OA samples, whereas stationary chondrocytes in clusters showed nuclear staining for p-CREB (C) (magnification × 40).
Discussion
In the last decades migration of chondrocytes and its possible function in vivo has been a controversial issue. However, Seol et al. recently observed a population of chondrogenic progenitor cells migrating towards damaged sites of cartilage in explants of articular cartilage [6]. Apparently, chondrogenic progenitor cells are derived from the superficial layers of articular cartilage and express CD166 [6,19]. Dysfunction of this process for instance by misdirection of chondrocytes or disruption of migration may be a possible cause of alterations in matrix composition and generation of inferior repair tissue in osteoarthritis (OA).
If there are migratory activities of chondrocytes during development, repair, and pathological conditions, they have to be directed by chemo-attractant and chemo-repulsive factors. Otherwise, misdirected chondrocytes or progenitor cells might contribute to alterations of the joint structure and disturbance of joint function such as already mentioned above. In a recent study, we reported strong induction of the repellent factor receptor DCC in OA chondrocytes indicating that altered regulation of DCC takes place in the course of OA [13]. The presence of binding sites for the transcription factors Sox9 and AP-2 in the DCC promoter of chondrocytes, the demonstration of binding of Sox9 and AP-2 to the DCC promoter, and the induction of DCC by both factors corroborate the finding of up-regulation of DCC during OA [13], since induction of Sox9 expression in OA was described by several groups [20]. In addition, also AP-2 binding sites were determined in several genes found to be up-regulated in OA [21]. The differential expression of DCC was reflected in an increased migratory activity of OA chondrocytes compared to normal chondrocytes after exposure to recombinant Netrin-1, a ligand of the DCC-receptor. Migration in response to Netrin-1 could also be induced in normal chondrocytes transfected with the DCC receptor [13].
In the present study, it was demonstrated that DCC is expressed in OA cartilage in spindle shaped cells in the superficial layers displaying a morphological phenotype suspicious of migratory activity as opposed to roundish stationary chondrocytes. DCC positive spindle shaped cells also expressed CD166, a marker of chondrogenic progenitor cells in immunofluorescence double stainings of OA cartilage, whereas roundish chondrocytes were mostly negative for CD166. CD166-positive cell were mainly located in the superficial and middle zone of cartilage and at the border of cartilage and synovium as reported by Pretzel et al [19]. In normal cartilage DCC was not detectable by immunofluorescence confirming our previous findings [13]. Furthermore, there were only very few CD166 positive cells in the superficial layers of normal cartilage compared to OA cartilage. Taking into account our previous in vitro data DCC may regulate migration of chondrocytes or chondrogenic progenitor cells in OA cartilage.
Since the cellular signaling transduction of DCC in chondrocytes is completely unknown so far, the down-stream signaling of DCC in chondrocytes was addressed in this study. DCC expression had no detectable effects on NFkB and AP-1 signaling, however, a strong reduction of CREB DNA-binding activity was observed. Also, DCC expression in chondrocytes leads to a significant reduction of CREB activity in luciferase reporter gene assays. Down-regulation of DCC expression by siRNA resulted in the induction of CREB signaling in OA chondrocytes and SW1353 cells in EMSAs and luciferase reporter gene assays. A quantitative cAMP-ELISA revealed a marked decrease of cAMP in DCC expressing chondrocytes in accordance with the reduced CREB activity described above. In addition, it was demonstrated that DCC-expressing chondrocytic progenitor cells show reduced p-CREB nuclear content in OA cartilage as opposed to normal cartilage corroborating our in vitro data. Interestingly, in cell clusters of OA cartilage chondrocytes DCC was only faintly detectable and a strong nuclear signal for p-CREB was observed. This finding is in accordance with reports that cell clusters contain proliferating chondrocytes that are unlikely to migrate [22,23].
The reported data demonstrate that DCC is regulated in chondrogenic progenitor cells in OA possibly influencing migration of these cells similarly to the already described effects of DCC on migration of chondrocytes in vitro [13]. Expression of DCC reduces cAMP levels, CREB DNA-binding activity and CREB activity in chondrocytes, whereas down-regulation of DCC results in the induction of CREB signaling. There are several reports that cAMP-signaling is involved in the regulation of cell migration. Reduction of cAMP levels promotes migratory activity of several cancer cell types [24-27]. There is also a report on induction of migration by down-regulation of CREB in glioma cells [28]. However, in other cell types induction of cAMP or CREB were associated with increased migration [29]. DCC signaling via cAMP and CREB may be pertinent to the regulation of migratory activities of chondrogenic progenitor cells. Netrin, a ligand of DCC that is expressed by chondrocytes and synovial cells may direct progenitor cells to sites of damage, since attractant effects of Netrin 1 on DCC expressing chondrocytes were demonstrated in vitro [13]. Moreover, the CREB KO mouse has a dwarf phenotype [30] indicating that CREB has an important role in the development of the skeleton possibly due to disordered direction of cell migration or differentiation of cartilage.
In summary, our results suggest that DCC is expressed in chondrocytic progenitor cells in OA and may be involved in cell migration of progenitor cells. Furthermore, expression of DCC reduces cAMP levels, CREB DNA-binding activity and CRE activity in chondrocytes, whereas down-regulation of DCC results in induction of CRE signaling.
In the light of new theories regarding a migratory capability of chondrocytes pertinent to cartilage remodeling these findings indicate that DCC may be involved in directing chondrogenic progenitor cells to sites of damage in OA possibly by regulation of CREB.
Acknowledgements
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG) to A.K.B. and T.S.
Disclosure of conflict of interest
There is no conflict of interest.
References
- 1.Morales TI. Chondrocyte moves: clever strategies? Osteoarthritis Cartilage. 2007;8:861–871. doi: 10.1016/j.joca.2007.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aszodi A. 1 integrins regulate chondrocyte rotation, G1 progression, and cytokinesis. Genes Dev. 2003;19:2465–2479. doi: 10.1101/gad.277003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim KW, Lim TH, Kim JG, Jeong ST, Masuda K, An HS. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine (Phila Pa 1976) 2003;10:982–990. doi: 10.1097/01.BRS.0000061986.03886.4F. [DOI] [PubMed] [Google Scholar]
- 4.Kim KW, Ha KY, Park JB, Woo YK, Chung HN, An HS. Expressions of membrane-type I matrix metalloproteinase, Ki-67 protein, and type II collagen by chondrocytes migrating from cartilage endplate into nucleus pulposus in rat intervertebral discs: a cartilage endplate-fracture model using an intervertebral disc organ culture. Spine (Phila Pa 1976) 2005;12:1373–1378. doi: 10.1097/01.brs.0000166155.48168.0e. [DOI] [PubMed] [Google Scholar]
- 5.Kambic HE, Futani H, McDevitt CA. Cell, matrix changes and alpha-smooth muscle actin expression in repair of the canine meniscus. Wound Repair Regen. 2000;6:554–561. doi: 10.1046/j.1524-475x.2000.00554.x. [DOI] [PubMed] [Google Scholar]
- 6.Seol D, McCabe DJ, Choe H, Zheng H, Yu Y, Jang K, Walter MW, Lehman AD, Ding L, Buckwalter JA, Martin JA. Chondrogenic progenitor cells respond to cartilage injury. Arthritis Rheum. 2012;11:3626–3637. doi: 10.1002/art.34613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert T, Denk A, Magdefrau U, Kaufmann S, Bastone P, Lowin T, Schedel J, Bosserhoff AK. Role of the netrin system of repellent factors on synovial fibroblasts in rheumatoid arthritis and osteoarthritis. Int J Immunopathol Pharmacol. 2009;3:715–722. doi: 10.1177/039463200902200317. [DOI] [PubMed] [Google Scholar]
- 8.Serafini T, Kennedy TE, Galko MJ, Mirzayan C, Jessell TM, Tessier-Lavigne M. The netrins define a family of axon outgrowth-promoting proteins homologous to C. elegans UNC-6. Cell. 1994;3:409–424. doi: 10.1016/0092-8674(94)90420-0. [DOI] [PubMed] [Google Scholar]
- 9.Yuan W, Zhou L, Chen JH, Wu JY, Rao Y, Ornitz DM. The mouse SLIT family: secreted ligands for ROBO expressed in patterns that suggest a role in morphogenesis and axon guidance. Dev Biol. 1999;2:290–306. doi: 10.1006/dbio.1999.9371. [DOI] [PubMed] [Google Scholar]
- 10.Hinck L. The versatile roles of “axon guidance” cues in tissue morphogenesis. Dev Cell. 2004;6:783–793. doi: 10.1016/j.devcel.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Miller LE, Weidler C, Falk W, Angele P, Schaumburger J, Scholmerich J, Straub RH. Increased prevalence of semaphorin 3C, a repellent of sympathetic nerve fibers, in the synovial tissue of patients with rheumatoid arthritis. Arthritis Rheum. 2004;4:1156–1163. doi: 10.1002/art.20110. [DOI] [PubMed] [Google Scholar]
- 12.Okubo M, Kimura T, Fujita Y, Mochizuki S, Niki Y, Enomoto H, Suda Y, Toyama Y, Okada Y. Semaphorin 3A is expressed in human osteoarthritic cartilage and antagonizes vascular endothelial growth factor 165-promoted chondrocyte migration: An implication for chondrocyte cloning. Arthritis Rheum. 2011;10:3000–3009. doi: 10.1002/art.30482. [DOI] [PubMed] [Google Scholar]
- 13.Schubert T, Kaufmann S, Wenke AK, Grassel S, Bosserhoff AK. Role of deleted in colon carcinoma in osteoarthritis and in chondrocyte migration. Rheumatology. 2009;11:1435–1441. doi: 10.1093/rheumatology/kep245. [DOI] [PubMed] [Google Scholar]
- 14.Denk AE, Kaufmann S, Stark K, Schedel J, Lowin T, Schubert T, Bosserhoff AK. Slit3 inhibits Robo3-induced invasion of synovial fibroblasts in rheumatoid arthritis. Arthritis Res Ther. 2010;2:R45. doi: 10.1186/ar2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong K, Hinck L, Nishiyama M, Poo MM, Tessier-Lavigne M, Stein E. A ligand-gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin-induced growth cone attraction to repulsion. Cell. 1999;7:927–941. doi: 10.1016/s0092-8674(00)80804-1. [DOI] [PubMed] [Google Scholar]
- 16.Ellmann L, Joshi MB, Resink TJ, Bosserhoff AK, Kuphal S. BRN2 is a transcriptional repressor of CDH13 (T-cadherin) in melanoma cells. Lab Invest. 2012;12:1788–1800. doi: 10.1038/labinvest.2012.140. [DOI] [PubMed] [Google Scholar]
- 17.Ruedel A, Stark K, Kaufmann S, Bauer R, Reinders J, Rovensky J, Blazickova S, Oefner PJ, Bosserhoff AK. N-cadherin promoter polymorphisms and risk of osteoarthritis. FASEB J. 2014;2:683–691. doi: 10.1096/fj.13-238295. [DOI] [PubMed] [Google Scholar]
- 18.Ruedel A, Hofmeister S, Bosserhoff AK. Development of a model system to analyze chondrogenic differentiation of mesenchymal stem cells. Int J Clin Exp Pathol. 2013;12:3042–3048. [PMC free article] [PubMed] [Google Scholar]
- 19.Pretzel D, Linss S, Rochler S, Endres M, Kaps C, Alsalameh S, Kinne RW. Relative percentage and zonal distribution of mesenchymal progenitor cells in human osteoarthritic and normal cartilage. Arthritis Res Ther. 2011;2:R64. doi: 10.1186/ar3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taupin D, Podolsky DK. Mitogen-activated protein kinase activation regulates intestinal epithelial differentiation. Gastroenterology. 1999;5:1072–1080. doi: 10.1016/s0016-5085(99)70010-7. [DOI] [PubMed] [Google Scholar]
- 21.Li X, Saint-Cyr-Proulx E, Aktories K, Lamarche-Vane N. Rac1 and Cdc42 but not RhoA or Rho kinase activities are required for neurite outgrowth induced by the Netrin-1 receptor DCC (deleted in colorectal cancer) in N1E-115 neuroblastoma cells. J Biol Chem. 2002;17:15207–15214. doi: 10.1074/jbc.M109913200. [DOI] [PubMed] [Google Scholar]
- 22.Johnson WE, Eisenstein SM, Roberts S. Cell cluster formation in degenerate lumbar intervertebral discs is associated with increased disc cell proliferation. Connect Tissue Res. 2001;3:197–207. doi: 10.3109/03008200109005650. [DOI] [PubMed] [Google Scholar]
- 23.Pfander D, Kortje D, Weseloh G, Swoboda B. Cell proliferation in human arthrotic joint cartilage. Z Orthop Ihre Grenzgeb. 2001;5:375–381. doi: 10.1055/s-2001-17977. [DOI] [PubMed] [Google Scholar]
- 24.Zimmerman NP, Roy I, Hauser AD, Wilson JM, Williams CL, Dwinell MB. Cyclic AMP regulates the migration and invasion potential of human pancreatic cancer cells. Mol Carcinog. 2013 doi: 10.1002/mc.22091. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Z, Tsuchiya H, Zhang Y, Hartnett ME, Wang L. MicroRNA-433 inhibits liver cancer cell migration by repressing the protein expression and function of cAMP response element-binding protein. J Biol Chem. 2013;40:28893–28899. doi: 10.1074/jbc.M113.502682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burdyga A, Conant A, Haynes L, Zhang J, Jalink K, Sutton R, Neoptolemos J, Costello E, Tepikin A. cAMP inhibits migration, ruffling and paxillin accumulation in focal adhesions of pancreatic ductal adenocarcinoma cells: effects of PKA and EPAC. Biochim Biophys Acta. 2013;12:2664–2672. doi: 10.1016/j.bbamcr.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stock AM, Powe DG, Hahn SA, Troost G, Niggemann B, Zanker KS, Entschladen F. Norepinephrine inhibits the migratory activity of pancreatic cancer cells. Exp Cell Res. 2013;12:1744–1758. doi: 10.1016/j.yexcr.2013.04.015. [DOI] [PubMed] [Google Scholar]
- 28.Tan X, Wang S, Yang B, Zhu L, Yin B, Chao T, Zhao J, Yuan J, Qiang B, Peng X. The CREB-miR-9 negative feedback minicircuitry coordinates the migration and proliferation of glioma cells. PLoS One. 2012;11:e49570. doi: 10.1371/journal.pone.0049570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steven A, Leisz S, Massa C, Iezzi M, Lattanzio R, Lamolinara A, Bukur J, Muller A, Hiebl B, Holzhausen HJ, Seliger B. HER-2/neu mediates oncogenic transformation via altered CREB expression and function. Mol Cancer Res. 2013;11:1462–1477. doi: 10.1158/1541-7786.MCR-13-0125. [DOI] [PubMed] [Google Scholar]
- 30.Corset V, Nguyen-Ba-Charvet KT, Forcet C, Moyse E, Chedotal A, Mehlen P. Netrin-1-mediated axon outgrowth and cAMP production requires interaction with adenosine A2b receptor. Nature. 2000;6805:747–750. doi: 10.1038/35037600. [DOI] [PubMed] [Google Scholar]


