Abstract
Objects: Neurite outgrowth inhibitor proteins (Nogos) comprise a family of three major members and are characterized by a conserved RHD domain. Among all the members, Nogo-B was identified to be significantly elevated and to play an important role in liver cirrhosis while Nogo-C was the shortest one and received little attention. The aim of this study is to investigate the relevance and mechanism of Nogo-C involved in Hepatocellular carcinoma (HCC). Methods: The expression of Nogo-C in paired HCC specimens was measured with quantitative RT-PCR. The function of Nogo-C over expressing in SMMC-7721 and WRL-68 HCC cell lines were estimated through cell proliferation assay and colony formation assay. A proteome-wide identification of Nogo-C-binding proteins was performed using affinity purification combined with a highly sensitive mass spectrometric technique. The protein interactions were confirmed using co-IP and immunofluorescence confocal assays. Results: Compared with the neighboring pathologically normal tissues, the expression of Nogo-C mRNA was extremely down-regulated in HCC specimens and was significantly related to greater tumor size and worse prognosis. Overexpression of Nogo-C in HCC cell lines resulted in an inhibition of cell growth. A total of 73 proteins were detected and considered in association with Nogo-C, among which B-raf and Nogo-B were validated. Conclusion: We identify Nogo-C as a tumor suppressor gene in HCC and B-raf as a novel interacting protein. These findings provide new directions for the mechanism research of Nogo family.
Keywords: Nogo-C, HCC, tumor suppressor gene, comparative proteomics, interactome, B-raf
Introduction
Reticulon (RTN) 4 is one member of the RTN protein family characterized by a homologous carboxy terminal tail containing an endoplasmic reticulum (ER) targeting motif. RTN4 has three major splice forms termed as neurite outgrowth inhibitor protein (Nogo)-A, -B, and -C, which only differ in their amino terminal sequence. Among them, Nogo-C bears the simplest structure of a unique eleven-amino-acid N-terminal tail and a reticulon homology domain (RHD), comparing with other two transcripts [1]. The three Nogo family members have a broad tissue expression pattern and multiple functions over them. Nogo-A expresses mainly in nervous tissues and behaves as a neurite outgrowth inhibitor [2]. Nogo-B is found in many tissues and regulates vascular remodeling in pathological vascular conditions caused by ischemia, atherosclerosis, and other insults [3-8]. Compared with the two well defined members, research on Nogo-C remains limited. Previous work identified that exogenous expression of Nogo-C in Schwann cells of transgenic mice induced a delayed axonal regeneration while similar expression in SMMC-7721 cells resulted in apoptosis via JNK-c-Jun dependent pathway accompanied with p53 activation [9,10], indicating its proapoptotic role under certain circumstances. However, given the high expression in skeletal muscle and nervous system, it seems that Nogo-C has ubiquitous roles other than participating in apoptosis [7,11].
Recent studies have shown diversified expression levels of Nogo family proteins in different tumors. The elevated expression of Nogo-A was detected correlated with the malignancy grading in oligodendroglial tumors, although its significance as a diagnostic marker was still argumentative [12-14]. It is also noteworthy that Nogo-B highly expresses in cervical cancer and induces cancer metastasis through Fibulin-5 [15]. However, current knowledge of expression and function of Nogo-C, as well as its possible mechanism in hepatocellular carcinoma (HCC) is still limited.
The co-IP protein identification technique is a new interaction detection experimental procedure based on the development of ultra-sensitive mass spectrometric techniques and it provides an important method for protein function exploring since it is crucial for all biological processes [16]. To unravel the mechanisms of Nogo-A and Nogo-B in regulating cell growth and migration, several in vitro approaches have been applied to identify Nogo protein complexes, including yeast two-hybrid interaction mating and AP-Nogo binding assay specifically searching for membrane proteins, in which the placental alkaline phosphatase (AP) is used as the affinity tag. Bcl-2, Bcl-xL, NIMP, membrane protein NgR and NgBR are identified as Nogo interaction proteins [17-19]. Yet, such large scale screening has not been done to Nogo-C.
In this study, we reported the down-regulated expression of Nogo-C and its correlations with clinic pathological features in HCC for the first time. Down-regulated expression was appeared in nearly 70% HCC specimens compared with the counterpart normal tissues, and tended to be correlated with greater tumor size and worse prognosis. In HCC cell lines, over-expression of Nogo-C resulted in lower tumor cell growth and colony formation. These results implicated the function of Nogo-C in inhibiting tumorigenesis. In addition, a comprehensively profile of Nogo-C interacting proteins were identified based on the proteomic research, among which Nogo-B and B-raf were confirmed to bind with Nogo-C. The evaluation of the data will allow us to better understand the function and mechanism of Nogo-C in inhibiting tumorigenesis.
Materials and methods
Cell lines and cell culture
In this study, liver tumor-derived cell lines SMMC-7721 and WRL-68 were used. Both cell lines were maintained in DMEM culture medium supplemented with 10% FBS in 5% CO2 at 37°C.
Human tissue samples
Surgical specimens were obtained from Zhongshan Hospital (Shanghai, China), approved by the Clinical Research Ethics Committee of Zhongshan Hospital of Fudan University. The tumor tissues and the neighboring pathologically nontumorous liver tissues were immediately frozen in liquid nitrogen after surgery and then stored at -80°C for further analysis.
Reverse transcription polymerase chain reaction and real time polymerase chain reaction (RT-PCR)
Total RNA was extracted from tissues using Trizol reagent (Invitrogen) and 1 ug of RNA were applied for reverse transcription (TAKARA). Real-time PCR analysis was conducted SYBR Green Supermix kit (Takara) with the ABI7300 detection system. Properly diluted cDNA was used in a 20 μl RT-PCR reaction in triplicate for each gene. Cycle parameters were 95°C for 30 s hot start and 40 cycles of 95°C for 5 s and 60°C for 30 s.
Primers for Nogo-C were forward, 5’-TGTCTCAGGGAGTAGGTTTGTG-3’, and reverse, 5’-TCCAGTACAGGAGGTCAACAA-3’. Primers for β-actin were forward, 5’-AATCGTGCGTGACATTAAGGAG-3’, and reverse, 5’-ACTGTGTTGGCGTACAGGTCTT-3’.
Cell proliferation and colony formation assay
SMMC-7721 and WRL-68 cell lines were transfection with either Nogo-C-pcDNA3.1 or empty pcDNA3.1 plasmids, using lipofectamine 2000 reagent (Invitrogen). For cell proliferation assay, all cells were plated at a density of 2,000 cells per well in 96-well plates. During a 5-day culture period, cells were subjected to cck-8 (Dojindo Laboratories, Tokyo) every day. The absorbance of each well (n=6) was measured at 1 h after incubation by a microtiter reader (Bio-Tek) at 450 nm. For colony formation assay, transfected hepatoma cells were plated at a density of 5 × 104 per well in 6-well plates. G418 was added into the medium 24 h later. Colonies were identified by crystal violet staining after about 10 to 14 days of culture.
SDS-PAGE and in-gel digestion
The protein mixtures affinity-purified with anti-Myc antibody were separated by SDS-PAGE. After SDS-PAGE, each gel lane was equally cut into 5 pieces, 1.5 mm in width. The proteins within gel slices were treated with DTT and iodoacetamide followed by in-gel digestion with trypsin.
Mass spectrometry (MS) analysis
The peptide mixtures of each piece were separation by RP chromatography (75 μm × 200 mm C18 column) and the peptides were eluted into a SYNAPT mass spectrometer coupled with a Nano LOCKSpray at a flow rate of 200 nL/min. NanoUPLC was used to deliver mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in ACN) at a linear gradient from 5% B to 50% B within 60 min, along with a gradient from 50% B to 90% B within 30 min and a hold at 90% B for 5 min.
Spectra were acquired in high-definition MSEmode switching every 1.2 seconds between low collision energy of 4 eV and high collision energy ramping from 15 eV to 40 eV. The acquired spectra were further processed with ProteinLynx Global Server (PLGS version 2.2.6, Waters, Milford, MA, USA) to reconstruct MS/MS spectra by combining all masses with identical retention times. The mass accuracy of the mass spectrometer was calibrated by the peak of m/z of 785.8426 Da [M+2H]2+ which was generated by glu-fibrinopeptide (GFP), when analyzed off-line at a concentration of 400 fmol/μl and a flow rate of 100 nl/min.
Mass data interpretation
The MS/MS spectra were searched against the human international protein index protein database (IPI ID: 2.66) downloaded from ftp://ftp.ebi.ac.uk/pub/databases/IPI/current/ [20]. The min fragment ion matches per peptide was set at three while the min fragment ion matches per protein was set at seven. The Min Peptide Matches Per Protein was configured to one. Two missed cleavage sites were allowed. Variable modifications of oxidation (M) and carbamidomethyl (C) were permitted. The false positive rate was fixed at 4%.
Coimmunoprecipitation (Co-IP) assay
Co-IP assay was taken to validate the proteins identified by MS. Plasmids encoding the identified candidate proteins and the bait Nogo-C were transfected into SMMC-7721 cells for 36 h before the cells were lysed using 1 × CLB. For co-IP, 10 μl protein A/G-Sepharose beads pre-absorbed with appropriate antibodies were utilized to isolate the immunocomplexes. The isolated beads were washed twice with CLB and twice with RIPA buffer (0.15 mM NaCl/0.05 mM Tris-HCl, pH 7.2/1% Triton X-100/1% sodium deoxycholate/0.1% SDS) [21]. Immunoblot (IB) analysis was conducted to probe the resolved complexes with indicated antibodies. Monoclonal anti-c-Myc antibody (clone 9E10; Chemicon) and monoclonal anti-HA antibody (clone 12CA5; Roche) were used for isolation and detection of the tagged Nogo-B, Nogo-C and B-Raf.
Immunofluorescence microscopy
Transfected SMMC-7721 cells were fixed with 4% paraformaldehyde for 15 min at room temperature, permeabilized with 0.1% Triton X-100. After blocking in phosphate-buffered saline (PBS) containing 5% BSA for 1 h, cells were immunostained with primary anti-Myc antibody (9E10) overnight at 4°C, followed by incubation with Cy3-conjugated anti-mouse immunoglobulin G antibody (Biological Detection Systems) at room temperature for 1 hours. To visualize nuclei, cells were incubated in 100 μg/ml 4,6-diamidino-2-phenylindole (DAPI) in PBS for 10 minutes. Coverslips were mounted and observed under Zeiss confocal microscope.
Bioinformatic and statistic data analyses
The Gene Ontology (GO) is the current standard for annotating gene products and proteins. The identified candidate proteins were annotated to the terms of molecular function and cellular component based on GO under standard procedure using Blast2Go [22] (http://www.blast2go.org/).
Non parametric test was used to determine the Spearman correlation between the Nogo-C expression level and the clinial characteristics (PRISM software version 5.0, GraphPad, San Diego, CA). A two-tailed Student’s t test was used to evaluate the significance between grouped data. P<0.05 was considered statistically significant.
Result
Nogo-C was frequently down-regulated in HCC
We first assessed Nogo-C expression pattern in several normal human tissues using semi-quantitative PCR. Nogo-C was relatively abundant in the brain, heart, kidney and liver tissues, with very low levels in fetal liver and other tissues (Figure 1A). In 10 pairs of HCC specimens with the matched non-tumor tissue counterparts, Nogo-C expression was extremely down-regulated (Figure 1B). To further explore the clinicopathological correlation of Nogo-C down-regulation in HCC tissues, the quantitative reverse transcriptase PCR assay was conducted to evaluate Nogo-C expression in other 76 HCC specimens. The result showed that the mRNA levels encoding Nogo-C was down-regulated (expression was decreased more than twofold) in 50 (65.8%) of 76 HCC specimens (Figure 1C). The expression level of Nogo-C in these HCC specimen appeared to be negatively correlated with HCC tumor size (r=-0.25, P=0.036) (Figure 1D). Patients with HCC with lower Nogo-C level had a shorter survival times than patients with HCC expressing high levels of Nogo-C (r=0.421, P<0.001, Figure 1E).
Figure 1.
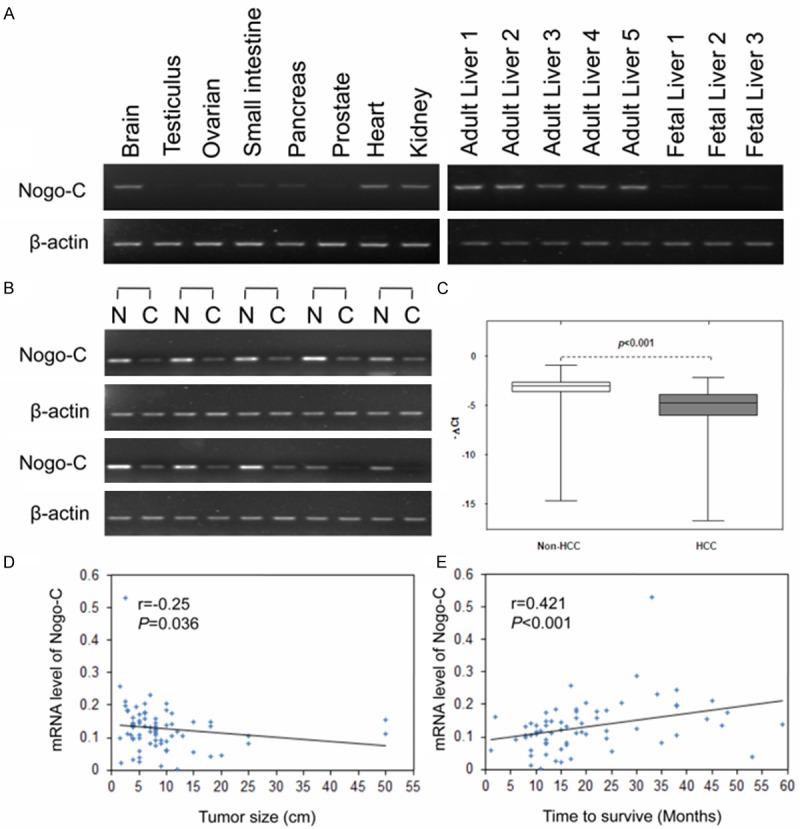
Down-regulated expression of Nogo-C in human hepatocellular carcinomas (HCC). (A) Semi-quantitative PCR analysis of Nogo-C expression in 9 human adult tissues and fatal liver. Expression of the human β-actin served as a loading control. (B) Nogo-C mRNA levels were analyzed in 6 paired HCCs with their corresponding non-cancerous specimens by semi-quantitative PCR. (C) Nogo-C mRNA expression levels in 76 paired HCC specimens with their corresponding neighboring nontumorous specimens measured by RT-PCR. (D and E) The Spearman correlation between tumor size (D) or between patient’s living time and Nogo-C mRNA expression level in HCC specimens (E).
Nogo-C suppressed HCC cell growth
To evaluate whether Nogo-C function as a tumor suppressor gene in HCC cells, we further observed the effect of Nogo-C expression on cell proliferation and colony formation. SMMC-7721 and WRL-68 cells were transiently transfected with Nogo-C-pcDNA3.1 and control vector. Cell growth and colony formation of these cells were suppressed by Nogo-C overexpression (Figure 2).
Figure 2.
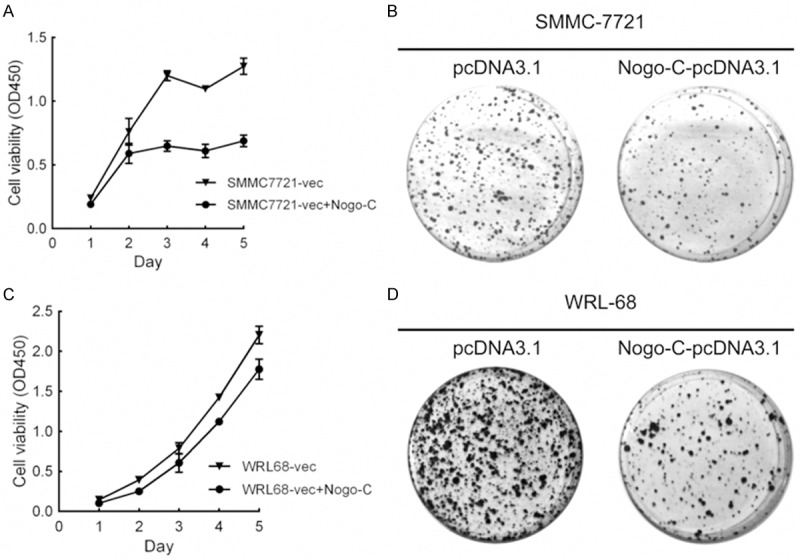
Overexpression of Nogo-C suppressed growth of HCC cell lines. (A and B) Exogenous Nogo-C was expressed in SMMC-7721 (A) and WRL-68 (B) cells transfected with the pcDNA3.1 vector. Empty vector were used as a control. The growth of these cells was analyzed using cck-8 kit. (C and D) Representative images of colony formation assay in SMMC-7721 (C) and WRL-68 (D).
Interactome analysis of Nogo-C
To identify Nogo-C interacting proteins, a proteomic analysis was carried out. The scheme of the strategy was shown in Figure 3A. Plasmids consisting of Nogo-C and the empty pCMV vectors were transfected into SMMC-7721 cells in parallel and the cell lysates were acquired separately. The transfection efficiency of the plasmids in both samples was evaluated using western blot (Figure 3B). Immunoprecipitations were done to both Nogo-C sample and pCMV sample using anti-Myc antibody. A combined one-dimensional PAGE with LC/MS/MS strategy was used to identify compartments of protein complexes. The vast majority of the sequence-matched spectra were of good quality and all the identified peptides were above the threshold score for identity with a confidence level of >95%. We assigned 2,261 distinct peptides with significant probability scores into 325 unique proteins in all, including 165 proteins identified in the Nogo-C sample and 160 proteins in the pCMV sample. All proteins were identified based on the presence of at least 2 peptide fragments. Ninety-one proteins identified in both samples and 69 proteins identified in the pCMV sample were excluded as non-specific binding proteins. Apart from the identified core protein Nogo-C, 73 proteins specifically detected in the Nogo-C sample were recognized as potential Nogo-C interacting proteins. Their information, including accession number (IPI#), protein name, molecular weight, number of detected peptides, PLGS score, PI (PH) and their molecular functions, were summarized in Table 1. It ought to be emphasized that the bait protein Nogo-C was only detectable in the Nogo-C sample, providing the evidence of the high efficiency of the immunopurification. Five peptides of the core protein Nogo-C were detected, with a total peptide sequence coverage of 43% (Figure 3C), and the illustration of the representative fragment of Nogo-C was shown in Figure 3D.
Figure 3.
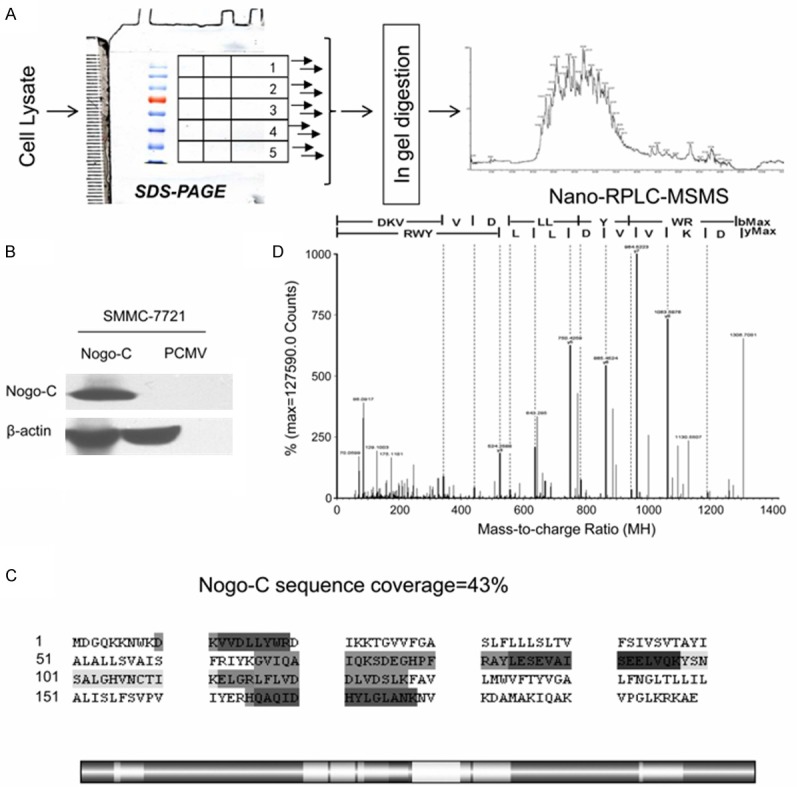
Proteomic analysis of Nogo-C binding proteins. A: The schematic flow chart of the MS-based proteomic analysis of the protein complexes. B: Ectogenous Myc-Nogo-C plasmids and the pCMV plasmids were transfected separately into SMMC-7721 cells. The presence or absence of Nogo-C on Western blots was revealed by probing with an anti-Myc antibody. β-actin was used as the loading control. C: Sequence coverage of Nogo-C (43%) from the 5 identified peptides (emphasized in gray) from the experiment. The distribution of the identified peptides of Nogo-C was shown in the strip chart in highlight. The dark gray section represented the overlapped sequences from identified proteins. D: One representative illustration of a MS/MS specific peptide of Nogo-C.
Table 1.
Summary of Nogo-C interacting proteins identified
| Accession | Annotationa | mW (Da) | Peptidesb | PLGS | pI (pH) | molecular functionc |
|---|---|---|---|---|---|---|
|
| ||||||
| Score | ||||||
| structural molecule | ||||||
| IPI00478908.3 | TUBA1C | 28545 | 4 | 24.9027 | 5.0944 | SCOC; GTP-A |
| IPI00007752.1 | TUBB2C | 49799 | 7 | 25.3933 | 4.602 | SCOC; GTP-A |
| IPI00387144.5 | TUBA1B | 57723 | 4 | 10.9827 | 4.7631 | SMA; GTP-A |
| IPI00791613.1 | TUBA8 | 51951 | 2 | 7.4946 | 4.9852 | SCOC; GTP-A |
| IPI00295857.7 | COPA | 138257 | 13 | 19.4444 | 7.4464 | SMA |
| IPI00646493.1 | COPA | 139235 | 5 | 7.1371 | 7.3354 | SMA |
| IPI00386712.1 | MYL4 | 21550 | 2 | 15.736 | 4.7834 | SCOM; AF-B |
| binding | ||||||
| IPI00011253.3 | RPS3 | 26671 | 6 | 27.1605 | 10.0402 | SCOR |
| IPI00878297.1 | RPS2 | 29984 | 4 | 24.9084 | 8.9835 | SCOR; FGF-B |
| IPI00877699.1 | TRIOBP | 28939 | 7 | 21.4844 | 5.5144 | MGRA-B |
| IPI00014149.3 | TTC35 | 34811 | 3 | 14.1414 | 6.1335 | binding |
| IPI00029266.1 | SNRPE | 10796 | 2 | 25 | 9.7804 | DR-B; RNA-S |
| IPI00644224.2 | HNRNPU | 86806 | 14 | 27.551 | 6.0108 | DRP-B |
| IPI00908896.1 | HNRNPH | 16838 | 2 | 30.1887 | 4.821 | RP-B |
| IPI00644079.2 | HNRNPU | 90527 | 14 | 1222.6204 | 5.6479 | RP-B |
| IPI00643152.2 | HSP70 | 77513 | 8 | 17.305 | 8.1035 | PA-B |
| IPI00910482.1 | HSP70 | 61492 | 10 | 445.444 | 5.2773 | PA-B |
| IPI00878876.1 | SNRPD3 | 13282 | 2 | 15.8333 | 9.2411 | RP-B |
| IPI00167147.1 | HNRNPU | 90235 | 11 | 25.3731 | 6.4883 | RP-B |
| IPI00219037.5 | H2AFX | 15135 | 2 | 11.1888 | 11.1559 | DP-B |
| IPI00642249.1 | AMOT | 68350 | 4 | 8.7102 | 8.016 | P-B |
| IPI00028888.1 | HNRNPD | 38410 | 3 | 9.5775 | 7.8979 | DP-B; TF |
| IPI00479191.2 | HNRNPH | 51197 | 5 | 32.4153 | 6.3431 | DRP-B |
| IPI00013877.2 | HNRNPH | 36903 | 3 | 11.2717 | 6.3999 | RP-B |
| IPI00479217.1 | HNRNPU | 88924 | 14 | 25.6824 | 5.4659 | DRPA-B |
| IPI00217030.10 | RPS4X | 29579 | 5 | 28.1369 | 10.5861 | D-B |
| IPI00916188.1 | NCL | 32427 | 4 | 21.8855 | 5.0956 | RP-B |
| IPI00604620.3 | NCL | 76568 | 4 | 139.2712 | 4.4004 | RP-B |
| IPI00402185.4 | SYNCRIP | 46299 | 3 | 9.5122 | 9.1055 | PS; RP-B |
| IPI00641719.1 | SURF4 | 21113 | 5 | 19.3548 | 6.0936 | P-B |
| IPI00399142.5 | SURF4 | 21115 | 2 | 21.8085 | 8.7072 | P-B |
| IPI00023673.1 | LGALS3BP | 65289 | 3 | 11.453 | 4.9438 | P-B |
| IPI00887555.1 | LGALS3BP | 61605 | 4 | 86.0234 | 5.1229 | P-B |
| IPI00889009.1 | RPL23A | 22681 | 3 | 11.1111 | 10.5355 | SCOR; DRP-B |
| IPI00455428.3 | RPS2 | 29745 | 4 | 13.8182 | 10.3555 | SCOR; FGF-B |
| IPI00888294.1 | RPS2 | 25528 | 4 | 18.4549 | 10.2367 | SCOR; FGF-B |
| IPI00187140.1 | RPS26 | 12993 | 2 | 18.2609 | 10.9547 | SCOR; RP-B |
| IPI00886830.1 | RPS16 | 18223 | 2 | 10.9756 | 9.4964 | SCOR; RP-B |
| IPI00397609.2 | RPS18 | 17627 | 5 | 28.9474 | 10.7721 | SCOR; R-B |
| IPI00909890.1 | FUS | 44785 | 8 | 24.9417 | 9.1479 | DRPI-B |
| IPI00260715.5 | FUS | 53344 | 7 | 675.9695 | 9.559 | DRPI-B |
| IPI00045801.3 | FIZ1 | 51960 | 2 | 5.0403 | 8.0577 | DPI-B |
| IPI00397701.3 | RPS16 | 16401 | 4 | 26.7123 | 10.3768 | RP-B |
| IPI00456429.3 | UBA52 | 14718 | 3 | 27.3438 | 10.2647 | P-B |
| IPI00789951.2 | CALML4 | 21459 | 3 | 15.8974 | 6.7083 | Ca2+-B |
| IPI00478437.3 | CALML4 | 21869 | 2 | 14.7959 | 7.4013 | Ca2+-B |
| IPI00014456.4 | STRN | 86078 | 4 | 6.5385 | 4.9459 | Ca2+-B; CA |
| IPI00219806.7 | S100A7 | 11463 | 2 | 21.7822 | 6.3404 | ZI/Ca2+-B |
| IPI00845339.1 | HSP70 | 69995 | 21 | 34.4774 | 5.3154 | DPA-B; UPL-B |
| IPI00005667.4 | N4BP1 | 100345 | 6 | 11.8304 | 5.0488 | MI-B |
| signal transducer | ||||||
| IPI00873380.1 | Nogo C | 22238 | 25 | 43.2161 | 9.5266 | ST; P-B |
| IPI00021766.5 | RTN4 | 129851 | 22 | 7.047 | 4.2241 | ST; P-B |
| IPI00298289.1 | RTN4 | 40292 | 10 | 26.2735 | 4.4905 | ST; P-B |
| IPI00219207.1 | RTN4 | 22381 | 27 | 34.6734 | 9.6482 | ST; P-B |
| IPI00477663.1 | RTN4 | 106294 | 21 | 6.9792 | 4.2167 | ST; P-B |
| IPI00894099.1 | RTN4 | 22791 | 9 | 28.5714 | 9.0452 | ST; P-B |
| IPI00478442.3 | RTN4 | 42247 | 7 | 18.3673 | 4.4485 | ST; P-B |
| IPI00894213.1 | RTN4 | 108382 | 3 | 5.2738 | 4.3154 | ST; P-B |
| IPI00220880.1 | RGS20 | 31465 | 4 | 24.9084 | 5.04 | ST; P-B |
| IPI00303797.3 | B-RAF | 84383 | 2 | 179.4723 | 7.2643 | MAPKKK |
| IPI00020898.1 | RPS6KA3 | 83683 | 5 | 12.8378 | 6.4204 | MAPKKK |
| transcription regulator activity | ||||||
| IPI00641665.1 | ILF2 | 12374 | 4 | 63.4783 | 4.8984 | TF |
| IPI00011274.3 | HNRPDL | 46409 | 5 | 14.5238 | 9.8221 | TF; DRP-B |
| IPI00798127.1 | UBC | 76981 | 3 | 4.9635 | 7.7573 | T; COR |
| IPI00798155.4 | UBC | 12229 | 4 | 29.2453 | 10.5756 | T; COR |
| IPI00556173.1 | ILF3 | 76454 | 5 | 9.3484 | 7.853 | TR; I-B |
| IPI00298788.4 | ILF3 | 95279 | 5 | 9.9553 | 8.9555 | TF |
| transporter | ||||||
| IPI00797148.1 | HNRNPA1 | 29368 | 7 | 33.3333 | 9.4607 | RNA TT |
| IPI00879501.2 | HNRNPA1 | 34204 | 5 | 16.875 | 9.2814 | RNA TT |
| IPI00644968.1 | HNRNPA1 | 34202 | 6 | 23.75 | 9.4684 | RNA TT |
| IPI00176692.7 | HNRNPA1 | 32142 | 5 | 495.4168 | 9.1154 | RNA TT |
| IPI00478539.2 | HNRNPA1 | 32360 | 5 | 21.5947 | 9.1631 | RNA TT |
| unknown function | ||||||
| IPI00902571.1 | TNRC18 | 169808 | 5 | 7.8076 | 7.1545 | unknown |
| IPI00186448.7 | TNRC18 | 64571 | 3 | 10.2564 | 10.2389 | unknown |
The gene name encoding for each protein identified;
Number of unique peptides found for each protein identified;
SCOC, structural constituent of cytoskeleton;
GTP-A, GTP activation; SMA, structural molecule activity; SCOM, structural constituent of muscle; SCOR, structural constituent of ribosome; RNA-B, RNA binding; FGF-B, FGF binding; MGRA-B, myosin/GTP-Rho/actin binding; DRP-B, DNA/RNA/protein binding; D-B, DNA binding; DR-B, DNA/RNA binding; RNA-S, RNA splicing; RP-B, RNA/protein binding; PA-B, protein/ATP binding; DP-B, DNA/protein binding; P-B, protein binding; TF, transcription factor; DRPA-B, protein/RNA/DNA/ATP binding; PS, protein stabilization; DRPI-B, DNA/RNA/protein/ion binding; DPI-B, protein kinase/DNA/ion binding; Ca2+-B, Ca2+ binding; CA, catalytic activity; ZI/Ca2+-B, zinc ion/Ca2+ binding; MI-B, metal ion binding; ST, signal transducer; MAPKKK, MAP kinase kinase kinase activity; COR, constituents of ribosome; TR, transcription repressor; I-B, ion binding; DPA-B, DNA/Protein/ATP binding; UPL-B, ubiquitin protein ligase binding; RNA TT, RNA transmembrane transporter; R-B, RNA binding; AF-B, actin filament binding.
To better characterize the Among the 73 proteins, 71 proteins (97.2%) were of known functions, which could be characterized into five groups according to their molecular functions. Seven proteins are structural molecules of cytoskeleton, including TUBA8, COPA and MYL4. Forty-three proteins are proteins binding with various components of cells, such as DNA, RNA, Protein, ion, Ca2+ or ATP, including TRIOBP, AMOT, SNRPE, SYNCRIP, SURF4, LGALS3BP, CALML4, STRN, S100A7, N4BP1 and HSPA1B. Ten proteins are signal transducer, including RTN4 family proteins, B-RAF and RGS20. Six proteins are transcription-related proteins, including RPS6KA3, UBC and ILF3. Five HNRNPA1 proteins are detected functioning as RNA transporter. The rest two TNRC18 proteins are of still unknown functions to our knowledge, and need more investigation. The highly diverse of interaction proteins and their multi functions indicated the complication of Nogo-C function. Other RTN4 family members were detected to bind with Nogo-C, demonstrating the previously reported interaction between Nogo-A, -B and -C through their hydrophobic reticulon domain.
Nogo-B and B-Raf interacted with Nogo-C
To confirm the interaction between Nogo-C and proteins identified by mass spectrometry, Nogo-B and B-Raf were chosen for validations. Their unique MS/MS sequencing peptides were shown in Figure 4.
Figure 4.
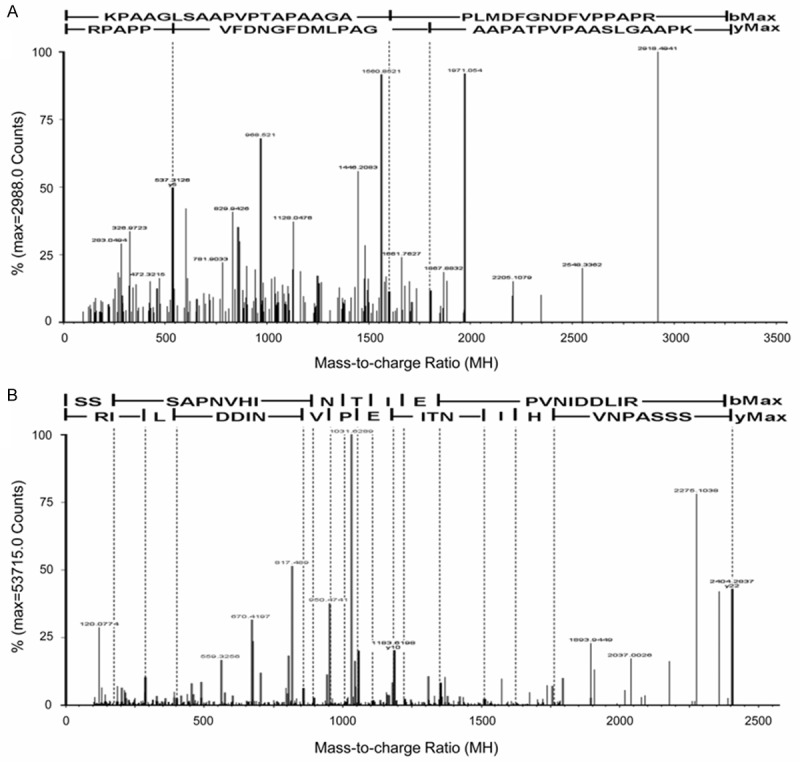
Peptides of selected proteins identified by MS/MS spectrum. A: The MS/MS sequencing of the specific peptide of Nogo-B: KPAAGLSAAPVPTAPAAGAPLMDFGNDFVPPAPR. B: The MS/MS sequencing of the specific peptide of B-RAF: SSSAPNVHINTIEPVNIDDLIR. Reversed database searching were performed to optimize the specificity of the data, and the results were shown under the sequence of each peptide.
Co-IP experiments were first performed for the verification of the possible interactions. The plasmids of the proteins to be validated were transfected into SMMC-7721 cells together with the Nogo-C plasmid. Western blots were performed to trace the proteins and as shown in Figure 5A and 5B, Myc-Nogo-C was detectable at approximately 25 KD after being immunoprecipitated with anti-HA monoclonal antibody. In the ‘reciprocal’ IP experiment, the protein of Myc-B-Raf was also detectable at approximately 85 KD with anti-Myc antibody.
Figure 5.
Verification of identified protein binding with Nogo-C. Co-immunoprecipitation and intracellular co-localization between identified proteins and Nogo-C were performed. A: Myc-Nogo-C and HA-Nogo-B. B: HA-Nogo-C and Myc-B-Raf. Both forward and reciprocal immunoprecipitations were performed using anti-HA or anti-Myc monoclonal antibody. C: Co-localization of exogenous Myc-Nogo-B and Nogo-C-GFP. D: Co-localization of exogenous Myc-B-Raf and Nogo-C-GFP. primary anti-Myc monoclonal antibody and Alexa Fluor 488-congulated secondary antibody were used. SMMC-7721 cells were observed by confocal laser scanning microscope. Bars represent 10 μm.
Then the subcellular localization of the protein complexes were observed in SMMC-7721 cells using confocal. The intracellular localization of Nogo-C-Nogo-B complex co-localized in cytoplasm (Figure 5C) while Nogo-C-B-Raf complex was in both cytoplasm and nucleus (Figure 5D), which might be affected by the dual localization of B-raf.
Discussion
Nogo-C is a member of Nogo family with no well-known functions. In this study, we identified Nogo-C as a tumor suppressor gene in HCC, and B-raf as a novel interacting protein of Nogo-C. This study is the first high-throughput functional proteomic analysis on Nogo-C. The validity of our data was supported by two evidences. First, the high peptide sequence coverage of Nogo-C in test samples convinces the specificity and efficiency of the immunopurification; Second, the identification of other members of Nogo family, coincident with the previously reported interaction among Nogo-A, -B, -C [23]. The remaining proteins are supposed to form complex with Nogo-C in either direct or indirect way. Function of Nogo-C could be analyzed according to these protein complexes.
B-Raf is one of the key moleculars consisting the mitogen-activated protein kinase (MAPK) pathway, and involved in the control of cell growth, proliferation, and migration in majority of the cancers [24]. Antagonism of B-raf using sorafenib is demonstrated to be effective in HCC [25]. For the first time, we demonstrated the interaction between Nogo-C and B-Raf and suggested the relationship between Nogo-C and RAF/MAPK pathway. It is possible for Nogo-C to modulate the kinase activity by altering its phosphorylation and tethering signaling components to a specific area of the cell like the previously reported interacting proteins; or to function as a physiological substrate of the kinase, similarly with its homolog Nogo-B [26].
Coatomer-alpha subunit (COPA) and Surf4 are ER-located proteins with dilysine motif identified in our results. Abundance of peptides identified and the overlapping localization with Nogo-C intensively suggested the possibility of interactions. Functioning as cargo receptors, both COPA and Surf4 not only mediate proteins sorting in anterograde and retrograde cellular transportation in the secretory pathway, but also stabilize the cellular membrane architecture in an oligomeric state [27-29]. Reticulons, including Nogo-A, have been reported to be a key component to structurally shape ER tubules [30,31]. Thus, it is highly possible for Nogo-C to function with the same region, forming complexes with the proteins identified, in either protein trafficking or ER tubule formation.
Disclosure of conflict of interest
None.
References
- 1.BI Anding YL, Yang J, Zhang M, Zhou Y, Zhao SY. Cloning and expression analysis of human reticulon 4c cDNA. Chinese Science Bulletin. 2000;45:1862–1869. [Google Scholar]
- 2.Yang YS, Strittmatter SM. The reticulons: a family of proteins with diverse functions. Genome Biol. 2007;8:234. doi: 10.1186/gb-2007-8-12-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bullard TA, Protack TL, Aguilar F, Bagwe S, Massey HT, Blaxall BC. Identification of Nogo as a novel indicator of heart failure. Physiol Genomics. 2008;32:182–189. doi: 10.1152/physiolgenomics.00200.2007. [DOI] [PubMed] [Google Scholar]
- 4.Paszkowiak JJ, Maloney SP, Kudo FA, Muto A, Teso D, Rutland RC, Westvik TS, Pimiento JM, Tellides G, Sessa WC, Dardik A. Evidence supporting changes in Nogo-B levels as a marker of neointimal expansion but not adaptive arterial remodeling. Vascul Pharmacol. 2007;46:293–301. doi: 10.1016/j.vph.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rodriguez-Feo JA, Hellings WE, Verhoeven BA, Moll FL, de Kleijn DP, Prendergast J, Gao Y, van der Graaf Y, Tellides G, Sessa WC, Pasterkamp G. Low levels of Nogo-B in human carotid atherosclerotic plaques are associated with an atheromatous phenotype, restenosis, and stenosis severity. Arterioscler Thromb Vasc Biol. 2007;27:1354–1360. doi: 10.1161/ATVBAHA.107.140913. [DOI] [PubMed] [Google Scholar]
- 6.Yu J, Fernandez-Hernando C, Suarez Y, Schleicher M, Hao Z, Wright PL, DiLorenzo A, Kyriakides TR, Sessa WC. Reticulon 4B (Nogo-B) is necessary for macrophage infiltration and tissue repair. Proc Natl Acad Sci U S A. 2009;106:17511–17516. doi: 10.1073/pnas.0907359106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oertle T, Schwab ME. Nogo and its paRTNers. Trends Cell Biol. 2003;13:187–194. doi: 10.1016/s0962-8924(03)00035-7. [DOI] [PubMed] [Google Scholar]
- 8.Zhang D, Utsumi T, Huang HC, Gao L, Sangwung P, Chung C, Shibao K, Okamoto K, Yamaguchi K, Groszmann RJ, Jozsef L, Hao Z, Sessa WC, Iwakiri Y. Reticulon 4B (Nogo-B) is a novel regulator of hepatic fibrosis. Hepatology. 2011;53:1306–15. doi: 10.1002/hep.24200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Tang X, Cao X, Chen H, Zhang X. Human Nogo-C overexpression induces SMMC-7721 cell apoptosis via a mechanism that involves JNK-c-Jun pathway. Biochem Biophys Res Commun. 2006;348:923–928. doi: 10.1016/j.bbrc.2006.07.166. [DOI] [PubMed] [Google Scholar]
- 10.Kim JE, Bonilla IE, Qiu D, Strittmatter SM. Nogo-C is sufficient to delay nerve regeneration. Mol Cell Neurosci. 2003;23:451–459. doi: 10.1016/s1044-7431(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 11.Magnusson C, Libelius R, Tagerud S. Nogo (Reticulon 4) expression in innervated and denervated mouse skeletal muscle. Mol Cell Neurosci. 2003;22:298–307. doi: 10.1016/s1044-7431(02)00036-2. [DOI] [PubMed] [Google Scholar]
- 12.Jung TY, Jung S, Lee KH, Cao VT, Jin SG, Moon KS, Kim IY, Kang SS, Kim HS, Lee MC. Nogo-A expression in oligodendroglial tumors. Neuropathology. 2011;31:11–9. doi: 10.1111/j.1440-1789.2010.01118.x. [DOI] [PubMed] [Google Scholar]
- 13.Marucci G, Di Oto E, Farnedi A, Panzacchi R, Ligorio C, Foschini MP. Nogo-A: a useful marker for the diagnosis of oligodendroglioma and for identifying 1p19q codeletion. Hum Pathol. 2012;43:374–80. doi: 10.1016/j.humpath.2011.05.007. [DOI] [PubMed] [Google Scholar]
- 14.Xiong NX, Zhao HY, Zhang FC, He ZQ. Negative correlation of Nogo-A with the malignancy of oligodendroglial tumor. Neurosci Bull. 2007;23:41–45. doi: 10.1007/s12264-007-0006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao W, Zhou S, Xu H, Li H, He G, Liu Y, Qi Y. Nogo-B promotes the epithelial-mesenchymal transition in SMMC-7721 cervical cancer cells via Fibulin-5. Oncol Rep. 2013;29:109–16. doi: 10.3892/or.2012.2069. [DOI] [PubMed] [Google Scholar]
- 16.Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 17.Fournier AE, GrandPre T, Strittmatter SM. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature. 2001;409:341–346. doi: 10.1038/35053072. [DOI] [PubMed] [Google Scholar]
- 18.Hu WH, Hausmann ON, Yan MS, Walters WM, Wong PK, Bethea JR. Identification and characterization of a novel Nogo-interacting mitochondrial protein (NIMP) J Neurochem. 2002;81:36–45. doi: 10.1046/j.1471-4159.2002.00788.x. [DOI] [PubMed] [Google Scholar]
- 19.Tagami S, Eguchi Y, Kinoshita M, Takeda M, Tsujimoto Y. A novel protein, RTN-XS, interacts with both Bcl-XL and Bcl-2 on endoplasmic reticulum and reduces their anti-apoptotic activity. Oncogene. 2000;19:5736–5746. doi: 10.1038/sj.onc.1203948. [DOI] [PubMed] [Google Scholar]
- 20.Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- 21.Jiang M, Gao Y, Yang T, Zhu X, Chen J. Cyclin Y, a novel membrane-associated cyclin, interacts with PFTK1. FEBS Lett. 2009;583:2171–2178. doi: 10.1016/j.febslet.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Gotz S, Garcia-Gomez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talon M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodd DA, Niederoest B, Bloechlinger S, Dupuis L, Loeffler JP, Schwab ME. Nogo-A, -B, and -C are found on the cell surface and interact together in many different cell types. J Biol Chem. 2005;280:12494–12502. doi: 10.1074/jbc.M411827200. [DOI] [PubMed] [Google Scholar]
- 24.Colombino M, Sperlongano P, Izzo F, Tatangelo F, Botti G, Lombardi A, Accardo M, Tarantino L, Sordelli I, Agresti M, Abbruzzese A, Caraglia M, Palmieri G. BRAF and PIK3CA genes are somatically mutated in hepatocellular carcinoma among patients from South Italy. Cell Death Dis. 2012;3:e259. doi: 10.1038/cddis.2011.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc JF, de Oliveira AC, Santoro A, Raoul JL, Forner A, Schwartz M, Porta C, Zeuzem S, Bolondi L, Greten TF, Galle PR, Seitz JF, Borbath I, Haussinger D, Giannaris T, Shan M, Moscovici M, Voliotis D, Bruix J. Sorafenib in advanced hepatocellular carcinoma. N Engl J Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 26.Rousseau S, Peggie M, Campbell DG, Nebreda AR, Cohen P. Nogo-B is a new physiological substrate for MAPKAP-K2. Biochem J. 2005;391:433–440. doi: 10.1042/BJ20050935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nyfeler B, Reiterer V, Wendeler MW, Stefan E, Zhang B, Michnick SW, Hauri HP. Identification of ERGIC-53 as an intracellular transport receptor of alpha1-antitrypsin. J Cell Biol. 2008;180:705–712. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nyfeler B, Zhang B, Ginsburg D, Kaufman RJ, Hauri HP. Cargo selectivity of the ERGIC-53/MCFD2 transport receptor complex. Traffic. 2006;7:1473–1481. doi: 10.1111/j.1600-0854.2006.00483.x. [DOI] [PubMed] [Google Scholar]
- 29.Nyfeler B, Michnick SW, Hauri HP. Capturing protein interactions in the secretory pathway of living cells. Proc Natl Acad Sci U S A. 2005;102:6350–6355. doi: 10.1073/pnas.0501976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK. The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem. 2008;283:18892–18904. doi: 10.1074/jbc.M800986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–586. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]



