Abstract
Protein phosphatase 1, regulatory subunit 13 like PPP1R13L, also coined iASPP, was found high expression in prostate cancer tissues and cell lines. In previous research, in vitro and in vivo RNAi mediated by artificial lentiviral shRNAs which proved that suppression of iASPP decrease the proliferation of cancer cells. Endogenous interference RNAs, microRNAs play key roles in cell proliferation by post-transcriptional regulation of gene expression. Natural base pair matched microRNA for iASPP is mir124, which was found high expression in growth factorloss prostate cancer cell lines. In this study we examined effect of mir124 upon iASPP and proliferation of prostate cells in vitro with lentiviral infection and use artificial shRNA as control. In vitro reporter assay confirmed that mir124 binding the 3’UTR of iASPP and suppress mRNA expression. Lentivirus mediated mir124 expression decreased the proliferation and viability of PC3 while endogenous iASPP were knocked down.
Keywords: Mir124, iASPP, prostate cancer, cell growth
Introduction
Prostate cancer (PCa) is the most commonly detected malignancy of the male and the second leading cause of cancer death [1,2]. Investigation of the molecular mechanisms that underlie the progression of PCa may help to develop new effective clinical therapies and consequently benefit patient. Our studies found that high expression of iASPP, Homo sapiens protein phosphatase 1, regulatory (inhibitor) subunit 13 like (PPP1R13L), in prostate tumor tissues. Down-regulation of endogenous iASPP expression by lentiviral shRNA inhibitor growth and decrease proliferation in the prostate cancer cell lines PC-3 and DU145 in vitro and significantly reduced the tumorigeneses potency of DU145 in xenograft model. Which suggested that iASPP could be a molecular target in prostate cancer therapy [3].
miRNAs (microRNAs) are small, non-coding RNA (,20-22 nucleotides) that negatively regulate gene expression at the post-transcriptional level [4]. Given the important roles of miRNAs in post-transcriptional regulation, identification of miRNAs targeting certain protein will further uncover the molecular mechanisms involved and intrigue novel method for therapy of prostate cancer. As an abundant class of non-coding RNAs, miRNAs often are evolutionarily conserved in metazoans and expressed in a cell and tissue specific manner. MicroRNAs exert their gene regulatory activity primarily by imperfect base pairing to the 3’UTR of their target mRNAs, leading to mRNA degradation or translational inhibition. They are involved in numerous cellular processes including proliferation, differentiation, apoptosis and metabolism [5]. Due to complicated combination relation of miRNA and UTR of mRNA, several online tools have been developed for prediction. These resources provide microRNA target predictions based on sequence complementarity to target sites with emphasis on perfect base-pairing in the seed region and sequence conservation, TargetScan [6], PicTar [7], Target Rank [8].
In prostate cancer, functional miRNA were screened by high throughput method as microarray or next generation sequencing (NGS). These transcriptomic level researches provide abundant data and highlight a number of miRNA expression markers. In spite of abundance of data from wide scale research, consequence of mRNA regulation requires substantial evidence in vitro. White et al have performed in LNCaP cells to identify mir125b a key regulator of cell growth [9].
To identify the iASPP targeting miRNA we primarily analyze the expression of candidate miRNA predicted by software in patient samples and investigate the inhibition of iASPP by introduction of exogenous miRNA expression.
Materials and methods
Cells culture and tissue samples
PC-3 and DU145, human prostate cancer cell lines, were purchased from ATCC (Manassas, VA, USA). PC-3 and DU145 cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) containing 10% fetal bovine serum (FBS), 2 mmol/l glutamine, 100 units/ml penicillin and 100 g/ml streptomycin and cultured in a humidified atmosphere of 95% air and 5% CO2 at 37°C. Five patients with prostate cancer were recruited from our outpatient and in-patient services at the Third Affiliated Hospital of Sun Yat-Sen University (Guangzhou, China). Their prostate tumor tissues and adjacent tissues were collected during the surgical procedures. Informed consent was obtained from individual subjects. The experimental protocols were approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-Sen University.
Construction of mir124 expression lentivirus vectors
To generate lentivirus expressing mature miRNA of mir124, the pre-miRNA sequence were synthesized and linked into vector, control construct (control RNAi) having no homology with human genome was created by a scrambled sequence (AAT GTA CTG CGC GTG GAG A). The sequences were cloned into the HpaI and XhoI sites of the pGCSIL-GFP (Neuronbiotech, Shanghai, China) to generate pGCSIL- GFP-miR124 or pGCSIL-GFP-Ctr, respectively. Viral shRNA to iASPP were produced as previously described [3].
Lentivirus production, titration, and infection
The plasmid encoding miR124 or control, together with plasmids, pHelper 1.0 and pHelper 2.0, containing the imperative elements for virus packaging, were co-transfected into 293T cells with lipofectamine 2000, according to the manufacturer’s instructions (Invitrogen) for the generation of miR124 or control, respectively. The culture supernatants containing lentivirus vectors were harvested and ultra-centrifuged. The virus titers of each preparation were determined. To perform lentiviral infections, the target cells were plated at 40%-50% confluence and incubated overnight (16 h). On the day of infections, the culture medium was replaced by the appropriately titered viral supernatant (1.5 ml/well) and incubated at 37°C for 10 h; afterwards, the viral supernatant was replaced with fresh media. Forty-eight hours later, infected cell populations were selected in puromycin (2 mg/ml). After 5 days of selection, shRNA knockdown efficiency was determined by quantitative real-time RT-PCR and western blot analysis.
RNA extraction and quantitative real-time RT-PCR
Approximately 1.0 × 106 PC-3 cells (uninfected or infected cells) were seeded into a six-well culture plate, respectively. Cells of each group were harvested after culture or 72 h. For small RNAs (~200 nt) were isolated with mirVanaTM PARIS TM Kit (Ambion) according to the manufacturer’s instructions. For RT reactions, 1 mg of small RNA was used for reverse transcription with miScript Reverse Transcription Kit (Qiagen), performed at 37°C for 60 min and a final incubation at 95°C for 5 min. MiRNA real-time RT-PCR was carried out by using the miScript SYBR Green PCR kit (Qiagen) on an Applied Biosystems 7000 real-time PCR machine (ABI). The PCR reaction was conducted at 95°C for 15 min, followed by 40 cycles of incubation at 94°C for 15 s, 55°C for 30 s, and 70°C for 30 s. Expression level of each miRNA was normalized by against U6snRNA levels. Total RNA was extracted from cells with Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Expression of iASPP mRNA was detected by quantitative real-time RT-PCR (qRT-PCR) using the standard SYBR Green RT-PCR Kit (Takara) according to the manufacturer’s instructions. Briefly, the cDNA was synthesized using the RevertAid First-Strand cDNA Synthesis kit (Fermentas, Lithuania), according to the manufacturer’s protocol. The cDNA was used as the template in an iQTM SYBR Green Supermix (Bio-Rad, Her-cules, CA) and in triplicate subjected to denaturation at 94°C for 1 min and 30 cycles of 94°C for 40 s and 60°C for 40 s, followed by extension at 72°C for 6 min. The specific primer pairs are as follows: iASPP (107 bp), sense: 5’-GGC GGT GAA GGA GAT GAA C-3’; antisense: 5’-TGA TGA GGA AAT CCA CGA TAG AG-3’; β-actin as an internal, primers, sense: 5’-GGC GGC ACC ACC ATG TAC CCT-3’; reverse: 5’-AGG GGC CGG ACT CGT CAT ACT-3’ (202 bp). The relative levels of iASPP mRNA transcripts were normalized to the control β-actin. Relative gene expression was quantified using the GraphPad Prism 4.0 software (GraphPad Software, San Diego, CA) and expressed as % of the control.
Western blotting
Different cultures in 35 mm dishes were lysed in 0.2 ml lysis buffer (0.1% SDS, 1% NP-40, 50 mM HEPES, pH 7.4, 2 mM EDTA, 100 mM NaCl, 5 mM sodium orthovanadate, 40 μM p-nitrophenyl phosphate, and 1% protease inhibitor mixture set I; Calbiochem). Lysates were centrifuged at 12,000 rpm for 15 min. The supernatant was collected and denatured. Proteins were separated in 10% SDS-PAGE gel and blotted onto polyvinylidene difluoride membrane. The blot was blocked for 1.5 h at room temperature in 5% BSA, followed by overnight incubation at 4°C with indicated antibodies. Membranes were rinsed and incubated for 1 h with the correspondent peroxidase-conjugated secondary antibodies. Chemiluminescent detection was performed with the ECL kit (Pierce).
MTT assay
Cell viability and proliferation were evaluated by a modified MTT and BrdU incorporation method. Cell viability of PC-3 with miR124 or control was assessed by MTT assay performing at five time points (on day 1, 2, 3, 4 and 5) after the seeding. Briefly, quantification of mitochondrial dehydrogenase activity was achieved via the enzymatic conversion of MTT [3-(4,5-dimethyldiazol-2-yl)-2,5-diphenyltetrazolium bromide] (Sigma-Aldrich) to a colored formazan product. The test cells in exponential growth were plated at a final concentration of 2 × 103 cells/well in 96-well culture plates for different culture time (1 day, 2 day, 3 day, 4 day, and 5 day, respectively). MTT (10 μl, 10 mg/ml) was then added. After an additional 4 h of incubation, the reaction was terminated by removal of the supernatant and addition of 100 μl DMSO to dissolve the formazan product. After 0.5 h, the optical density (OD) of each well was measured at 570 nm using ELISA reader (EL x 808 Bio-Tek Instruments, USA).
BrdU incorporation assay
DNA synthesis in proliferating cells was determined by measuring BrdU incorporation assay. BrdU assays were performed on 24 h and 48 h after PC-3 treated with miR124 or control vector. The cells were seeded in 96-well culture plates at a density of 2 × 103 cells/well for 24 h or 48 h, and were incubated with a final concentration of 10 μM BrdU (BD Pharmingen, San Diego, CA) for 2 h to 24 h. At the end of incubation period, medium was removed and the cells were fixed (30 min, RT) and labeled with peroxidase-coupled anti-BrdU-antibody (Sigma-Aldrich). After incubation (60 min, RT), the cells were washed three times with PBS. Thereafter, peroxidase substrate (tetramethylbenzidine) was added for 30 min and the absorbance values were measured at 490 nm using an ELISA reader. Background BrdU immunofluorescence was determined in cells not exposed to BrdU but stained with the BrdU antibody.
Colony formation assay
The effect of iASPP silence on the colony formation of PC-3 cells was analyzed by colony formation assay. PC-3 cell line, control RNAi, or iASPP RNAi cells at 200 cells per six-well culture plate were cultured in 10% FBS DMEMat 37°C, 5% CO2 for 2 weeks. The cell colonies were washed twice with PBS, fixed by 4% PFA for 15 min and stained with Gimsa form 20 min, and then washed twice by ddH2O. Individual clones with more than 50 cells were counted. The parent cells value was set to 100 and the clone forming efficiency for other type of cells was calculated.
3’UTR reporter assay
UTR binding reporter assays were performed in PC3 cells. pMIR-REPORT vectors harboring 350 bp fragment of iASPP-3’UTR behind initiated from stop codon with wild type (WT) miR-124 binding sites (199-194) or mutated (MUT) miR-124 binding sites were produced by cloning the synthesized fragments into the Hind III and Spe I restriction sites of pMIRREPORT: Cells were transfected with (1) miR-124 or celmiR-67-negative-control mimics (50 nM), (2) pMIRREPORT vectors containing WT or MUT miR-124 binding sites (400 ng) and (3) pRL-SV40 (Promega) expressing Renilla luciferase (400 ng) for normalization. Cells were grown in high-glucose DMEM supplemented with 10% fetal bovine serum, and luciferase measurements were performed 48 hours post-transfection using the Dual-Luciferase Reporter Assay System (Promega).
Statistical analysis
Data were expressed as mean ± SD. Statistical analysis was performed using SPSS software (Release 11.0, SPSS Inc.). The difference between two groups was analyzed by the Student’s t-test. A value of p < 0.05 was considered as statistical significance.
Result
MicroRNA124 binds to 3’UTR of iAsPP
The combining sites of iASPP 3’UTR with Mir124 were predicted by Targetscan and Pictar [6,7]. A sequential replacement of 8 base pair region was performed to produce mutant vector. Then we constructed mir124 expression vector and produce lentiviral particles. 293T cells were infected by mir124 or negative control virus at MOI=1 prior to reporter vectors transfection. Luciferase expression measurement indicated significant decrease of iASPP 3’UTR tailed vector transfected 293T cell while no change observed in mutant group. This validate the binding of mir124 and iASPP mRNA in vitro and confirmed the down-regulation effect of microRNA (Figure 1).
Figure 1.

A. Illustration of iASPP 3’UTR mutation in vector construction. B. Luciferase expression of 293T cells under mir124 regulation upon iASPP 3’UTR. 293T cells were transfected with Lucifurase reporter 96 h post infection with lentiviral negative control or mir124 vectors. Wild type (WT) and mutant (MUT) indicated reporter harbored 3’UTR of iASPP.
MicroRNA124 presented low expression in prostate cancer
To verify the expression pattern of mir124, we perform qPCR test on cancer cell lines of PC3 and DU145, in complement of 5 patient samples. PC3 cell infected with viral mir124 expression vector were set as a positive control for expression. The expression level of mir124 was normalized to housekeeping microRNA U6. Result showed considerably low expression of mir124 in both cell lines and tumor tissues (Table 1).
Table 1.
Relative expression of mir124 in cell lines and patients tissues
| samples | PC3 | PC3-mir124 | DU145 | P1 | P2 | P3 | P4 | P5 |
|---|---|---|---|---|---|---|---|---|
| Fold | 0.04 | 133 | 0.13 | 0.15 | 0.08 | 0.17 | 0.03 | 0.02 |
| ± SD | 0.00 | 2.8 | 0.05 | 0.01 | 0.02 | 0.03 | 0.00 | 0.01 |
± SD indicates standard differences; PC3-mir124 indicates RNA extraction from PC3 cell 96 h post infection of lentiviral mir124 vectors.
MicroRNA124 suppress prostate cancer cell growth
The over expression of mir124 were performed on PC3 cells. The iASPP expression were then determined, and a validated shRNA target was adopted in the test. qPCR and western blot convinced that both mRNA and protein level were knocked down (Figure 2).
Figure 2.

Expression level of iASPP were down-regulated by mir124 expression in PC3 cells. CON blank control, NC, negative control, shRNA, viral shRNA vector target at iASPP, mir124, mir124 expression lentivirus.
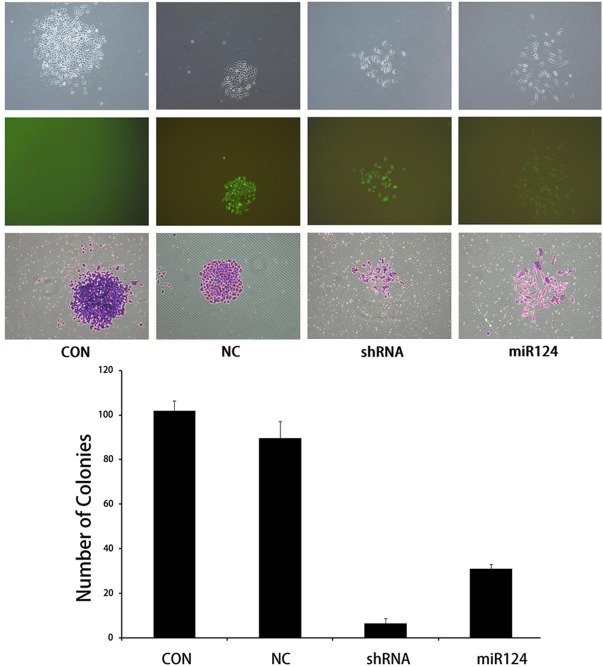
Function assay were performed on PC3 cells infected by negative control, mir124 or shRNA virus. MTT and BrdU method were used to analyze proliferation of cells. Result revealed that both mir124 and specific RNAi decreased the cells growth of PC3 significantly (Figure 3). In consistent with these results, clone formation ability of PC3 was also blocked with infection treatments (Figure 4).
Figure 3.

Proliferation of PC3 cells were impaired with mir124 expression, (A) Cell proliferation test by MTT method. (B) Cell viability determined by BrdU test; CON blank control, NC, negative control, shRNA, viral shRNA vector target at iASPP, mir124, mir124 expression lentivirus.
Figure 4.
Clone formation of PC3 cells were impaired with mir124 expression. CON blank control, NC, negative control, shRNA, viral shRNA vector target at iASPP, mir124, mir124 expression lentivirus.
Discussion
iASPP play a role in prostate cancer cells as we described previously [3], when knocked down by RNAi in PC3 and DU145 cells, proliferation and clone formation ability were dramatically decreased. In addition, in vivo convinced that block of iASPP impaired tumorigenesis of prostate cancer cells. These results were consistent with the research on leukemia, breast cancer and non-small cell lung cancer, which identified iASPP high expression and suppress p53 pathway [10-16]. Besides, investigation of genotype mapping of breast cancer patient suggested that iASPP might balance the p53 and NF-kB pathway, therefore, control the growth of cells [17-19]. These proofs implied that a delicate iASPP regulation was involved in tumorigenesis or apoptosis.
MicroRNAs are a class of functional diverse regulators diverse cellular through RNA interference-based mechanisms on post-transcription level. miRNAs are driven by polyperase III promoter and transcribed as primary RNA (pri-miRNAs) then processed in the nucleus to smaller precursor hairpin structures (premiRNAs), exported to the cytoplasm where they are processed further by the Dicer nuclease to become mature, functional miRNAs approximately 21 nucleotides in length. Mature miRNAs are then incorporated into the RNA-induced silencing complex, which facilitates their interaction with, and inhibition of, target messenger RNAs (mRNAs) by binding 3’UTRs [20]. Human microRNA 124 was chosen in this research to knock down iASPP expression. The luciferase reporter assay validated the binding and repressing of mir124 upon iASPP in 293T cells. Meanwhile, qPCR determination of cell lines and patient tissue indicated considerable low expression level. Molecular and function test confirmed that both shRNA and mir124 down regulated iASPP expression and blocked cell growth of cancer cell line PC3. Collecting the points, mir124 is proved a potential target for cancer therapy.
Mir124 was found playing a role in glioblastoma differentiation especially intracellular growth factor are absent [21]. Likewise, mir124 up-regulation was also found in trained LNCaP cells which obtained androgen-independent growth [22]. To sum up views, an intrinsic mechanism might be drawn between growth factor depletion and mir124 regulated cell growth, which requires further investigation in vivo and clinical research.
Acknowledgements
This study was supported by Science and Technology Planning Project of Guangdong Province, China (2011B080701075) and Fundamental Research Funds for the Central Universities, China (12ykpy40).
Disclosure of conflict of interest
None.
References
- 1.Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N Engl J Med. 2004;351:1488–1490. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- 2.Schroder FH. Progress in understanding androgen-independent prostate cancer (AIPC): a review of potential endocrine-mediated mechanisms. Eur Urol. 2008;53:1129–1137. doi: 10.1016/j.eururo.2008.01.049. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Xiao HJ, Chen J, Tao X, Cai LH. Inhibitory member of the apoptosis-stimulating protein of p53 (ASPP) family promotes growth and tumorigenesis in human p53-deficient prostate cancer cells. Prostate Cancer Prostatic Dis. 2011;14:219–224. doi: 10.1038/pcan.2011.25. [DOI] [PubMed] [Google Scholar]
- 4.Rana TM. Illuminating the silence: understanding the structure and function of small RNAs. Nat Rev Mol Cell Biol. 2007;8:23–36. doi: 10.1038/nrm2085. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 6.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 7.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M, Rajewsky N. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894–1910. doi: 10.1261/rna.768207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ZJ, Zhang Y, Zhang XB, Yang X. Abnormal mRNA expression of ASPP members in leukemia cell lines. Leukemia. 2004;18:880. doi: 10.1038/sj.leu.2403300. [DOI] [PubMed] [Google Scholar]
- 11.Lossos IS, Natkunam Y, Levy R, Lopez CD. Apoptosis stimulating protein of p53 (ASPP2) expression differs in diffuse large B-cell and follicular center lymphoma: correlation with clinical outcome. Leuk Lymphoma. 2002;43:2309–2317. doi: 10.1080/1042819021000040017. [DOI] [PubMed] [Google Scholar]
- 12.Liu ZJ, Cai Y, Hou L, Gao X, Xin HM, Lu X, Zhong S, Gu SZ, Chen J. Effect of RNA interference of iASPP on the Apoptosis in MCF-7 breast cancer cells. Cancer Invest. 2008;26:878–882. doi: 10.1080/07357900801965042. [DOI] [PubMed] [Google Scholar]
- 13.Agirre X, Román-Gómez J, Jimenez-Velasco A, Garate L, Montiel-Duarte C, Navarro G, Vazquez I, Zalacain M, Calasanz MJ, Heiniger A, Torres A, Minna JD, Prósper F. ASPP1, a common activator of TP53, is inactivated by aberrant methylation of its promoter in acute lymphoblastic leukemia. Oncogene. 2006;25:1862–1870. doi: 10.1038/sj.onc.1209236. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Wang M, Zhou C, Chen S, Wang J. The expression of iASPP in acute leukemias. Leuk Res. 2005;29:179–183. doi: 10.1016/j.leukres.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Chen J, Xie F, Zhang L, Jiang WG. iASPP is over-expressed in human non-small cell lung cancer and regulates the proliferation of lung cancer cells through a p53 associated pathway. BMC Cancer. 2010;10:694. doi: 10.1186/1471-2407-10-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bergamaschi D, Samuels Y, O’Neil NJ, Trigiante G, Crook T, Hsieh JK, O’Connor DJ, Zhong S, Campargue I, Tomlinson ML. iASPP oncoprotein is a key inhibitor of p53 conserved from worm to human. Nat Genet. 2003;33:162–167. doi: 10.1038/ng1070. [DOI] [PubMed] [Google Scholar]
- 17.Yang JP, Hori M, Sanda T, Okamoto T. Identification of a novel inhibitor of nuclear factor-κB, RelA-associated inhibitor. J Biol Chem. 1999;274:15662–15670. doi: 10.1074/jbc.274.22.15662. [DOI] [PubMed] [Google Scholar]
- 18.Takada N, Sanda T, Okamoto H, Yang JP, Asamitsu K, Sarol L, Kimura G, Uranishi H, Tetsuka T, Okamoto T. RelA-associated inhibitor blocks transcription of human immunodeficiency virus type 1 by inhibiting NF-kappaB and Sp1 actions. J Virol. 2002;76:8019–8030. doi: 10.1128/JVI.76.16.8019-8030.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nexø BA, Vogel U, Olsen A, Nyegaard M, Bukowy Z, Rockenbauer E, Zhang X, Koca C, Mains M, Hansen B, Hedemand A, Kjeldgaard A, Laska MJ, Raaschou-Nielsen O, Cold S, Overvad K, Tjønneland A, Bolund L, Børglum AD. Linkage disequilibrium mapping of a breast cancer susceptibility locus near RAI/PPP1R13L/iASPP. BMC Med Genet. 2008;9:56. doi: 10.1186/1471-2350-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng Y. Principles of micro-RNA production and maturation. Oncogene. 2006;25:6156–6162. doi: 10.1038/sj.onc.1209908. [DOI] [PubMed] [Google Scholar]
- 21.Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, Vandenberg SR, Ginzinger DG, James CD, Costello JF, Bergers G, Weiss WA, Alvarez-Buylla A, Hodgson JG. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:1741–7015. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu G, Wu J, Zhou L, Chen B, Sun Z, Zhao F, Tao Z. Characterization of the small RNA transcriptomes of androgen dependent and independent prostate cancer cell line by deep sequencing. PLoS One. 2010;5:e15519. doi: 10.1371/journal.pone.0015519. [DOI] [PMC free article] [PubMed] [Google Scholar]