Abstract
MAGE-D4 is a novel member of MAGE super-family. It has preliminarily been demonstrated that MAGE-D4 mRNA is not expressed in majority of normal tissues except for brain and ovary in which only trace amount of MAGE-D4 mRNA can be detected, but predominantly expressed in glioma. MAGE-D4 protein expression and its immunogenicity in glioma have not been elucidated well. This study was designed to analyze MAGE-D4 expression both at mRNA and protein level, characteristic of humoral immune response, and their relationships with glioma patients’ clinicopathological parameters. Recombinant MAGE-D4 protein and antiserum were generated. Quantitative RT-PCR analysis revealed that MAGE-D4 mRNA expression was overall up-regulated in 41 glioma specimens compared with that in 14 normal brain tissues. Immunohistochemistry analysis showed that 78% (21/27) glioma tissues expressed MAGE-D4 protein, which was predominantly located in the cytoplasm of tumor cells, but absent in any neuroglia cell of normal brain tissues. ELISA analysis demonstrated that humoral response against MAGE-D4 was detected in 17% (7/41) of glioma patients’ sera but not in 77 healthy donors. No apparent correlation was observed between the expression and immunogenicity of MAGE-D4 with clinicopathological parameters of glioma. In summary, these results indicate that MAGE-D4 is highly expressed in glioma and can develop specifically humoral response in glioma patients, which supports that it may be a promising biomarker for glioma diagnosis and immunotherapy.
Keywords: Glioma, tumor antigen, melanoma-associated antigen D4, gene expression, immunogenicity
Introduction
Glioma is the most common malignant neoplasm of central nervous system, approximately accounting for 50% of primary brain tumors. The poor prognosis of patients with malignant glioma has not significantly improved despite developments of surgery, radiation therapy and chemotherapy in recent decades. Therefore, it is very necessary to search for more effective treatment strategies.
The discovery of human tumor antigens that are exclusively or preferential expressed in tumors and can coordinately elicit cellular and humoral immune responses, provides powerful tools for early diagnosis and target-specific immunotherapy of tumors. The fact that central nervous system is not a strictly immune-privileged site as people previously thought makes the immunotherapy of glioma feasible [1].
MAGE-D4, originally termed MAGE-E1, is a novel member of MAGE (Melanoma associated antigen) super-family identified by serial analysis of gene expression (SAGE) technique. Three isoforms (MAGE-D4a, -b, and -c) were produced by alternative splice in the process of gene transcription, which possess NH2-terminal 348 amino acids. MAGE-D4b is identical to MAGE-D4a in sequence except for 2 amino acids absence in the COOH terminus, which reveals that both of them may have the similar functions, while MAGE-D4c lacks a large portion of the COOH terminus and has special 66 amino acids in the end [2]. MAGE-D4 is almost not expressed in normal tissues except for brain and ovary in which only trace amount of MAGE-D4 mRNA can be detected, but predominantly expressed in glioma [3]. It has been reported that MAGE-D4 mRNA expression was up-regulated 2.6 to 15 folds in glioblastoma, relatively to cultured human astrocytes [2]. Then, another study [4] got the similar results, in which 2 cases of glioblastoma were identified with high expression of MAGE-D4 by both oligonucleotide microarray analysis and qRT-PCR analysis. In addition high MAGE-D4 expression was also found in poorly differentiated tumors and in patients with lymph node metastasis of non-small cell lung cancer (NSCLC), indicating that MAGE-D4 may play a role in proliferation of some malignant cells [5,6]. To some extent, these preliminary results show that MAGE-D4 may be a potentially promising target for the diagnosis and treatment of tumors.
However, researches about MAGE-D4 gene expression in glioma were only based on mRNA level with limited number and types of detected cases. MAGE-D4 protein expression has not been elucidated. Moreover, it is still unclear whether immune responses against MAGE-D4 of patients with glioma could be induced, and its roles in tumorigenesis also remain largely unknown.
Thus, in the present study, the expression of MAGE-D4 mRNA and protein was analyzed in glioma. And then, specific IgG antibody to MAGE-D4 from sera of glioma patients was detected. Finally, possible relationships between MAGE-D4 expression and antibody response with clinicopathological parameters, such as tumor stage and histological type, were also investigated.
Materials and methods
Tissues and serum samples
Tissues and serum samples were available from the First Affiliated Hospital of Guangxi Medical University, approved by Hospital Ethic Review with informed consent from patients. 41 glioma tissue samples (6 anaplastic astrocytomas, 4 oligodendrogliomas, 20 diffuse astrocytomas and 11 glioblastomas; patient age range, 2-63 years; average patient age, 33.4 years; 29 men and 12 women) and 14 normal brain tissue samples collected from autopsies of tumor-free patients (patient age range, 17-72 years; average patient age, 35.8 years; 7 men and 7 women) were analyzed. The tissues were surgically removed and immediately frozen in liquid nitrogen, and then were stored at -80°C. Serum samples were obtained from 41 glioma patients of the initial diagnosis of disease (3 pilocytic astrocytomas, 22 diffuse astrocytomas, 5 anaplastic astrocytomas, 2 ependymomas, and 9 glioblastomas; patient age range, 3-65 years; average patient age, 35.4 years; 25 men and 16 women), and sera of 77 healthy donors from routine physical examination were used as controls. 27 paraffin-embedded tissue sections (3 pilocytic astrocytomas, 13 diffuse astrocytomas, 5 anaplastic astrocytomas, 2 ependymomas, and 4 glioblastomas; patient age range, 3-60 years; average patient age, 39.7 years; 13 men and 14 women) were processed for immunohistochemistry analysis. Tumors were classified according to the World Health Organization (WHO) criteria as follows: glioblastoma (WHO grade IV), anaplastic astrocytoma and anaplastic ependymoma (WHO grade III), diffuse astrocytoma and ependymoma (WHO grade II), or pilocytic astrocytoma (WHO grade I) [7].
Total RNA extraction and real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted from frozen tissues using TRIzol Reagent (Invitrogen, USA), and cDNA was synthesized with 2.5 μg total RNA by using RevertAidTM First Strand cDNA Synthesis Kit (MBI Fermentas, Canada). The quality and integrity of cDNA were tested as described previously [8]. Real-time quantitative RT-PCR reaction was performed on the iCycler iQTM Multi-Color QRT-PCR Detection System (Bio-Rad, USA). The gene-specific primers and Taqman probe for MAGE-D4 were synthesized in accordance with previous report [2]: the forward primer sequence was 5’-CCAGCTTCTTCTCCTGGATC-3’ and the reverse primer sequence was 5’-GTAACACTGATACCCAAAACATG-3’. The Taqman probe was 5’ FAM-CGGCTTCTTCCTGTCAGTCGGAGGT-3’ TAMRA. Thermal cycle conditions for MAGE-D4 amplification were initial denaturation at 95°C for 2 min, then 45 cycles at 95°C for 5 sec and 60°C for 20 sec. All experiments were done in triplicate and the parameter threshold cycle (CT) was calculated. Recombinant plasmids containing MAGE-D4 and hypoxanthine phosphoribosyl transferase 1 (HPRT1) were serially diluted to construct the standard curve. The relative MAGE-D4 expression level of each sample was normalized to the endogenous HPRT1 amount.
Generation of recombinant MAGE-D4 protein and antiserum
MAGE-D4 coding region was amplified from human glioma tissue cDNA by PCR with high-fidelity PrimeSTARTM HS DNA polymerase (TaKaRa, Tokyo) and cloned into prokaryotic expression plasmid pMAL-c2 (New England Biolabs, USA) to generate recombinant fusion protein. MAGE-D4 protein with MBP (maltose binding protein) tag was purified by amylose resin affinity column according to the manufacturer’s instructions (New England Biolabs, USA) and identified by tandem mass spectrometry/mass spectrometry (MS/MS).
20-week-old New Zealand white female rabbits were subcutaneously injected with MAGE-D4 fusion protein (250 μg/kg) in complete Freund’s adjuvant (Sigma, USA) for the first immunization. Then 6 sequential immunizations were performed with 125 μg/kg of MAGE-D4 fusion protein mixed with incomplete Freund’s adjuvant (Sigma, USA) at 2-week interval. Antiserum was collected and purified by Affi-Prep protein A (Bio-Rad, USA) and antigen affinity columns using Affigel-15 gel (Bio-Rad, Hercules, CA) in accordance with the manufacturer’s instructions. The titer and specificity of MAGE-D4 antiserum were examined by indirect ELISA and Western blot, respectively.
Immunohistochemistry
After deparaffinized and rehydrated by standard method, the sections were heated in ethylene diamine tetraacetic acid (EDTA, pH 8.0) for antigen retrieval and then treated in 3% H2O2 to block endogenous peroxidase activity. The sections were incubated with MAGE-D4 antiserum (1:1000 dilution) at room temperature for 1 h, followed by applying horseradish peroxidase-conjugated goat anti-rabbit IgG (Long Island Biotech, China) as the secondary antibody and 3,3’-diaminobenzidine (DAB) (Maixin Biotec, China) as the chromogen. Preimmune rabbit serum was used as negative control and a known glioma tissue section of MAGE-D4-positive expression was served as positive control.
Immunoreactivity of MAGE-D4 protein was assessed by two independent pathologists who were blinded to the clinical diagnosis and recorded semi-quantitatively according to the staining intensity and the percentage of positive cells. The staining intensity was defined as follows: 1 point, weak staining; 2 points, moderate staining; 3 points, strong staining. The percentage of positive cells was defined as follows: 0 point, 0-5%; 1 point, 5%-25%; 2 points, 26%-50%; 3 points, 51-75%; 4 points, >75%. Then, MAGE-D4 protein expression was judged by the sum of both points: negative (overall score=‘-’), 0-1 point; weak expression (overall score=‘+’), 2-3 points; moderate expression (overall score=‘++’), 4-5 points; strong expression (overall score=‘+++’), 6-7 points [5,9].
Enzyme-linked immunosorbent assay analysis (ELISA)
Purified recombinant MAGE-D4 protein (1 μg/ml) was coated with 100 μl in 0.05 mol/l carbonate buffer (pH 9.6) into 96-well microtiter plates (Greiner bio-one, USA) at 4°C overnight. MBP protein was used as a blank control [10,11]. The plates were blocked with 5 % nonfat dry milk/PBS at 37°C for 1 h. Serum sample diluted at 1:800 was added with 100 μl into the well and incubated at 37°C for 1 h. A sensitive streptavidin-biotinylated horseradish peroxidase complex (KPL, USA) diluted at 1:5000 was used as the secondary antibody. Then, the wells were reacted with tetramethyl-benzidine substrate (100 μl) and terminated by using 3 mol/l sulfuric acid. The optical density (OD) was read immediately at 450 nm with 630 nm as reference filter. All samples were tested thrice respectively with two different plates. An OD value that exceeds three standard deviations (SDs) above the mean OD value of sera from healthy donors was defined as positive.
Statistical analysis
Statistical analysis was performed by SPSS 16.0 for Windows software package and two-sided P value less than 0.05 was considered statistically significant. Mann-Whitney U test was used to evaluate MAGE-D4 mRNA expression level between groups. Possible relationships between gene expression and antibody response with clinicopathological parameters were assessed by performing Fisher’s exact probability test or χ2 test.
Results
Expression of MAGE-D4 mRNA in glioma and normal brain tissues
Expression of MAGE-D4 mRNA was evaluated in 41 glioma specimens and 14 normal brain tissues by real-time quantitative RT-PCR. The median value of MAGE-D4/HPRT1 of glioma tissues was 29.00, which was significantly higher than that in normal brain tissues (4.16, p<0.0001) (Figure 1A). For different types of glioma analyzed, glioblastoma showed the highest median value of MAGE-D4/HPRT1 (31.90), diffuse astrocytoma was second (29.80), followed by oligodendroglioma (23.95) and anaplastic astrocytoma (9.74), respectively (Figure 1B). Statistical analysis revealed that there were significant differences between glioblastoma (p=0.003), diffuse astrocytoma (p<0.0001), oligodendroglioma (p=0.038) and normal brain. As shown in Figure 1C, high expression of MAGE-D4, which was defined as to exceeding three times above the median value of MAGE-D4/HPRT1 of normal brain tissues, was found in 79% (15/19) of diffuse astrocytoma, 73% (8/11) of glioblastoma, 50% (2/4) of oligodendroglioma, and 43% (3/7) of anaplastic astrocytoma, respectively. The ratio of high expression in glioblastoma and diffuse astrocytoma was compared with that in normal brain tissue (p=0.002 and p<0.0001). Overall, 68% (28/41) of glioma showed high expression of MAGE-D4 mRNA, whereas only 7.1% (1/14) of normal brain tissues was found high expression (p<0.0001). No apparent correlation was found between the relative expression level and high expression ratio of MAGE-D4 with age, gender, WHO grade, and histological type of glioma (data not shown).
Figure 1.
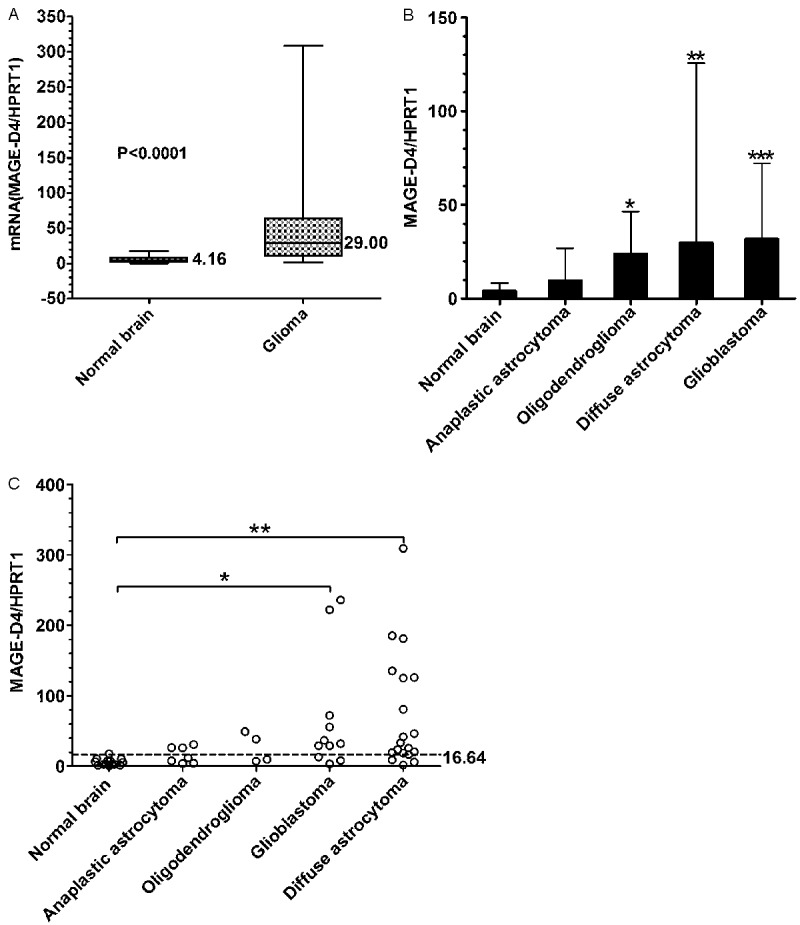
MAGE-D4 mRNA expression analyzed by real-time quantitative RT-PCR. A: The median values of MAGE-D4 in 14 normal brain tissues and 41 glioma tissues (p<0.0001, Mann-Whitney U test). B: MAGE-D4 expression in different types of glioma including 11 glioblastomas, 20 diffuse astrocytomas, 4 oligodendrogliomas and 6 anaplastic astrocytomas. *, p=0.038; **, p<0.0001; ***, p=0.003 (compared with normal brain, Mann-Whitney U test). C: MAGE-D4 high expression was defined as exceeding three times above the median value of MAGE-D4/HPRT1 of normal brain tissues. The cutoff value is 16.64 for MAGE-D4 high expression above the line. The ratios of MAGE-D4 high expression of different types of glioma tested were compared with normal brain (*, p=0.002, **, p<0.0001, Fisher’s exact probability test).
Verification of recombinant MAGE-D4 protein and antiserum
The frequency of E. coli rare codons in MAGE-D4 coding sequence is as high as 10.52%, which makes it almost can not be expressed in TB1 E. coli usually recommended by NEB Inc. Rosetta (DE3) (Novagen, Germany) is a BL21 derivative strain carrying pRARE2 plasmid which encodes tRNAs rarely produced in E. coli. So, co-expressing the pRARE2 plasmid with eukaryotic gene in the prokaryotic system can significantly improve the expression of recombinant protein [12,13]. In this study, we also found that recombinant plasmid induced in Rosetta was able to markedly increase the production of MAGE-D4 protein. Purified recombinant MAGE-D4 protein was confirmed with MS/MS analysis (data not shown).
The titer of purified antiserum checked by ELISA can reach more than 1:32000. Determination of MAGE-D4 antiserum specificity was accomplished by Western blot. As shown in Figure 2A, the antiserum could detect recombinant MAGE-D4 protein with around 140 kDa molecular weight that was higher than the predictive value (124 kDa). The slower electrophoretic mobility may be due to abnormal spatial conformation of MAGE-D4 protein caused by abundance of proline (7.02%) in composition [14]. Whereas the pre-immune rabbit serum failed to detect the recombinant MAGE-D4 protein, which further supported that the antiserum used is MAGE-D4 specific.
Figure 2.
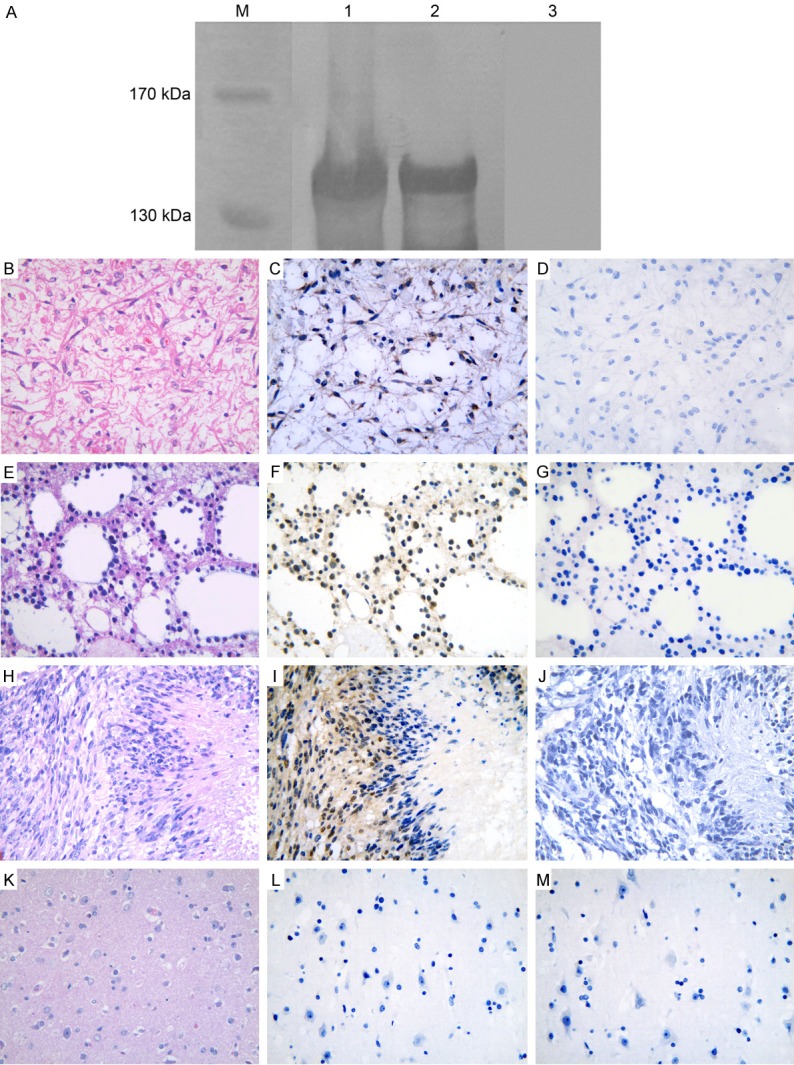
Specificity analysis of MAGE-D4 antiserum and immunohistochemical staining of MAGE-D4 protein. A: The antiserum can detect the precipitate from induced bacteria containing pMAL-c2/MAGE-D4 plasmid (Lane 1) and purified recombinant MAGE-D4 protein (Lane 2). The molecular weight of immune reactive band was around 140 kDa, which was higher than the predictive value (124 kDa) of MAGE-D4 fusion protein. The pre-immune serum failed to bind with the recombinant MAGE-D4 protein (Lane 3). Lane M, protein marker. C: Cytoplasmic expression of MAGE-D4 in diffuse astrocytoma. F and I: Nucleus and cytoplasm co-expression of MAGE-D4 in diffuse astrocytoma and glioblastoma, respectively. L: MAGE-D4 was negative in normal brain tissue. B, E, H and K: The corresponding tissue sections were stained with hematoxylin-eosin. D, G, J and M: The pre-immune rabbit serum was used as negative control.
Expression of MAGE-D4 protein in glioma
To investigate the presence of MAGE-D4 protein in tissues, immunohistochemistry was carried out with MAGE-D4 antiserum. In glioma, 78% (21/27) expressed MAGE-D4 protein, which showed intratumoral variability and was predominantly located in the cytoplasm of tumor cells (Figure 2C). Only two glioma tissues showed nuclear staining accompanying with cytoplasmic staining (Figure 2F and 2I). Strong, moderate and weak expression of MAGE-D4 protein in glioma was 11% (3/27), 45% (12/27), and 22% (6/27), respectively. In normal brain tissues, no positive reaction was observed in any neuroglia cells (Figure 2L). MAGE-D4 protein expression with clinicopathological features of glioma was summarized in Table 1. No significant correlation was found between the MAGE-D4 expression and clinicopathological parameters (data not shown).
Table 1.
MAGE-D4 expression and immunogenicity with clinicopathological characteristics in glioma
| Case no. | Histology | Age | Sexa | IHCb | ELISAc |
|---|---|---|---|---|---|
| WHO grade IV | |||||
| 1 | Glioblastoma | 59 | M | + | + |
| 2 | Glioblastoma | 36 | F | +++ | - |
| 3 | Glioblastoma | 60 | F | +++ | - |
| 4 | Glioblastoma | 48 | F | - | - |
| WHO grade III | |||||
| 5 | Anaplastic astrocytoma | 43 | M | ++ | - |
| 6 | Anaplastic astrocytoma | 37 | M | +++ | - |
| 7 | Anaplastic astrocytoma | 53 | F | ++ | - |
| 8 | Anaplastic astrocytoma | 31 | M | ++ | - |
| 9 | Anaplastic astrocytoma | 52 | M | - | - |
| 10 | Anaplastic ependymoma | 52 | M | ++ | + |
| WHO grade II | |||||
| 11 | Diffuse astrocytoma | 11 | F | ++ | + |
| 12 | Diffuse astrocytoma | 24 | F | ++ | + |
| 13 | Diffuse astrocytoma | 54 | M | + | + |
| 14 | Diffuse astrocytoma | 52 | F | ++ | - |
| 15 | Diffuse astrocytoma | 18 | F | +++ | - |
| 16 | Diffuse astrocytoma | 34 | F | ++ | - |
| 17 | Diffuse astrocytoma | 30 | M | - | - |
| 18 | Diffuse astrocytoma | 52 | F | - | - |
| 19 | Diffuse astrocytoma | 52 | M | - | - |
| 20 | Diffuse astrocytoma | 53 | F | ++ | - |
| 21 | Diffuse astrocytoma | 30 | M | +++ | - |
| 22 | Diffuse astrocytoma | 58 | M | + | - |
| 23 | Diffuse astrocytoma | 53 | M | - | - |
| 24 | Ependymoma | 16 | F | ++ | - |
| WHO grade I | |||||
| 25 | Pilocytic astrocytoma | 44 | M | ++ | + |
| 26 | Pilocytic astrocytoma | 3 | F | ++ | - |
| 27 | Pilocytic astrocytoma | 16 | F | +++ | - |
M, male; F, female.
MAGE-D4 protein expression was judged according to the staining intensity and the percentage of positive cells: -, negative; +, weak expression; ++, moderate expression; +++, strong expression.
The seroreactivities of MAGE-D4 are described as -, negative; +, positive.
Antibody response to MAGE-D4 in glioma patients
To analyze whether immune response against MAGE-D4 in glioma patients could be induced, sera from 41 patients and 77 healthy donors were detected by ELISA using recombinant MAGE-D4 protein. 7 of 41 (17%) patients with glioma developed humoral response, whereas all of healthy donors were negative for serum MAGE-D4 antibody (Figure 3A). The titration curves were illustrated for MAGE-D4 antibody-positive and -negative serum from selected individuals (Figure 3B). No antibody response against MBP protein was detected in sera from MAGE-D4 seropositive patients. Of 41 sera screened, 27 tissue samples were collected simultaneously from glioma patients used to assess for MAGE-D4 protein expression. As shown in Table 1, glioma tissues from 6 of serum antibody-positive patients were verified to express MAGE-D4 protein. Statistical analysis suggested that no apparent correlation was found between the immunogenicity of MAGE-D4 and clinicopathological parameters of glioma (Table 2).
Figure 3.
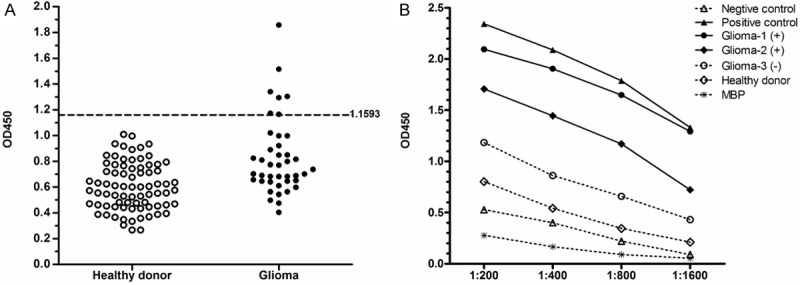
Analysis of MAGE-D4 specific IgG antibodies in glioma patients. A: OD450 results in serum from glioma patients and healthy donors. The cutoff value is 1.1593 for positive above or negative below the line. B: The titration curves of a serially diluted serum from glioma patients and healthy donors.
Table 2.
Correlation of serum anti-MAGE-D4 antibody with the clinical characteristics of glioma
| Characteristics (no. of patients) | MADE-D4 antibody positive (%) | P value |
|---|---|---|
| Sex | ||
| Male (25) | 5 (20.0) | 0.685 |
| Female (16) | 2 (12.5) | |
| Histology | ||
| Pilocytic astrocytoma (3) | 1 (33.3) | 0.499 |
| Diffuse astrocytoma (22) | 4 (18.2) | |
| Anaplastic astrocytoma (5) | 0 (0.0) | |
| Ependymoma (2) | 1 (50.0) | |
| Glioblastoma (9) | 1 (11.1) | |
| WHO grade | ||
| I-II (27) | 5 (18.5) | 1.000 |
| III-IV (14) | 2 (14.3) | |
| Total (41) | 7 (17.1) |
Discussion
In previous reports it was known that MAGE-D4 was over-expressed in some malignances including non-small cell lung cancer, squamous cell carcinoma of oral and esophagus [15,16], breast cancer [17], hepatocellular carcinoma [18], glioma, and only was detected trace amount of expression in brain and ovary, but not in other normal tissues. With this expressing profile it seems to be a potential candidate for immunotherapy and/or diagnosis of tumors.
A study showed that MAGE-D4 mRNA was expressed in a variety of brain tumors with different frequencies as follows: 100% (3/3) in oligodendroglioma, 71% (5/7) in glioblastoma, 67% (2/3) in anaplastic astrocytoma, 50% (2/4) in diffuse astrocytoma and 50% (1/2) in ependymoma [19]. But unfortunately, all of these studies about glioma mentioned above presented limited cases without clinicopathological data analysis. Therefore, in our study, the number of glioma as well as normal brain tissues was greatly increased. Our results again confirmed that the MAGE-D4 mRNA was overall up-regulated in glioma compared with normal brain tissues. The average mRNA expression level of MAGE-D4 is the highest in glioblastoma, followed by diffuse astrocytoma, oligodendroglioma and anaplastic astrocytoma, respectively. Considering the threshold of antigen expression level was critical for tumor cell recognized by cytotoxic T lymphocyte, we arbitrarily set a value for evaluation of high expression of MAGE-D4, as exceeding three times above the median value of MAGE-D4/HPRT1 of normal brain tissues. With the criteria it was found that there were around 80% to 30% of different types of glioma with high expression of MAGE-D4. In contrast to other MAGE family members (e.g. MAGE-A3) [20], expression level of MAGE-D4 was quite high in our study, especially in glioblastoma and diffuse astrocytoma, which supported MAGE-D4 as a potentially valuable target for immunotherapy of glioma.
According to the patterns of eukaryote gene expression, the mRNA level does not always accurately reflect the protein level, which may be due to mRNA stability, post-transcriptional modification, translational regulation and proteasomal degradation [21,22]. Whether the transcript can be translated into the corresponding protein with biological function should be investigated. In this regard, obviously, it is more important to perform protein analysis rather than mRNA analysis. So immunohistochemistry was put into practice to obtain information of MAGE-D4 protein expression in glioma, which had never been mentioned in pervious reports. Although the samples used in our study were not able to be done parallel tests with both qRT-PCR and immunohistochemical staining due to some objective causes, we did demonstrate there were more than 70% of glioma tissues expressing MAGE-D4 protein, which was frequently homogeneous and mainly located in the cytoplasm of tumor cells.
Localization of protein in the cells is closely related to their biological functions. Immunofluorescence microscopy was performed to reveal that MAGE-D4 was concentrated in the central spindle and in the midbody from telophase to the postmitotic phase, which indicated that MAGE-D4 might exist in a cell cycle specific manner and play some roles in cell division [5]. Recently, MAGE-D4 reportedly promotes cell growth, evades apoptosis, and associates with lymph node metastasis and poor disease specific survival in oral squamous cell carcinoma [23]. Moreover, MAGE-D4 was found to participate in the breast cancer tumorigenesis by affecting adhesion, migration and invasion [17]. However, information about the function of MAGE-D4 and its role in tumorigenesis of glioma is very limited at present, so further researches would be required.
The level of tumor antigen expression was considered to be very important for the success of the immunotherapy [24]. Based on the immunostaining intensity presented in the sections it was 11% of glioma with strong MAGE-D4 protein expression, most of them were moderate and weak expression. Some studies have uncovered that CpG island demethylation of the promoter regions might cause abnormal activation of a variety of MAGE genes in tumors [25-28]. Regarding to MAGE-D4, a putative CpG island in the promoter region was identified from -320bp to 720bp position near its transcription start site, suggesting that MAGE-D4 gene over-expression in glioma may be regulated by such a CpG demethylation mechanism [2]. A follow-up should be done to further verify this assumption.
Many MAGE family members and other tumor-associated antigens, which can elicit humoral immunity, have been identified in cancer patients [29-31]. Specific serum auto-antibodies are stable and can be detected in patients with subclinical stage of tumor, which may provide important serum markers for early diagnosis of disease [32]. In recent years, there are several tumor antigens detected by serological screening from glioma patients. These antigens include SOX5, SOX6 and PHF3 etc [33-35]. Presence of antibody against some of these antigens has been shown with correlation of survival in glioma patients [34]. As of yet, little information is available regarding immunogenicity of MAGE-D4 in patients with glioma. Therefore, we established an indirect ELISA assay to evaluate the presence of IgG antibodies against MAGE-D4 protein in glioma. Our preliminary results of serological analysis confirmed that MAGE-D4 antigen could stimulate the generation of auto-antibodies in about one-fifth of glioma patients and its immunogenicity had a good correlation with the expression of MAGE-D4 protein in glioma tissues, revealing that preferential, high expression of this antigen may give rise to the specific immune response in glioma patients. Although brain is generally considered as an immune-privileged organ, MHC expression is up-regulated and then the immune response is initiated in the case of inflammation, degenerative disease and tumors in the brain [1]. Some tumor antigens that induce antibody responses in cancer patients were likely to have elicited simultaneous T cell responses, including MAGE-A1, MAGE-A3, NY-ESO-1, and SSX [36]. Immunogenicity of MAGE-D4 suggests that B cells have been activated with the assistance of T cell recognition [37,38], and that the first MAGE-D4 T-cell epitope (HLA-A*25/MHC I) was identified by mass spectrometry [4]. MAGE-D4 is definitely not suitable for antibody-based therapy as not being a cell surface antigen, so it has potential for T cell-mediated therapies. Next, whether cellular immune response to MAGE-D4 can be induced in sero-positive glioma patients needs more experimental evidence.
In summary, our study revealed that the mRNA and protein of MAGE-D4 were highly expressed in the majority of different types of glioma. It was the first report that glioma patients can develop humoral response against MAGE-D4. These data support MAGE-D4 as a promising biomarker for glioma diagnosis and immunotherapy.
Acknowledgements
We thank Ms Fang Chen (Department of Histology and Embryology, Guangxi Medical University) for excellent technical assistance. This work was supported by National Natural Science Foundation of China (No. 30760055, No. 81060207, No. 81360371, No. 81360374), Natural Science Foundation of Guangxi (No. 0832144, No. 2011GXNSFA018275), Science and Technology Research Foundation of Guangxi Colleges and Universities (No. 2013YB052), Open Foundation of Medical experimental Center of Guangxi (No. KFJJ2010-33) and Research Fund for the Doctoral Program of Ministry of Education (No. 20114503120010).
Disclosure of conflict of interest
All the authors declare no editorial or financial conflicts of interest.
References
- 1.Yang MY, Zetler PM, Prins RM, Khan-Farooqi H, Liau LM. Immunotherapy for patients with malignant glioma: from theoretical principles to clinical applications. Expert Rev Neurother. 2006;6:1481–1494. doi: 10.1586/14737175.6.10.1481. [DOI] [PubMed] [Google Scholar]
- 2.Sasaki M, Nakahira K, Kawano Y, Katakura H, Yoshimine T, Shimizu K, Kim SU, Ikenaka K. MAGE-E1, a new member of the melanoma-associated antigen gene family and its expression in human glioma. Cancer Res. 2001;61:4809–4814. [PubMed] [Google Scholar]
- 3.Kawano Y, Sasaki M, Nakahira K, Yoshimine T, Shimizu K, Wada H, Ikenaka K. Structural characterization and chromosomal localization of the MAGE-E1 gene. Gene. 2001;277:129–137. doi: 10.1016/s0378-1119(01)00698-9. [DOI] [PubMed] [Google Scholar]
- 4.Krämer BF, Schoor O, Krüger T, Reichle C, Müller M, Weinschenk T, Hennenlotter J, Stenzl A, Rammensee HG, Stevanovic S. MAGED4-expression in renal cell carcinoma and identification of an HLA-A*25-restricted MHC class I ligand from solid tumor tissue. Cancer Biol Ther. 2005;4:943–948. doi: 10.4161/cbt.4.9.1907. [DOI] [PubMed] [Google Scholar]
- 5.Ito S, Kawano Y, Katakura H, Takenaka K, Adachi M, Sasaki M, Shimizu K, Ikenaka K, Wada H, Tanaka F. Expression of MAGE-D4, a novel MAGE family antigen, is correlated with tumor-cell proliferation of non-small cell lung cancer. Lung Cancer. 2006;51:79–88. doi: 10.1016/j.lungcan.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 6.Ma QY, Pang LW, Chen ZM, Zhu YJ, Chen G, Chen J. The significance of MAGED4 expression in non-small cell lung cancer as analyzed by real-time fluorescence quantitative PCR. Oncol Lett. 2012;4:733–738. doi: 10.3892/ol.2012.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luo G, Huang S, Xie X, Stockert E, Chen YT, Kubuschok B, Pfreundschuh M. Expression of cancer-testis genes in human hepatocellular carcinomas. Cancer Immun. 2002;2:11. [PubMed] [Google Scholar]
- 9.Thaker PH, Deavers M, Celestino J, Thornton A, Fletcher MS, Landen CN, Kinch MS, Kiener PA, Sood AK. EphA2 expression is associated with aggressive features in ovarian carcinoma. Clin Cancer Res. 2004;10:5145–5150. doi: 10.1158/1078-0432.CCR-03-0589. [DOI] [PubMed] [Google Scholar]
- 10.Zhou SF, Xie XX, Bin YH, Lan L, Chen F, Luo GR. Identification of HCC-22-5 tumor-associated antigen and antibody response in patients. Clin Chim Acta. 2006;366:274–280. doi: 10.1016/j.cca.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 11.Zhou SF, Mo FR, Bin YH, Hou GQ, Xie XX, Luo GR. Serum immunoreactivity of SMP30 and its tissues expression in hepatocellular carcinoma. Clin Biochem. 2011;44:331–336. doi: 10.1016/j.clinbiochem.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 12.Ivanov AV, Korovina AN, Tunitskaya VL, Kostyuk DA, Rechinsky VO, Kukhanova MK, Kochetkov SN. Development of the system ensuring a high-level expression of hepatitis C virus nonstructural NS5B and NS5A proteins. Protein Expr Purif. 2006;48:14–23. doi: 10.1016/j.pep.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 13.Huang CJ, Chen RH, Vannelli T, Lee F, Ritter E, Ritter G, Old LJ, Batt CA. Expression and purification of the cancer antigen SSX2: a potential cancer vaccine. Protein Expr Purif. 2007;56:212–219. doi: 10.1016/j.pep.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Wurtz T, Kruger A, Christersson C, Lundmark C. A new protein expressed in bone marrow cells and osteoblasts with implication in osteoblast recruitment. Exp Cell Res. 2001;263:236–242. doi: 10.1006/excr.2000.5114. [DOI] [PubMed] [Google Scholar]
- 15.Cheong SC, Chandramouli GV, Saleh A, Zain RB, Lau SH, Sivakumaren S, Pathmanathan R, Prime SS, Teo SH, Patel V, Gutkind JS. Gene expression in human oral squamous cell carcinoma is influenced by risk factor exposure. Oral Oncol. 2009;45:712–719. doi: 10.1016/j.oraloncology.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 16.Oya H, Kanda M, Takami H, Hibino S, Shimizu D, Niwa Y, Koike M, Nomoto S, Yamada S, Nishikawa Y, Asai M, Fujii T, Nakayama G, Sugimoto H, Fujiwara M, Kodera Y. Overexpression of melanoma-associated antigen D4 is an independent prognostic factor in squamous cell carcinoma of the esophagus. Dis Esophagus. 2013 doi: 10.1111/dote.12156. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 17.Germano S, Kennedy S, Rani S, Gleeson G, Clynes M, Doolan P, McDonnell S, Hughes L, Crown J, O’Driscoll L. MAGE-D4B is a novel marker of poor prognosis and potential therapeutic target involved in breast cancer tumorigenesis. Int J Cancer. 2012;130:1991–2002. doi: 10.1002/ijc.26200. [DOI] [PubMed] [Google Scholar]
- 18.Takami H, Kanda M, Oya H, Hibino S, Sugimoto H, Suenaga M, Yamada S, Nishikawa Y, Asai M, Fujii T, Nomoto S, Kodera Y. Evaluation of MAGE-D4 expression in hepatocellular carcinoma in Japanese patients. J Surg Oncol. 2013;108:557–62. doi: 10.1002/jso.23440. [DOI] [PubMed] [Google Scholar]
- 19.Lee MH, Son EI, Kim E, Kim IS, Yim MB, Kim SP. Expression of cancer-testis genes in brain tumors. J Korean Neurosurg Soc. 2008;43:190–193. doi: 10.3340/jkns.2008.43.4.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sahin U, Koslowski M, Türeci O, Eberle T, Zwick C, Romeike B, Moringlane JR, Schwechheimer K, Feiden W, Pfreundschuh M. Expression of cancer testis genes in human brain tumors. Clin Cancer Res. 2000;6:3916–3922. [PubMed] [Google Scholar]
- 21.Chen G, Gharib TG, Huang CC, Taylor JM, Misek DE, Kardia SL, Giordano TJ, Iannettoni MD, Orringer MB, Hanash SM, Beer DG. Discordant protein and mRNA expression in lung adenocarcinomas. Mol Cell Proteomics. 2002;1:304–313. doi: 10.1074/mcp.m200008-mcp200. [DOI] [PubMed] [Google Scholar]
- 22.Mathieu MG, Linley AJ, Reeder SP, Badoual C, Tartour E, Rees RC, McArdle SE. HAGE, a cancer/testis antigen expressed at the protein level in a variety of cancers. Cancer Immun. 2010;10:2. [PMC free article] [PubMed] [Google Scholar]
- 23.Chong CE, Lim KP, Gan CP, Marsh CA, Zain RB, Abraham MT, Prime SS, Teo SH, Silvio Gutkind J, Patel V, Cheong SC. Over-expression of MAGED4B increases cell migration and growth in oral squamous cell carcinoma and is associated with poor disease outcome. Cancer Lett. 2012;321:18–26. doi: 10.1016/j.canlet.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Yan Y, Phan L, Yang F, Talpaz M, Yang Y, Xiong Z, Ng B, Timchenko NA, Wu CJ, Ritz J, Wang H, Yang XF. A novel mechanism of alternative promoter and splicing regulates the epitope generation of tumor antigen CML66-L. J Immunol. 2004;172:651–660. doi: 10.4049/jimmunol.172.1.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu G, Fang J, He Y. 5’ CpG island methylation analysis identifies the MAGE-A1 and MAGE-A3 genes as potential markers of HCC. Clin Biochem. 2006;39:259–266. doi: 10.1016/j.clinbiochem.2006.01.014. [DOI] [PubMed] [Google Scholar]
- 26.Kim KH, Choi JS, Kim IJ, Ku JL, Park JG. Promoter hypomethylation and reactivation of MAGE-A1 and MAGE-A3 genes in colorectal cancer cell lines and cancer tissues. World J Gastroenterol. 2006;12:5651–5657. doi: 10.3748/wjg.v12.i35.5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Honda T, Tamura G, Waki T, Kawata S, Terashima M, Nishizuka S, Motoyama T. Demethylation of MAGE promoters during gastric cancer progression. Br J Cancer. 2004;90:838–843. doi: 10.1038/sj.bjc.6601600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wischnewski F, Pantel K, Schwarzenbach H. Promoter demethylation and histone acetylation mediate gene expression of MAGE-A1, -A2, -A3, and -A12 in human cancer cells. Mol Cancer Res. 2006;4:339–349. doi: 10.1158/1541-7786.MCR-05-0229. [DOI] [PubMed] [Google Scholar]
- 29.Gerhardt A, Usener D, Keese M, Sturm J, Schadendorf D, Eichmuller S. Tissue expression and sero-reactivity of tumor-specific antigens in colorectal cancer. Cancer Lett. 2004;208:197–206. doi: 10.1016/j.canlet.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 30.Türeci O, Mack U, Luxemburger U, Heinen H, Krummenauer F, Sester M, Sester U, Sybrecht GW, Sahin U. Humoral immune responses of lung cancer patients against tumor antigen NY-ESO-1. Cancer Lett. 2006;236:64–71. doi: 10.1016/j.canlet.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Zhang SC, Huang P, Zhao YX, Liu SY, He SJ, Xie XX, Luo GR, Zhou SF. Soluble expression of recombinant human SMP30 for detecting serum SMP30 antibody levels in hepatocellular carcinoma patients. Asian Pac J Cancer Prev. 2013;14:2383–2386. doi: 10.7314/apjcp.2013.14.4.2383. [DOI] [PubMed] [Google Scholar]
- 32.Lu H, Goodell V, Disis ML. Humoral immunity directed against tumor-associated antigens as potential biomarkers for the early diagnosis of cancer. J Proteome Res. 2008;7:1388–1394. doi: 10.1021/pr700818f. [DOI] [PubMed] [Google Scholar]
- 33.Ueda R, Yoshida K, Kawase T, Kawakami Y, Toda M. Preferential expression and frequent IgG responses of a tumor antigen, SOX5, in glioma patients. Int J Cancer. 2007;120:1704–1711. doi: 10.1002/ijc.22472. [DOI] [PubMed] [Google Scholar]
- 34.Ueda R, Iizuka Y, Yoshida K, Kawase T, Kawakami Y, Toda M. Identification of a human glioma antigen, SOX6, recognized by patients’ sera. Oncogene. 2004;23:1420–1427. doi: 10.1038/sj.onc.1207252. [DOI] [PubMed] [Google Scholar]
- 35.Pallasch CP, Struss AK, Munnia A, König J, Steudel WI, Fischer U, Meese E. Autoantibodies against GLEA2 and PHF3 in glioblastoma: tumor-associated autoantibodies correlated with prolonged survival. Int J Cancer. 2005;117:456–459. doi: 10.1002/ijc.20929. [DOI] [PubMed] [Google Scholar]
- 36.Caballero OL, Chen YT. Cancer/testis (CT) antigens: potential targets for immunotherapy. Cancer Sci. 2009;100:2014–2021. doi: 10.1111/j.1349-7006.2009.01303.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Zhang Y, Mandal A, Zhang J, Giles FJ, Herr JC, Lim SH. The spermatozoa protein, SLLP1, is a novel cancer-testis antigen in hematologic malignancies. Clin Cancer Res. 2004;10:6544–6550. doi: 10.1158/1078-0432.CCR-04-0911. [DOI] [PubMed] [Google Scholar]
- 38.Luo C, Xiao X, Liu D, Chen S, Li M, Xu A, Liu J, Gao S, Wu S, He D. CABYR is a novel cancer-testis antigen in lung cancer. Clin Cancer Res. 2007;13:1288–1297. doi: 10.1158/1078-0432.CCR-06-1742. [DOI] [PubMed] [Google Scholar]


