Abstract
Single nucleotide polymorphisms (SNPs) of VKORC (1173T/C, rs9934438) and CYP2C9 (1075A/C, rs1057910) are major contributory factors on the sensitivity of warfarin in Chinese. Analysis of the two genomic loci could help warfarin treatment individual from bleeding or thrombosis events. An assay with the advantages of simplicity, speed, high sensitivity and low cost for genotyping is calling for clinical laboratories. High resolution method (HRM) meets these callings, but no study with large sample tested its performance in genotyping of rs9934438 and rs1057910. In this study, we identified polymorphisms of rs9934438 and rs1057910 in 255 unrelated Chinese heart valve replacement patients of Han ethnic group from West China Hospital. The two genomic loci were genotyped by HRM using LightCycler® 480 High Resolution Melting Master on LightCycler® 480 Real-Time PCR instruments (Roche Diagnostics), and all amplified PCR products were sent for direct DNA sequencing. The genotyping of rs1057910 between HRM and sequencing showed perfect 100% concordance. While the concordance of rs9934438 between HRM and sequencing was 99.2%. Unexpected mutation interfered genotyping results of HRM when genotyping rs9934438. HRM is a valuable technique for genotype detection of rs9934438 and rs1057910 to assess individual sensitivity to warfarin, where DNA sequencing is added inevitably sometimes.
Keywords: HRM, genotyping, warfarin, CYP2C9, VKORC
Introduction
Warfarin is the most widely used antithrombotic drug. Due to a narrow therapeutic range and wide interindividual variability in drug sensitivity, poor warfarin control with risk of thrombosis and bleeding events is highest during the first months of treatment [1-3]. The genetic polymorphisms of the vitamin K epoxide reductase [VKORC1] and cytochrome P450 2C9 [CYP2C9] loci have been increasingly acknowledged as major contributory factors of enhanced warfarin sensitivity [4-6]. CYP2C9 encode the enzyme (cytochrome P450 2C9) primarily responsible for the metabolism of the more active S-enantiomer of warfarin, and VKORC1 encode the subunit 1 of the vitamin K epoxide reductase complex, the target of warfarin [7,8]. Single nucleotide polymorphisms (SNPs) of rs9934438 and rs1057910 within genes VKORC1 and CYP2C9, respectively, had a significant impact on the sensitivity of warfarin among Chinese [9,10]. Several small randomized controlled trials (RCTs) have yielded promising results that patient receiving genotype-guided dosing required fewer and smaller dose adjustments, fewer INR determinations, and resulted in faster stable does acquirement and reduction in hospitalization [11,12].
Successful implementation of genotype-guided dosing into clinical practice will depend on the use of methods available for rapid and easy genotyping in clinical settings. The clinical laboratory community has made substantial progress in preparing for warfarin pharmacogenetic testing, as US FDA-cleared assays are available to the clinical laboratory community for warfarin pharmacogenetic testing [13]. However, obstacles remain to testing expansion. US FDA-cleared assays are allele-specific probes analysis based on PCR, which is relatively expensive. Other assays like restriction enzyme digestion [14,15] and denaturing high-performance liquid chromatography (DHPLC) [16] are often preceded by laboratories. But they are also expensive and/or time-consuming. Considering the increasing number of requests for VKORC1 and CYP2C9 gene tests, the demand for a fast, reliable and low costing technique is high.
High-resolution melting curve (HRM) analysis is a potentially useful method for fast genotyping and high-throughput mutation scanning of disease-related genes in genome diagnostics [17]. This technique uses a fluorescent DNA binding dye instead of costly labeled probes for genotyping. In the presence of saturating concentrations of DNA binding dyes, fluorescence intensity decreases as the double stranded DNA becomes single stranded following the rise of temperature and the dye is released [18]. The melting profile of the PCR product depends on its GC content, length, sequence, and heterozygosity, and mutations in the sequence will give rise to heteroduplexes that change the shape of the melting curve when compared to the wild-type (wt) melt profile [17]. HRM is a technique used for identifying genetic variations in nucleic acid sequences without the requirement of any post-PCR handling.
However, more than one SNP nearing these two loci in 100 bp, as well as CYP2C9 having high degree of sequence similarity with other genes make HRM in debate to apply in genotyping rs9934438 and rs1057910. In the present study, we evaluated the HRM technique to detect genotypes of two genomic loci rs9934438 and rs1057910 in Chinese populations on platform of 96-well LightCycler® 480 Real-Time PCR system. To validate the sensitivity and specificity, 255 samples were tested and all sequenced.
Materials and methods
Subjects and DNA samples
Patients were from the inpatient unit of the Cardiothoracic Surgery Department of West China Hospital, who were going to receive warfarin therapy. Because the great majority of patients were Han origin, we excluded patients of other minor origins from analysis. Finally 255 patients (31.4% male, mean age 49.3 years, range 20-74 years) were enrolled and 2 ml of peripheral venous blood was drawn with EDTA as an anticoagulant. All consent forms were received. Genomic DNA was extracted using a QIAamp® DNA Blood mini kit (Qiagen).
HRM assays and analysis
Amplification of rs9934438 and rs1057910 variants was carried out in a 96-well plate in the LightCycler® 480 Real-Time PCR System (Roche Diagnostics) under the same reaction system and conditions. The PCR reaction mixture was prepared in 20 μl total volume by adding 10 ng of genomic DNA, 10 μl Master Mix and 2.4 μl MgCl2 (High Resolution Melting Master, Roche), 5 pmol of forward and reverse primers and 4.8 μl dH2O. Thermocycling conditions for PCR included one cycle denaturation at 95°C for 10 min and 55 cycles consisted of denaturation at 95°C for 15 s, annealing at 63°C for 10 s and extension at 72°C for 10 s, and cooling at 40°C for 30 s. All primers were selected using the Primer5 software (CA, USA), synthesized by Introvogen (Shanghai, China). Primer sequences were as follows: VKORC-F, 5’-CCGAGAAAGGTGATTTCCAA-3’, VKORC-R, 5’-TGACATGGAATCCTGACGTG-3’, CYP2C9-F, 5’-CACGAGGTCCAGAGATAC-3’, CYP2C9-R, 5’-GGAATGAGATAGTTTCTGAATTTAAT-3’.
Determination of genotypes
Melting of the PCR products was monitored by plotting the changes in fluorescence that occurred by gradual temperature-dependent releasing of a saturating double-strand DNA binding dye. Heterozygous DNA samples formed heteroduplexes, resulting in a different shape of the melting curve compared with a homozygous sample. Different genotypes of homozygous DNA samples, in contrast, were detected by a melting temperature (Tm) shift rather than an altered curve shape.
Direct DNA sequencing
To confirm the quality and accuracy of genotyping data from HRM assay, sequencing analyses were also carried out in the 255 samples. These amplicons form HRM were sent for direct DNA Sequence by a company (Introvogen Shanghai, China), who use the ABI 3130xl capillary sequencer (Applied Biosystems, Foster City, CA). The sequence detection was conducted using Chromas program (Technelysium Pty. Ltd., Helensvale, Queensland, Australia).
Results
Genotyping of rs9934438 and rs1057910
As shown in Figure 1, three genotypes (common homozygote TT, heterozygote TC and rare homozygote CC) for SNP of VKORC1 rs9934438 were clearly distinguished. The Tm value of the homozygote TT (approximately 86.0°C) was shifted to the left compared to that of the homozygote CC (86.5°C), due to the presence of the GC base pair which had a higher melting temperature vs. the AT base pair. The temperature-shifted (temp-shifted) melting peak of the heterozygote TC was between those of homozygote TT and CC. The heterozygote TC samples were unambiguously assigned because of different melting curve shapes from the homozygote ones. Interestingly, in addition to the three groups of three genotypes for SNP rs9934438, we observed a fourth cluster different from these melting curves which were termed “unknown” (Figure 1C and 1D).
Figure 1.
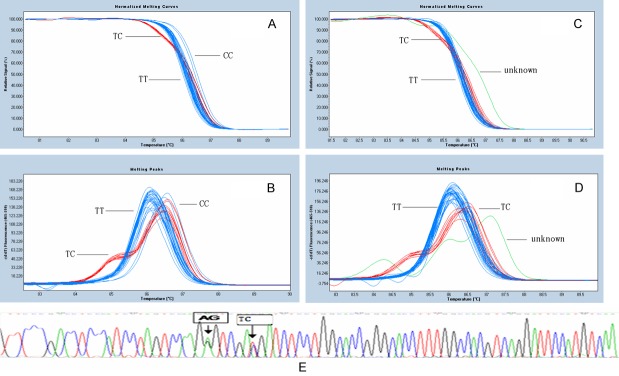
Melting curves of VKORC genotyping by HRM. Melting curves for three different genotypes of VKORC1 rs9934438, shown in (A) and (B); profile with a single-peak at 86.0°C representing homozygote of TT, profile with a single-peak at 86.7°C representing homozygote of CC, and profile with a double-peak at 85.0°C and 86.5°C representing heterozygote of TC. While profile with tri-peak labeled as “unknown” genotype, shown in (C) and (D). Direct DNA sequencing showed “unknown” representing TC genotype of rs9934438 and AG genotype of rs188090042, shown in (E).
In the case of CYP2C9, it is necessary to discriminate the signals from those for the highly related subtypes CYP2C8 (cytochrome P450, family 2, subfamily C, polypeptide 8), CYP2C18 (cytochrome P450, family 2, subfamily C, polypeptide 18), and CYP2C19 (cytochrome P450, family 2, subfamily C, polypeptide 19), because all 4 genes have a high degree of sequence similarity [19]. We engineered the folding primer to be the SNP-discrimination primer, and its 3’ terminal nucleotide recognized both Wt and Mt alleles. The normalized melting curves of CYP2C9 rs1057910 clearly differentiated into two groups, homozygote AA and heterozygote AC, as shown in Figure 2A and 2B. There was no CC genotype in the 255 patients.
Figure 2.
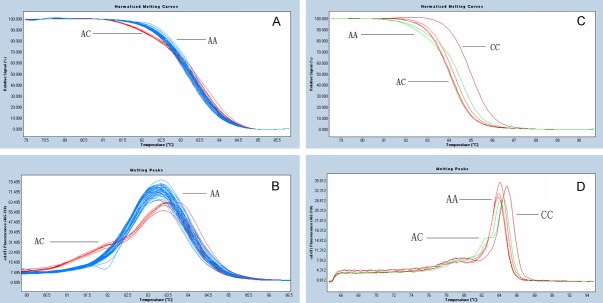
Melting curves of CYP2C9 genotyping by HRM. Melting curves for two different genotypes of CYP2C9 rs1057910, shown in (A) and (B); profile with a single-peak at 83.50°C representing homozygote of AA, and profile with a double-peak at 82.00°C and 83.50°C representing heterozygote of AC. We did not detect any CC genotype of CYP2C9 in 255 Chinese patients, but a confirm CC genotype sample was got to prove this HRM methods can differentiate it from AA genotype as shown in (C) and (D).
Validation of HRM
In the experiment, a total of 255 genomic DNA samples for each group of VKORC1 and CYP2C9 genes with genotypes obtained by melting curve analysis were sequenced. We compared the data obtained from HRM with those of direct DNA sequencing, which has been considered the gold standard for genotyping, and the results indicate 100% concordance between DNA sequence analysis and HRM genotyping for CYP2C9, while the accuracy for VKORC1 was 99.2% by HRM genotyping.
Sequence results showed the two samples which were typed “unknown” by HRM genotyping were heterozygote TC genotype of VKORC. In addition to the recognized rs9934438, we identified another heterozygote of rs188090042 (T > C) in the amplicon. Actually, the “unknown” was TC genotype of rs188090042 and TC genotype of rs9934438 (shown in Figure 1E).
As no CC genotype of CYP2C9 was detected in the 255 patients, a confirmed CC genotype sample was got and validated homozygote CC and AA genotype can be differentiated by HRM. The Tm value of the homozygote AA (approximately 83.5°C) was shifted to the left compared to that of the homozygote CC (84.5°C), shown in Figure 2C and 2D.
Allele frequencies of rs9934438 and rs1057910
The genotype distributions for the two SNPs in our study were all in Hardy–Weinberg equilibrium (P > 0.05). The minor allele frequencies of rs9934438 and rs1057910 were 10.98% and 6.27% respectively for Chinese Han population (Table 1).
Table 1.
Frequency of CYP2C9 rs1057910 and VKORC rs9934438
| VKORC1 | Total | ||||
|---|---|---|---|---|---|
|
|
|||||
| TT | TC | CC | |||
| CYP | AA | 177 | 44 | 2 | 223 |
| AC | 25 | 6 | 1 | 32 | |
| Total | 202 | 50 | 3 | 255 | |
Discussion
Considering the well known contribution of rs9934438 in VKORC1 and rs1057910 in CYP2C9 to individual warfarin dose variants, the demand for diagnostic based genetic testing is on the rise and the availability of rapid, efficient, and reliable technologies for genetic based testing is therefore critical for clinical patient management [5]. With the benefits of simplicity, speed, high sensitivity and low cost, HRM meets these demands rightly. Although many laboratories use HRM for genotyping of common DNA variants and have shown successful clinical use [17,20], few studies with application on HRM genotyping of rs9934438 and rs1057910 were reported [21,22]. We presented the first assessment study with 255 samples, all of which had sent for DNA sequencing.
In our study, using LightCycler® 480 Gene Scanning software v1.2 (Roche Diagnostics), heterozygote TC of VKORC was easily identified by the altered shape of the melting curves and homozygote TT or CC were distinguished by a change in melting temperature (Tm) that differed by approximately 0.50°C; heterozygote AC of CYP2C9 was easily distinguished with homozygote CC by the altered shape of the melting curves. And validating by sequence analysis, we demonstrated that HRM had high sensitivity and accuracy in genotyping of rs9934438 and rs1057910, above 99% concordance with DNA sequencing.
Though it is not 100% concordance with DNA sequencing when genotyping rs9934438 of VKORC with HRM, it was also proved to be sensitive in rs9934438 genotyping. A totally different melting curve was observed when unexpected mutation showed up in one amplicon simultaneously. This is often unavoidable when variants are closely located [20]. Of the 255 samples, only two carried the other variant rs188090042 and the melting curves of the two samples were identified. Data from NCBI SNP database suggest the MAF (minor allele frequency) of these closing SNPs is no more than 0.3%. The potential risk of false genotyping was extremely low.
The 100% concordance was observed in genotyping rs1057910 of CYP2C9 with HRM when compared with DNA sequencing results. But the limitation of the study was that of the 255 genotypes obtained, no CC genotype of CYP2C9 was detected. Homozygote clusters can result in incorrect calls, if the differences between homozygote are small [23]. In our other studies, of 260 more samples (data not shown), only one CC genotype were found, which made us can prove that CC genotype can be differentiated from AA genotype by HRM. Though more CC genotypes would be powerful to prove that HRM has high sensitivity to differentiate homozygote of CYP2C9, the frequency of CC genotype was extremely low. In fact, the frequency of C allele was higher in our study than that in other studies of China (6.27% VS 3.61%) [24].
HRM as a diagnostic assay for genotyping rs9934438 and rs1057910 was at risk. However, applying HRM as a screening test and gene sequencing as back up method may realize the goal of economic, rapid, accurate detection. Estimated turnaround time, including time for DNA extraction and a run size of 45 specimens, was 3-4 hours in our study. And the cost of one specimen was no more than $10. If DNA sequencing is added, 6 more hours and $10 for one specimen will need. While the cost of one specimen is $400-500 and the turn around time is 2-4 hours with US FDA cleared commercial warfarin genotyping kits [13,25]. For developing country, the cost burden may stop patients benefiting from pharmacogenetic guided treatment.
When concerned about the extended turnaround time, we must notice that warfarin do not anticoagulate blood immediately. Instead, onset of its effect requires about a day before remaining active clotting factors have had time to naturally disappear in metabolism, and the duration of action of a single dose of warfarin is 2 to 5 days. So, additional 6 more hours would be kind of acceptable, when DNA sequencing was unavoidable for some rare unexpected mutations.
In summary, we provided new evidence for applying HRM method to detect genotype of CYP2C9 * 3 (1075A/C, rs1057910) and VKORC1 (1173T/C, rs9934438) in Chinese population and it is worth noting that HRM as an indirect genotyping method, other mutations on the amplicon may interfere with the detection of target SNP genotyping. Applying HRM as a screening test and gene sequencing as back up method to detect genotype of rs1057910 and rs9934438 may realize the goal of economic, rapid, and accurate detection.
Disclosure of conflict of interest
None.
References
- 1.Gomes T, Mamdani MM, Holbrook AM, Paterson JM, Hellings C, Juurlink DN. Rates of hemorrhage during warfarin therapy for atrial fibrillation. CMAJ. 2013;185:E121–127. doi: 10.1503/cmaj.121218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma C, Zhang Y, Xu Q, Yang J, Zhang Y, Gao L, Xu B, Wang H, Li Y, Lu C, Yin T. Influence of warfarin dose-associated genotypes on the risk of hemorrhagic complications in Chinese patients on warfarin. Int J Hematol. 2012;96:719–728. doi: 10.1007/s12185-012-1205-8. [DOI] [PubMed] [Google Scholar]
- 3.Wieloch M, Sjalander A, Frykman V, Rosenqvist M, Eriksson N, Svensson PJ. Anticoagulation control in Sweden: reports of time in therapeutic range, major bleeding, and thrombo-embolic complications from the national quality registry AuriculA. Eur Heart J. 2011;32:2282–2289. doi: 10.1093/eurheartj/ehr134. [DOI] [PubMed] [Google Scholar]
- 4.Skov J, Bladbjerg EM, Leppin A, Jespersen J. The influence of VKORC1 and CYP2C9 gene sequence variants on the stability of maintenance phase warfarin treatment. Thromb Res. 2013;131:125–129. doi: 10.1016/j.thromres.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Fung E, Patsopoulos NA, Belknap SM, O’Rourke DJ, Robb JF, Anderson JL, Shworak NW, Moore JH. Effect of Genetic Variants, Especially CYP2C9 and VKORC1, on the Pharmacology of Warfarin (vol 38, pg 893, 2012) Semin Thromb Hemost. 2013;39:112–112. doi: 10.1055/s-0032-1328891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.International Warfarin Pharmacogenetics Consortium. Klein TE, Altman RB, Eriksson N, Gage BF, Kimmel SE, Lee MT, Limdi NA, Page D, Roden DM, Wagner MJ, Caldwell MD, Johnson JA. Estimation of the Warfarin Dose with Clinical and Pharmacogenetic Data. New Engl J Med. 2009;360:753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rost S, Fregin A, Ivaskevicius V, Conzelmann E, Hortnagel K, Pelz HJ, Lappegard K, Seifried E, Scharrer I, Tuddenham EGD, Muller CR, Strom TM, Oldenburg J. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature. 2004;427:537–541. doi: 10.1038/nature02214. [DOI] [PubMed] [Google Scholar]
- 8.Rettie AE, Wienkers LC, Gonzalez FJ, Trager WF, Korzekwa KR. Impaired (S)-warfarin metabolism catalysed by the R144C allelic variant of CYP2C9. Pharmacogenetics. 1994;4:39–42. doi: 10.1097/00008571-199402000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Tan SL, Li Z, Song GB, Liu LM, Zhang W, Peng J, Zhang T, Jia FF, Zhou G, Zhou HH, Zhou XM. Development and comparison of a new personalized warfarin stable dose prediction algorithm in Chinese patients undergoing heart valve replacement. Pharmazie. 2012;67:930–937. [PubMed] [Google Scholar]
- 10.Miao L, Yang J, Huang C, Shen Z. Contribution of age, body weight, and CYP2C9 and VKORC1 genotype to the anticoagulant response to warfarin: proposal for a new dosing regimen in Chinese patients. Eur J Clin Pharmacol. 2007;63:1135–1141. doi: 10.1007/s00228-007-0381-6. [DOI] [PubMed] [Google Scholar]
- 11.Fung E, Patsopoulos NA, Belknap SM, O’Rourke DJ, Robb JF, Anderson JL, Shworak NW, Moore JH. Effect of genetic variants, especially CYP2C9 and VKORC1, on the pharmacology of warfarin. Semin Thromb Hemost. 2012;38:893–904. doi: 10.1055/s-0032-1328891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang M, Lang X, Cui S, Fei K, Zou L, Cao J, Wang L, Zhang S, Wu X, Wang Y, Ji Q. Clinical application of pharmacogenetic-based warfarin-dosing algorithm in patients of Han nationality after rheumatic valve replacement: a randomized and controlled trial. Int J Med Sci. 2012;9:472–479. doi: 10.7150/ijms.4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stack G. Pathology consultation on warfarin pharmacogenetic testing. Am J Clin Pathol. 2011;135:13–19. doi: 10.1309/AJCPAO82OTNPUBLW. [DOI] [PubMed] [Google Scholar]
- 14.You JHS, Wong RSM, Waye MMY, Mu Y, Lim CK, Choi KC, Cheng G. Warfarin dosing algorithm using clinical, demographic and pharmacogenetic data from Chinese patients. J Thromb Thrombol. 2011;31:113–118. doi: 10.1007/s11239-010-0497-x. [DOI] [PubMed] [Google Scholar]
- 15.Sconce EA, Khan TI, Wynne HA, Avery P, Monkhouse L, King BP, Wood P, Kesteven P, Daly AK, Kamali F. The impact of CYP2C9 and VKORC1 genetic polymorphism and patient characteristics upon warfarin dose requirements: proposal for a new dosing regimen. Blood. 2005;106:2329–2333. doi: 10.1182/blood-2005-03-1108. [DOI] [PubMed] [Google Scholar]
- 16.Huang Y, Yang JF, Qi XL, Wang YQ, Wang WZ, Chen B. [Association between genetic polymorphisms of CYP2C19 and CYP2C9 and phenytoin serum concentration] . Zhonghua Yi Xue Za Zhi. 2004;84:1686–1689. [PubMed] [Google Scholar]
- 17.van der Stoep N, van Paridon CD, Janssens T, Krenkova P, Stambergova A, Macek M, Matthijs G, Bakker E. Diagnostic guidelines for high-resolution melting curve (HRM) analysis: an interlaboratory validation of BRCA1 mutation scanning using the 96-well LightScanner. Hum Mutat. 2009;30:899–909. doi: 10.1002/humu.21004. [DOI] [PubMed] [Google Scholar]
- 18.Graham R, Liew M, Meadows C, Lyon E, Wittwer CT. Distinguishing different DNA heterozygotes by high-resolution melting. Clin Chem. 2005;51:1295–1298. doi: 10.1373/clinchem.2005.051516. [DOI] [PubMed] [Google Scholar]
- 19.Aomori T, Yamamoto K, Oguchi-Katayama A, Kawai Y, Ishidao T, Mitani Y, Kogo Y, Lezhava A, Fujita Y, Obayashi K, Nakamura K, Kohnke H, Wadelius M, Ekstrom L, Skogastierna C, Rane A, Kurabayashi M, Murakami M, Cizdziel PE, Hayashizaki Y, Horiuchi R. Rapid single-nucleotide polymorphism detection of cytochrome P450 (CYP2C9) and vitamin K epoxide reductase (VKORC1) genes for the warfarin dose adjustment by the SMart-amplification process version 2. Clin Chem. 2009;55:804–812. doi: 10.1373/clinchem.2008.115295. [DOI] [PubMed] [Google Scholar]
- 20.Tindall EA, Petersen DC, Woodbridge P, Schipany K, Hayes VM. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments. Hum Mutat. 2009;30:876–883. doi: 10.1002/humu.20919. [DOI] [PubMed] [Google Scholar]
- 21.Cui GL, Ding H, Xu YJ, Chen C, Wang DW. [Development of the high resolution melting method for genotyping CYP2C9*3 (1075A/C, rs1057910) and VKORC1 (-1639A/G, rs9923231)] . Zhonghua Xin Xue Guan Bing Za Zhi. 2012;40:477–481. [PubMed] [Google Scholar]
- 22.Huang SW, Li Q, Zhu SY, Li L, Xiong F, Jia YK, Xu XM. SYBR Green-based real-time PCR assay for detection of VKORC1 and CYP2C9 polymorphisms that modulate warfarin dose requirement. Clin Chem Lab Med. 2009;47:26–31. doi: 10.1515/CCLM.2009.008. [DOI] [PubMed] [Google Scholar]
- 23.Cui G, Zhang L, Xu Y, Cianflone K, Ding H, Wang DW. Development of a high resolution melting method for genotyping of risk HLA-DQA1 and PLA2R1 alleles and ethnic distribution of these risk alleles. Gene. 2013;514:125–130. doi: 10.1016/j.gene.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 24.Liu Y, Zhong SL, Yang M, Tan HH, Fei HW, Chen JY, Yu XY, Lin SG. [Distribution of variant alleles association with warfarin pharmacokinetics and pharmacodynamics in the Han population in China] . Beijing Da Xue Xue Bao. 2011;43:798–803. [PubMed] [Google Scholar]
- 25.Patrick AR, Avorn J, Choudhry NK. Cost-effectiveness of genotype-guided warfarin dosing for patients with atrial fibrillation. Circ Cardiovasc Qual Outcomes. 2009;2:429–436. doi: 10.1161/CIRCOUTCOMES.108.808592. [DOI] [PubMed] [Google Scholar]


