Abstract
Tibial plateau fractures are often the result of blunt trauma and are associated with severe soft-tissue injury. Operative management of high-energy fractures remains difficult and challenging because the injuries often associated with serious complications. MicroRNAs (miRNAs) are the class of short noncoding single-stranded RNA molecules that negatively regulate gene expression. miRNAs contribute to every step of osteogenesis from embryonic bone development to maintenance of adult bone tissue, and disturbed miRNAs expression are identified related to osteoporosis, osteosarcoma, post-traumatic arthritis and bone remodeling. But our understandings about the roles of miRNAs in tibial plateau fractures repairing process are rare. In this study, we first detect seven candidate miRNAs expression in the SF cells of the mouse model. The results indicated that miR-9 and miR-181a were down-regulated significantly five days after injury. By using dual luciferase assay and western blot, we confirmed that the expression of Cbl is repressed by miR-9 and miR-181a. Meanwhile, the amount of ubiquitinated Bim was raised and the total Bim was reduced by miRNA inhibitors. Further functional study indicated that reduced miR-9 and miR-181a expression can active RAW264.7 cells migration ability and raise the primary mouse osteoclasts survival rate in vitro. To our understood, this is the first study about the function of disturbed miRNAs in the tibial plateau fracture mouse model, and may expand our understanding about post tibial plateau fracture recover and post-traumatic sequelae generation.
Keywords: microRNA, tibial plateau fracture, Cbl, E3 ubiquitin ligase, osteoclast
Introduction
MiRNAs are a group of approximately 20-24 nucleotides single strand RNA molecules that negatively regulate gene expression. These small RNA molecules repress gene expression by binding the 3’UTR of mRNA and block protein translation. MiRNAs play very important roles in maintaining normal human body physiology conditions, and abnormal miRNA expressions have been found related to many human diseases spanning from psychiatric disorders to malignant cancers [3,10,12]. miRNAs contribute to every step of osteogenesis from embryonic bone development to maintenance of adult bone tissue, by regulating the growth, differentiation and functional. miRNAs also play important roles in osteal or articular injury and repair process.
Tibial plateau fracture is a kind of bone fracture or break in the continuity of the bone occurring in the proximal part of the tibia or shinbone; affecting the knee joint, stability and motion. The tibial plateau is a critical weight-bearing area located on the upper extremity of the tibia and is composed of two slightly concave condyles separated by an intercondylar eminence and the sloping areas in front and behind it. Although tibial plateau fractures only constitute 1% of all fractures, operative management of high-energy fractures remains difficult and challenging because the injuries often associated with serious complications [2]. As osteoclasts are key players in joint and bone destruction, it is of critical importance to understand their modes of migration, differentiation and function in vivo [8,11].
Till now, our understanding of how miRNAs control the regulatory interplay between the different cell types in the bone remodeling unit is minimal. In addition, the roles of miRNAs in the differentiation and recruitment of mature cells derived from mesenchymal stem cells or hematopoietic stem cells remains to be established. The studies of miRNAs in tibial plateau fractures repair are rare.
In this study, we first detected seven candidate miRNAs expression in the synovial fluid cells of tibial plateau fracture mouse [9]. We found that the miR-9 and miR-181a were down-regulated. Confirmed by dual luciferase assay and western blot, we identified Cbl is a target gene of miR-9 and miR-181a. Up-regulated Cbl promote RAW264.7 cells migration ability and raise the primary mouse osteoclasts (OCs) survival rate in vitro, which may play important roles in tibial plateau fractures repair.
Materials and methods
Mouse tibial plateau fracture model
18 mice (male, C57BL/6, 8 wks) were housed until 16 weeks of age, as described previously [4], animals were anesthetized (pentobarbital, i.p. 60 mg/kg) and placed in a custom cradle with the left hindlimb in neutral position (90° flexion). A custom indenter applied a 10 N pre-load to the anterior aspect of the left proximal tibial plateau, followed by compression applied at a rate of 20 N/s. The energy of fracture was calculated from load-displacement curves for each joint. We got 18 high energy fracture model mice.
All mice underwent high resolution digital radiographs (Model MX-20 Digital, Faxitron) within 1 hr to confirm the presence of an articular fracture. No fixation or surgical intervention was employed. Animals were given analgesic (buprenorphine, s.q. 0.1 mg/kg, bid) for 48 hrs following fracture inductions and allowed immediate ad libitum weight bearing and motion. Mice were sacrificed at 0 (n=6), 3 (n=6), 5 (n=6) days post-fracture per group. Contralateral limbs were used as controls.
Synovial fluid (SF) samples were obtained from stifle joints. The samples were centrifuged to separate cells and debris from the SF (1000 g, 10 min at 4 °C). The purified SF was stored at -20 °C, and the SF cell pellet was used for total RNA extraction.
RNA extraction and miRNAs expression detection
Quantitative RT-PCR analysis was used to determine the relative expression level of 7 candidate miRNAs. Total RNA was extracted from the cells and purified synovial fluid by using Trizol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. The expression levels of miRNAs were detected by TaqMan miRNA RT-Real Time PCR. Single-stranded cDNA was synthesized by using TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and then amplified by using TaqMan Universal PCR Master Mix (Applied Biosystems, Foster City, CA, USA) together with miRNA-specific TaqMan MGB probes (Applied Biosystems, Foster City, CA, USA). The U6 snRNA was used for normalization. Each sample in each group was measured in triplicate and the experiment was repeated at least three times.
Dual luciferase assay
A 2508 bp Cbl 3’UTR segment, which contains miR-9 and miR-181a target sites, was cloned into downstream of firefly luciferase coding region in pmirGLO vector (Promega, Madison, WI, USA) to generate luciferase reporter vector. For luciferase reporter assays, HEK293T cells were seeded in 48-well plates. MiRNA mimics, inhibitors, sequence scrambled RNA controls were co-transfected with luciferase reporter vectors by using lipofectamine 2000 (Invitrogen, Carlsbad, CA USA). Two days later, cells were harvested and assayed with the Dual-Luciferase Assay (Promega, Madison, WI USA). Each treatment was performed in triplicate in three independent experiments. The results were expressed as relative luciferase activity (Firefly LUC/Renilla LUC).
Western blotting
Protein extracts were boiled in SDS/β-mercaptoethanol sample buffer, and 30 μg samples were loaded into each lane of 8% polyacrylamide gels. The proteins were separated by electrophoresis, and the proteins in the gels were blotted onto PVDF membranes (Amersham Pharmacia Biotech, St. Albans, Herts, UK) by electrophoretic transfer. The membrane was incubated with rabbit anti-Cbl polyclonal antibody (Abcam, Cambridge, MA, USA) or goat anti-Bim polyclonal antibody (Abcam, Cambridge, MA, USA) or mouse anti-β-actin monoclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, CA, USA) 4 °C overnight. The specific protein-antibody complex was detected by using horseradish peroxidase conjugated IgG. Detection by the chemiluminescence reaction was carried using the ECL kit (Pierce, Appleton, WI, USA). The β-actin signal was used as a loading control.
Immunoprecipitation
Cell cultures were washed twice with ice-cold PBS and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Triton X-100, 12 mM β-glycerophosphate, 5 mM EGTA, 0.5% deoxycholate, 3 mM DTT, 10 mM NaF, 1 mM Na3VO4, 2 mM leupeptin, 20 mg/ml aprotinin, and 1 mM PMSF). After 30 min on ice, cell lysates were cleared by centrifugation at 12,000 g for 20 min. The protein concentration in each sample was quantified by the Bradford method, and immunoprecipitation was performed by incubating 800 μl of lysate with 20 μl Flag tag antibody coated beads 4 h at 4 °C. The immunocomplexes were washed with washing buffer (20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EGTA, 2 mM DTT, and 1 mM PMSF) twice.
Detected the SF Cbl and Bim concentration by enzyme-linked immunosorbent assay
SF Cbl and Bim levels were detected by using sandwich ELISA method. The relative concentrations were compared using relative OD value. The results were analyzed by using Mann-Whitney u test. p<0.05 was considered statistically significant.
In vitro migration assays
A typical Transwell assay (Costar, 6.5 mm diameter, 8 μm pore size) was used. 3×104 cells in 200 μL serum-free medium were seeded to the top chamber and 500 μL medium with high concentration of serum was added to the bottom. After 12 h, Filters were then submerged in 4% PFA for 15 min and cells on the upper surface were removed by cotton swabs. The cells on the lower surface were stained with hematoxylin-eosin. Cells that had migrated through the membranes were quantified by determination of the cell number in five randomly chosen visual fields at 200× magnification.
Culture of osteoclast-like cells
Osteoblasts obtained from the calvaria of newborn mice and bone marrow cells obtained from the tibiae of male mice were co-cultured in α-minimal essential medium (α-MEM) containing 10% fetal bovine serum, 1α,25-dihydroxyvitamin D3(1α,25(OH)2 D3) (10-8 M) and prostaglandin E2 (PGE2) (10-6 M) in 100-mm diameter dishes coated with collagen gels (Nitta Gelatin Co., Osaka, Japan). OCLs were formed within 6 days in culture and were removed from the dishes by treatment with 0.2% collagenase (Wako Pure Chemical Co.). The purity of OCLs in this fraction (crude OCL preparation) was about 5%. To further purify the OCLs, the crude OCL preparation was replated on culture dishes. After an 8 h culture, osteoblasts were removed with PBS containing 0.001% Pronase E (Calbiochem, La Jolla, CA) and 0.02% EDTA for 10 min at 37 °C according to the method described previously [5,6].
Plasmids and virus infection
Retroviral vectors, pGLVU6-miR-9In-GFP and pGLVU6-miR-181aIn-GFP were constructed by inserting 3 miR-9 or miR-181a binding sites and empty pGLVU6-GFP vector was used as control. OCs were incubated with 2 ml of virus stock for 4 h in the presence of polybrene (1 μg/ml), and then, the medium was changed to normal α-MEM medium with 10% fetal calf serum and M-CSF for survival assay.
Survival of OCs
The survival rate was measured as reported [7]. OCs were purified 6 h after the infection and some of the cultures were subjected to tartrate-resistant acid phosphatase (TRAP) staining. Cell viability/survival is expressed as morphologically intact TRAP-positive multinucleated cells. Other cultures were further incubated for the indicated times, and then the number of living OCs was counted. The number of viable cells remaining at the different time points is shown as a percentage of the cells at time zero.
Statistical analysis
Data were analyzed by using SPSS Statistical Package version 16. Independent two group’s analyses are used t-test. P<0.05 was considered statistically significant.
Results
miR-9 and miR-181a were down-regulated in the synovial fluid cells
To explore the roles of microRNAs in the tibial plateau fracture microenvironment, we first detect the expression of seven candidate microRNAs, which were confirmed regulated bone formation and homeostasis, in stifle joints cells. As shown in Figure 1, miR-9 and miR-181a were significantly reduced in 3 days and 5 days after fracture occur, which means these two miRNAs may play important roles in the bone remodeling.
Figure 1.
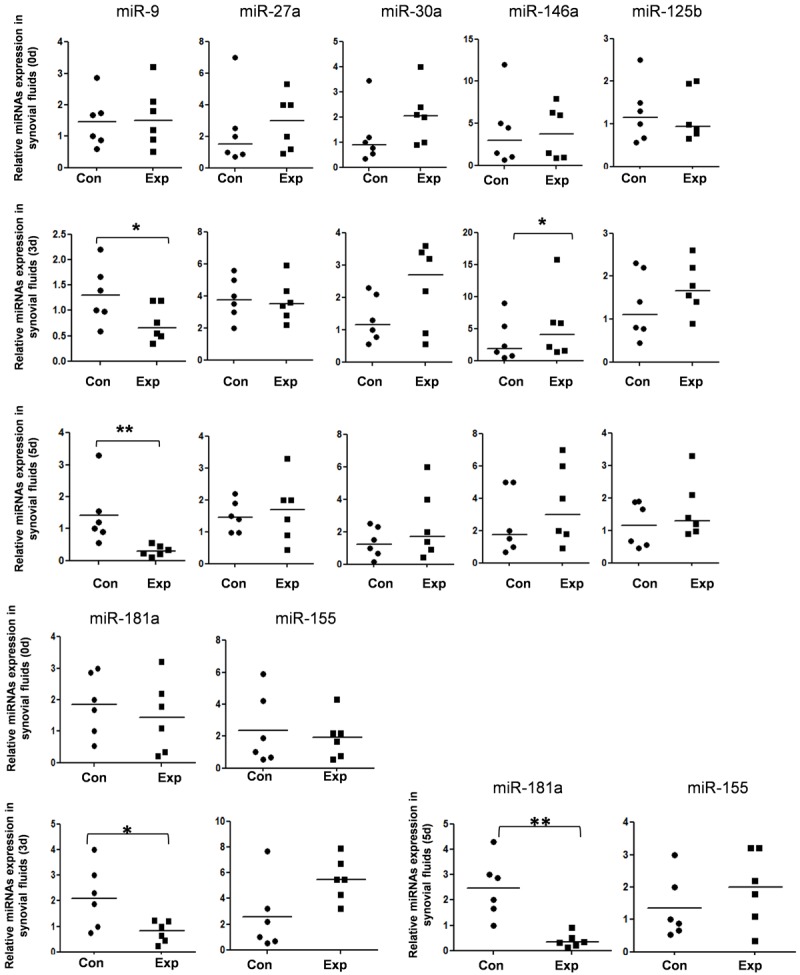
Disturbed microRNAs expression in the synovial fluid of tibial plateau fracture mouse. The expression level of seven candidate miRNAs in the synovial fluid cells of tibial plateau fracture mouse were detected by TaqMan miRNA RT-Real Time PCR. Statistical analyses were performed to analyze the overall trend of each miRNA in all groups. U6 serves as an internal reference among different samples and helps normalize for experimental error. *P<0.05, **P<0.01.
miR-9 and miR-181a repress Cbl expression by directly binding to specific sites of the 3’UTR
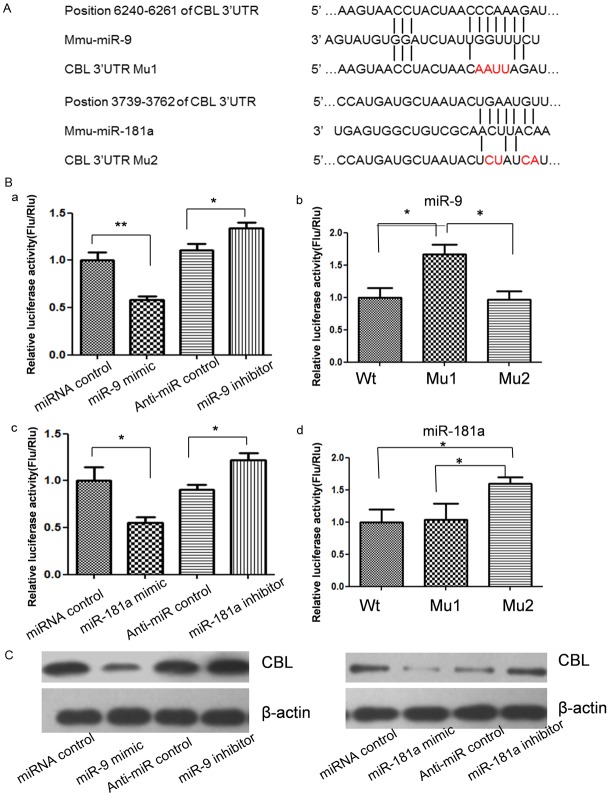
miRNA is a kind of important post transcription negative regulators for protein coding genes. So, to explore the relations between reduced miR-9 and miR-181a expression and their target genes overexpression, we first predict miR-9 and miR-181a targets by using online bioinformatics tools TargetScan. Surprisingly, miR-9 and miR-181a target Cbl, an important E3 ubiquitin ligase for bone formation and homeostasis regulation, directly according the results of online prediction. To validate whether Cbl is indeed the target gene of miR-9 and miR-181a, a 2508 bp segment of mouse Cbl 3’-UTR containing miRNAs binding sites was cloned into the downstream of the firefly luciferase reporter gene in the pGL3 control vector (designated as pGL3-Cbl) for the dual luciferase assay (Figure 2A). HEK293T cells were co-transfected with pGL3-Cbl and miR-9 and miR-181a mimics or inhibitor (Figure 2B). Compared with the miRNA control, the luciferase activity was significantly suppressed by the miR-9 and miR-181a, about 41.2% (P<0.01) and 43.5% (P<0.05). Furthermore, the luciferase activity was significantly up-regulated by the miR-9 and miR-181a inhibitor compared with the anti-miR control, about 19.7% (P<0.05) and 17.4% (P<0.05). These results indicate that miR-9 and miR-181a targets the 3’-UTR of Cbl, leading to the change of firefly luciferase translation.
Figure 2.
The expression of Cbl was suppressed by miR-9 and miR-181a. A: Schematic diagram for constructing the predict miR-9 and miR-181a binding sites into pmirGLO vector. B: Cbl is the target gene of miR-9 and miR-181a. RAW264.7 cells were co-transfected with miRNA control, miR-9/181a mimic, anti-miR control or miR-9/181a inhibitor and pmirGLO-Cbl for dual-luciferase assay. When 4 nucleotides of the binding sites of miR-9 or miR-181a in the 3’-UTR of Cbl were mutated (pmirGLO-Cbl-Mu1 and pmirGLO-Cbl-Mu2), the luciferase activities were significantly decreased in RAW264.7 cells co-transfected with miR-9 mimic and wild type Cbl 3’UTR vector and pmirGLO-Cbl-Mu2 compared with pmirGLO-Cbl-Mu1. The luciferase activities were significantly decreased in RAW264.7 cells co-transfected with miR-181a mimic and wild type Cbl 3’UTR vector and pmirGLO-Cbl-Mu1 compared with pmirGLO-Cbl-Mu2. C: Cbl protein level in miRNA mimics or inhibitors-treated RAW264.7 cells was detected by western blot.
Seed sequence mutation clone was also used to further confirm the binding site for miR-9 and miR-181a (Figure 2A). Putative miR-9 and miR-181a binding regions in the 3’-UTR of Cbl with 4 mutant nucleotides (designated as pGL3-Cbl-Mu1 and pGL3-Cbl-Mu2) were transfected into HEK293T cells with miR-9 or miR-181a mimics respectively, Cbl wild type vector pGL3-Cbl-Wt was used as control. The histogram in Figure 2B (right) showed that the enzyme activity was reduced about 60.3% in cells co-transfected with miR-9 mimics and pGL3-Cbl-Wt compared with pGL3-Cbl-Mu1 (P<0.05). The enzyme activity was reduced about 68.7% in the cells co-transfected with miR-181a mimics and pGL3-Cbl-Wt compared with pGL3-Cbl-Mu2 (P<0.05). These data indicate that miR-9 and miR-181a may suppress gene expression through binding to seed sequence at the 3’-UTR of Cbl.
MiR-9 and miR-181a regulates endogenous Cbl expression in RAW264.7 cells
Although Cbl was identified as a target gene for miR-9 and miR-181a, it was unknown whether miR-9 and miR-181a could regulate endogenous Cbl expression. RAW264.7 cells were transfected with miR-9 and miR-181a mimics or inhibitors to see whether the dysregulation of miR-9 and miR-181a expression affected endogenous Cbl expression. Compared with corresponding control, the levels of Cbl protein were significantly suppressed by miR-9 and miR-181a mimics and up-regulated by miR-9 and miR-181a inhibitors (Figure 2C).
miR-9 and miR-181a down-regulation enhanced Bim ubiquitination and degradation
The Cbl family proteins are evolutionarily conserved negative regulators of activated tyrosine kinase-coupled receptors, act as E3 ubiquitin ligases, and are involved in osteoclasts function. There are reports that Cbl can increase Bim ubiquitination dependent degradation and enhance osteoclasts survival and activation. To identify the effect of miR-9 and miR-181a expression on osteoclasts survival, we first examined the association of miRNA deregulation and Bim ubiquitination. 48 hours after transfection, we first detect the amount of miR-9 and miR-181a in RAW264.7.7 cells by qRT-PCR. As shown in Figure 3B, the expression of miR-9 and miR-181a were up-regulated to 58.3 and 38.7 fold by miRNA mimics and reduced to 33.8% and 40.3% percent by miRNA inhibitors. The result of immunoprecipitation showed that up-regulated miR-9 and miR-181a repressed the amount of ubiquitinated Bim. When the expression of miR-9 and miR-181a reduced, ubiquitinated Bim was increased associated with enhanced degradation of Bim in RAW264.7 cells (Figure 3A).
Figure 3.
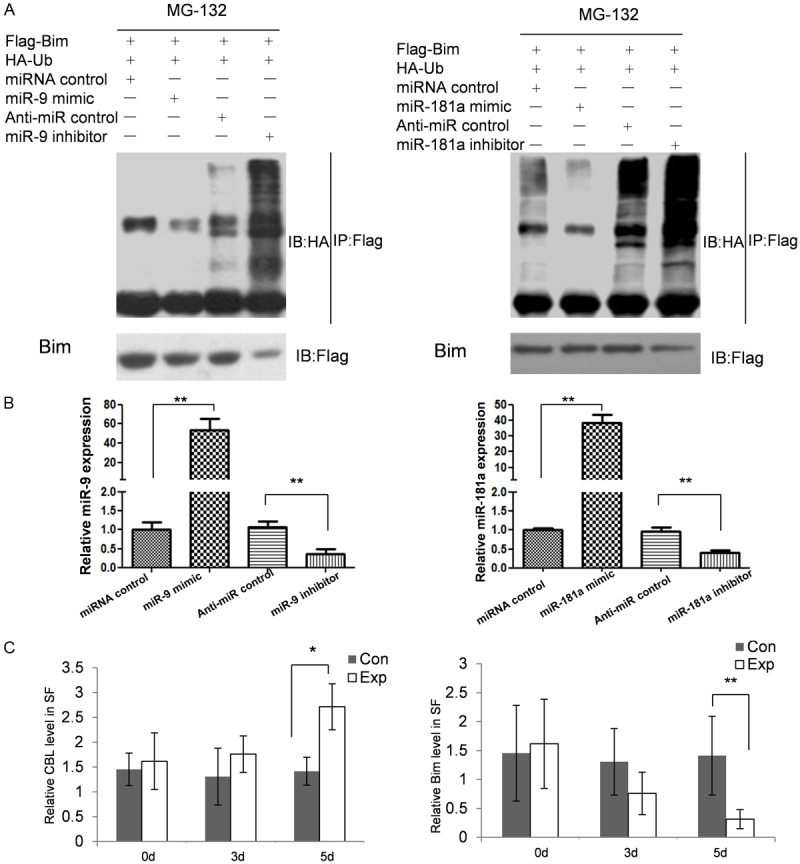
miR-9 and miR-181a regulate Bim ubiquitination-dependent degradation. A: Ubiquitination of Bim in RAW264.7 cells. Proteins immunoprecipitated with anti-Flag antibody were immunoblotted with an anti-HA antibody. Ubiquitinated Bim was detected as upper-shifted bands in anti-ubiquitin blotting. Ubiquitinated Bim was detected when cells were treated with MG132, and marked enhancement of its ubiquitination and reduction of total amount was induced by miR-9 and miR-181a transfection. B: qRT-PCR was used to detect the overexpression and downregulation of miR-9 and miR-181a. C: The amounts of soluble Cbl and Bim in the synovial fluid samples by using enzyme-linked immunosorbent assay.
Detected the Cbl and Bim concentration in the synovial fluid
To examine if the expression repression relationship was exist in vivo, we detected the amounts of soluble Cbl and Bim in the synovial fluid samples by using enzyme-linked immunosorbent assay. As shown in Figure 3C, in the tibial plateau fracture groups, the expressions of Cbl and Bim were no significant changed in 0 day and 3 day post fracture. When the time extended to 5 days, the time in which the amounts of miR-9 and miR-181a significantly reduced, the concentration of Cbl was raised accompanied with reduced Bim concentration.
miR-9 and miR-181a modulate migration capacity of RAW264.7 cells in vitro
In order to further research the roles of miR-9 and miR-181a in controlling the metastasis of osteoclast precursor cells, we analyzed the effects of miR-9 and miR-181a on the migratory behavior of RAW264.7 cells (Figure 4A). The results showed that the migration capacity of RAW264.7 cells transfected with miR-9 and miR-181a mimics were significantly lower than that transfected with miR control (P<0.05). The migration capacity were significantly suppressed in RAW264.7 cells transfected with miR-9 and miR-181a inhibitor compared with anti-miR control (P<0.05). These findings suggest that the level of miR-9 and miR-181a may be closely associated with the metastasis of osteoclast precursor cells.
Figure 4.
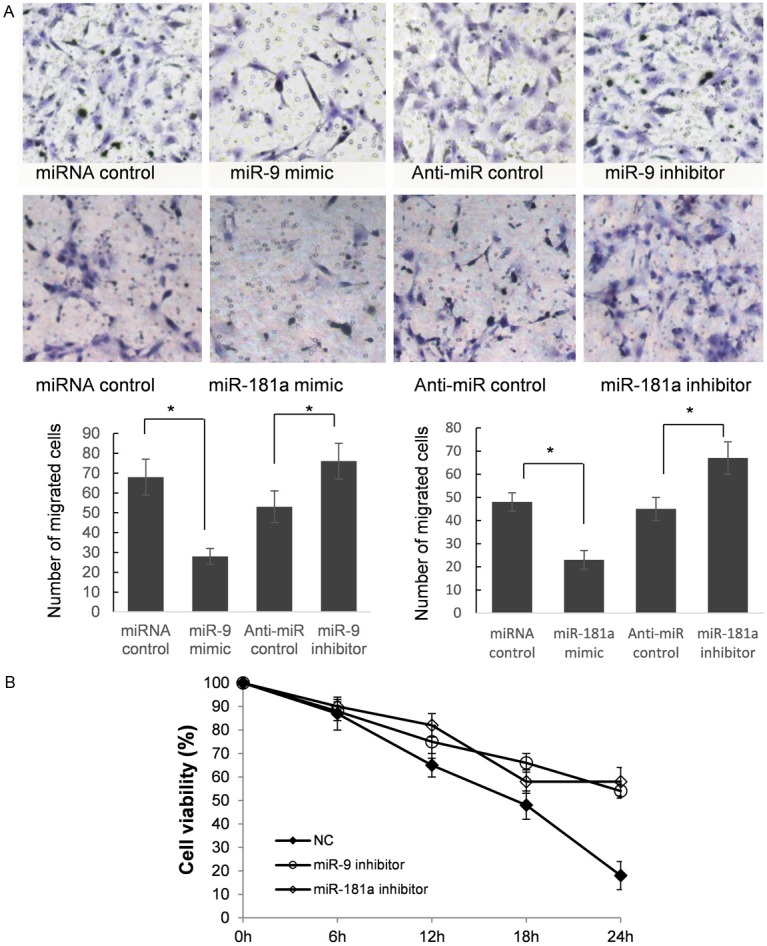
Reduction of miR-9 and miR-181a can enhance RAW264.7 cells migration and raise mouse primary OCs survival rate. A: RAW264.7 cells were transfected with pre-miR control, miR-9/181a mimics, anti-miR control or miR-9/181a inhibitor, respectively. Cells were harvested 48 h after transfection and recounted to 0.5×105 cells/ml in every group to seed Transwells for cell migration assay. At time of harvest, the cells on top of the membranes were removed, and the cells on the bottoms of the membranes were stained with hematoxylin and eosin. The cell migration was quantified by counting the amount of cells passing through the membrane from five different fields per sample at 200× selected in a random manner after 12 h of incubation. A show the representative photomicrographs of cells passing through the membrane at ×200 original magnification. Data are expressed as the mean numbers of independent triplicate experiments. *P<0.05. B: OCs were purified 6 h after the infection and some of the cultures were subjected to tartrate-resistant acid phosphatase (TRAP) staining. Cell viability/survival is expressed as morphologically intact TRAP-positive multinucleated cells. Other cultures were further incubated for the indicated times, and then the number of living OCs was counted. The number of viable cells remaining at the different time points is shown as a percentage of the cells at time zero.
miR-9 and miR-181a down-regulation increase the survival rate of OCs
Bim belongs to the BCL-2 protein family and was approved to be a pro-apoptotic gene in osteoclasts and their precursors [1]. To detect the impact of down regulated miR-9 and miR-181a on osteoclasts survival, we isolated mouse OCs. MiR-9 and miR-181a knock down were processed by using miRNA sponge strategy; M-SCF was added to increase OCs survival rate. As shown in Figure 4B, there are more than 50% OCs survived in the 24 h after infection, the survival rates of which were significantly higher than control group.
Discussion
High-energy tibial plateau fractures are often the result of blunt trauma and are associated with severe soft-tissue injury. The available surgical options do not always guarantee a favorable outcome. Operative management of high-energy fractures remains difficult and challenging because the injuries often associated with serious complications, such as knee stiffness, ankylosis, deep infection, post-traumatic arthritis and nonunion and so on. MiRNA is a kind of short RNA molecules that regulate normal human body physiology conditions, and reports indicated that miRNAs play an important role in the process of bone formation and homeostasis. To explore the function of miRNAs in the process of post tibial plateau fractures bone remodeling and sequelae formation, we first detect seven candidate miRNAs expression in the SF cells of the mouse model. The results indicated that miR-9 and miR-181a were down-regulated significantly 5 days after injury.
MiRNAs regulate normal cell physiology by repressing protein coding genes, so we first predict the target genes of miR-9 and miR-181a. Surprisingly, we found Cbl, an important E3 ubiquitin ligases for bone resorption, is a putative target gene of miR-9 and miR-181a. By using dual luciferase assay and western blot, we confirmed that the expression of Cbl is repressed by miR-9 and miR-181a. Akiyama T and colleagues reported that Cbl can regulate osteoclast apoptosis by promote pro-apoptotic gene Bim ubiquitin dependent degradation [1]. So we detect the effect of disturbed miR-9 and miR-181a expression on Bim ubiquitination. As expected, the amount of ubiquitinated Bim was raised the total Bim was reduced by miRNA inhibitors.
Finally, we detect the biological function of altered miR-9 and miR-181a expression on osteoclasts and RAW264.7 cells. Murine macrophage RAW264.7 cells can be differentiated to OCs by RANKL stimulation, so the effect on RAW264.7 cell can partially represent the effect on OC precursor cells. The enhanced RAW264.7 cell migration by miRNAs inhibitor suggest that miR-9 and miR-181a may inhibit OC precursor cells in vivo and may be associated with joint and bone destruction. What’s more miR-9 and miR-181a inhibitors can raise mouse primary OCs survival rate the result of which was coincidence with our prediction above.
As osteoclasts are key players in joint and bone destruction, it is of critical importance to understand their modes of migration, differentiation and function in vivo. In this study, we detect the expression of seven bone formation and remodeling related miRNAs in the tibial plateau fracture mouse model and found miR-9 and miR-181a were significantly reduced in the SF cells. Further functional study indicated that reduced miR-9 and miR-181a expression can active RAW264.7 cells migration ability and raise the primary mouse OCs survival rate in vitro by targeting Cbl, a E3 ubiquitin ligase critical for bone formation.
To our understood, this is the first study about the function of disturbed miRNAs in the tibial plateau fracture mouse model, and may expand our understanding about post tibial plateau fracture recover and post-traumatic sequelae generation.
Disclosure of conflict of interest
All the authors declare that this work has no competing interest.
References
- 1.Akiyama T, Bouillet P, Miyazaki T, Kadono Y, Chikuda H, Chung UI, Fukuda A, Hikita A, Seto H, Okada T, Inaba T, Sanjay A, Baron R, Kawaguchi H, Oda H, Nakamura K, Strasser A, Tanaka S. Regulation of osteoclast apoptosis by ubiquitination of proapoptotic BH3-only Bcl-2 family member Bim. EMBO J. 2003;22:6653–6664. doi: 10.1093/emboj/cdg635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chan YS. Arthroscopy-assisted surgery for tibial plateau fractures. Chang Gung Med J. 2011;34:239–247. [PubMed] [Google Scholar]
- 3.Farazi TA, Hoell JI, Morozov P, Tuschl T. MicroRNAs in human cancer. Adv Exp Med Biol. 2013;774:1–20. doi: 10.1007/978-94-007-5590-1_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furman BD, Strand J, Hembree WC, Ward BD, Guilak F, Olson SA. Joint degeneration following closed intraarticular fracture in the mouse knee: a model of posttraumatic arthritis. J Orthop Res. 2007;25:578–592. doi: 10.1002/jor.20331. [DOI] [PubMed] [Google Scholar]
- 5.Jimi E, Ikebe T, Takahashi N, Hirata M, Suda T, Koga T. Interleukin-1 alpha activates an NF-kappaB-like factor in osteoclast-like cells. J Biol Chem. 1996;271:4605–4608. doi: 10.1074/jbc.271.9.4605. [DOI] [PubMed] [Google Scholar]
- 6.Jimi E, Nakamura I, Ikebe T, Akiyama S, Takahashi N, Suda T. Activation of NF-kappaB is involved in the survival of osteoclasts promoted by interleukin-1. J Biol Chem. 1998;273:8799–8805. doi: 10.1074/jbc.273.15.8799. [DOI] [PubMed] [Google Scholar]
- 7.Jimi E, Shuto T, Koga T. Macrophage colony-stimulating factor and interleukin-1 alpha maintain the survival of osteoclast-like cells. Endocrinology. 1995;136:808–811. doi: 10.1210/endo.136.2.7835314. [DOI] [PubMed] [Google Scholar]
- 8.Kikuta J, Ishii M. Osteoclast migration, differentiation and function: novel therapeutic targets for rheumatic diseases. Rheumatology. 2013;52:226–234. doi: 10.1093/rheumatology/kes259. [DOI] [PubMed] [Google Scholar]
- 9.Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maes OC, Chertkow HM, Wang E, Schipper HM. MicroRNA: Implications for Alzheimer Disease and other Human CNS Disorders. Curr Genomics. 2009;10:154–168. doi: 10.2174/138920209788185252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szekanecz Z, Koch AE. Macrophages and their products in rheumatoid arthritis. Curr Opin Rheumatol. 2007;19:289–295. doi: 10.1097/BOR.0b013e32805e87ae. [DOI] [PubMed] [Google Scholar]
- 12.Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F, Xie X, Zhou C, Lu W. Suppressed miR-424 expression via upregulation of target gene Chk1 contributes to the progression of cervical cancer. Oncogene. 2013;32:976–987. doi: 10.1038/onc.2012.121. [DOI] [PubMed] [Google Scholar]