Abstract
The early identification of individuals at risk for type 2 diabetes (T2D) enables prevention. Recent genome-wide association studies (GWAS) have added at least 40 genetic variants to the list of already well characterized T2D risk predictors, including family history, obesity, and elevated fasting plasma glucose levels. Although these variants can significantly predict T2D alone and as a part of genotype risk scores, they do not yet offer clinical discrimination beyond that achieved with common clinical measurements. Future progress on at least two research fronts may improve the predictive performance of genotype information. First, expanded GWAS efforts in non-European populations will allow targeted sequencing of risk loci and the identification of true causal variants. Second, studies with longer prediction time horizons may demonstrate that genotype information performs better than clinical risk predictors over a longer period of the life course. At present, however, genetic testing cannot be recommended for clinical T2D risk prediction in adults.
Keywords: type 2 diabetes genetics, risk prediction models, genotype risk scores
Introduction
The Human Genome Project listed among its benefits the “earlier detection of genetic predispositions to disease”.1 The decade that followed the sequencing of the human genome has seen the generation of enough genetic data to occupy the scientific community for decades more. Before the current genomic era, the prevalent genetic paradigm concerned monogenic and highly penetrant diseases such as cystic fibrosis and Huntington disease. Nonetheless, a considerable body of evidence had also suggested a large heritable component to complex diseases such as coronary artery disease, obesity, and diabetes mellitus. Now, large genome-wide association studies (GWAS) have begun to uncover some of the genetic determinants of these complex diseases. Current genetic discoveries challenge us to find their appropriate place in disease risk prediction, particularly in the context of other already well characterized clinical disease predictors.
Type 2 diabetes mellitus (T2D) exemplifies the complex interactions between environmental and genetic determinants. The increasing prevalence2 of T2D and its associated comorbidities and healthcare expenditures make it a public health priority. Fortunately, the incidence of T2D can be delayed or prevented by maintaining healthy lifestyle behaviors.3–5 However, the increasingly obesogenic global environment6,7 is at odds with people’s attempts at lifestyle modification. The identification of population subgroups at particularly high risk for T2D might facilitate the targeting of prevention efforts to those who might benefit from them the most. Risk prediction has this identification and prevention as its goals.
It is reasonable to ask whether recent genetic associations improve the T2D risk prediction already achieved with traditional risk predictors alone. This review will summarize the data that address this question. We will review the known genetic and non-genetic risk predictors of T2D and compare the prediction models that incorporate them. We will then identify current limitations and future directions for the field of T2D genetics. In this review, we will use the term risk predictor to indicate a characteristic significantly associated with a future outcome (here, incident T2D). In contrast, a risk factor may be associated with a current (prevalent) and/or incident outcome.
Genetic associations with type 2 diabetes
The pace of accumulation of known T2D variants has mirrored the pace of advances in genetic technology. In 2003, linkage analysis (see Glossary) in samples of European ancestry found a T2D-associated variant on chromosome 10q,8 later identified in the TCF7L2 gene, which encodes a transcription factor in the Wnt signaling pathway.9 To date, it remains the variant with the largest effect size; each copy of the T allele (see Glossary) at rs7903416 carries a 1.4–1.5 increased odds of type 2 diabetes.10 Also by 2003, candidate gene studies (see Glossary) had associated T2D with variants in PPARG and KCNJ11, both genes encoding targets of diabetes medications.11,12 The arrival of GWAS, however, considerably lengthened the list of T2D-associated variants. The sequencing of the entire human genome had made possible the development of genotyping platforms consisting of only a few hundred-thousand single-nucleotide polymorphisms (SNPs) that could statistically capture the majority of genetic variation in the human genome. By genotyping large samples of cases and controls on these platforms, investigators could scan the entire genome in a non-biased way for novel genetic associations with T2D.13 The first T2D GWAS in 2007 confirmed the association with TCF7L2 and identified new T2D associations with variants at SLC30A8 and HHEX.14 The same year, other groups replicated these findings and discovered additional associations at CDKAL1, IGF2BP2, and CDKN2A/B.15–17 When research groups merged to form the Diabetes Genetics Replication and Meta-Analysis (DIAGRAM) consortium, the increased statistical power enabled the discovery of variants with smaller effects sizes.18 The most recent publication by the DIAGRAM group has brought the number of independent replicated genetic T2D associations to at least 38,19 with most variants increasing an individual’s relative odds of T2D by a modest 5–15% (Table 1).13
Table 1.
Thirty-eight genetic variants associated with type 2 diabetes (T2D) at genome-wide significance.
| Locus | Marker | Chr | Type of variant | Risk allele/ other allele |
Risk allele frequency |
OR for T2D (95% CI) |
|---|---|---|---|---|---|---|
| KCNQ1 | rs223789274 a | 11 | Intronic | C/T | 0.61b | 1.40 (1.34–1.47) |
| TCF7L2 | rs79031469 | 10 | Intronic | T/C | 0.25 | 1.37 (1.28–1.47)18 |
| DUSP9 | rs594532619 | X | 8 kb upstream | G/A | 0.12 | 1.27 (1.18–1.37) |
| CDKN2A/B | rs1081166115–17 | 9 | 125 kb upstream | T/C | 0.79 | 1.20 (1.14–1.25) |
| IRS1 | rs294364175 | 2 | 502 kb upstream | C/T | 0.61 | 1.19 (1.13–1.25) |
| IGF2BP2 | rs440296015–17 | 3 | Intronic | T/G | 0.29 | 1.17 (1.10–1.25)18 |
| FTO | rs805013615, 16 | 16 | Intronic | A/C | 0.45 | 1.15 (1.09–1.22)18 |
| KCNJ11/ABCC8 | rs5219c/rs75711012 c | 11 | Missense:Glu23Lys/Ala1369Ser | T/C | 0.50 | 1.15 (1.09–1.21)17 |
| G/T | 0.40 | |||||
| THADA | rs757859718 | 2 | Missense: Thr1187Ala | T/C | 0.92 | 1.15 (1.10–1.20) |
| CENTD2 | rs155222419 | 11 | 5′ UTR | A/C | 0.88 | 1.14 (1.11–1.17) |
| PPARG | rs180128211 | 3 | Missense: Pro12Ala | C/G | 0.92 | 1.14 (1.08–1.20)15–17 |
| HHEX | rs111187514 | 10 | 7.7 kb downstream | C/T | 0.56 | 1.13 (1.08–1.17)17 |
| NOTCH2 | rs1092393118 | 1 | Intronic | T/G | 0.11 | 1.13 (1.08–1.17) |
| WFS1 | rs180121419 | 4 | Intron-exon junction | G/A | 0.27 | 1.13 (1.07–1.18) |
| ADCY5 | rs1170806776 | 3 | Intronic | A/G | 0.78 | 1.12 (1.09–1.15) |
| CDKAL1 | rs775484015–17 | 6 | Intronic | C/G | 0.31 | 1.12 (1.08–1.16) |
| HNF1B | rs75721077, 78 | 17 | Intronic | A/G | 0.43 | 1.12 (1.07–1.18)19 |
| SLC30A8 | rs1326663414 | 8 | Missense: Arg325Trp | C/T | 0.75 | 1.12 (1.07–1.16)19 |
| CDC123/CAMK1D | rs1277979018 | 10 | Intergenic region | G/A | 0.23 | 1.11 (1.07–1.14) |
| RBMS1/ITGB6 | rs759373079 | 2 | Intronic | C/T | 0.57 | 1.11 (1.08–1.16) |
| TLE4 | rs1329213619 | 9 | 234 kb upstream | C/T | 0.93 | 1.11 (1.07–1.15) |
| HMGA2 | rs153134319 | 12 | 43 kb upstream | C/G | 0.10 | 1.10 (1.07–1.14) |
| JAZF1 | rs86474518 | 7 | Intronic | T/C | 0.52 | 1.10 (1.07–1.13) |
| ADAMTS9 | rs460710318 | 3 | 38 kb upstream | C/T | 0.81 | 1.09 (1.06–1.12) |
| MTNR1B | rs1083096376 | 11 | Intronic | G/C | 0.30 | 1.09 (1.06–1.12) |
| TSPAN8/LGR5 | rs796158118 | 12 | Intronic | C/T | 0.23 | 1.09 (1.06–1.12) |
| BCL11A | rs24302119 | 2 | 99 kb downstream | A/G | 0.46 | 1.08 (1.06–1.10) |
| KCNQ1 | rs23136219 a | 11 | Intronic | G/A | 0.52 | 1.08 (1.06–1.10) |
| ZBED3 | rs445705319 | 5 | 41 kb upstream | G/A | 0.26 | 1.08 (1.06–1.11) |
| GCK | rs460751776 | 7 | 36 kb upstream | A/G | 0.20 | 1.07 (1.05–1.10) |
| KLF14 | rs97228319 | 7 | 47 kb upstream | G/A | 0.55 | 1.07 (1.05–1.10) |
| OASL/HNF1A | rs795719719 | 12 | 20 kb downstream | T/A | 0.85 | 1.07 (1.05–1.10) |
| PRC1 | rs804268019 | 15 | Intronic | A/C | 0.22 | 1.07 (1.05–1.09) |
| PROX1 | rs34087476 | 1 | 2 kb upstream | C/T | 0.50 | 1.07 (1.05–1.09) |
| DGKB/TMEM195 | rs219134976 | 7 | Intergenic region | T/G | 0.47 | 1.06 (1.04–1.08) |
| GCKR | rs78009476 | 2 | Intronic | C/T | 0.62 | 1.06 (1.04–1.08) |
| TP53INP1 | rs89685419 | 8 | Intronic | T/C | 0.48 | 1.06 (1.04–1.09) |
| ZFAND6 | rs1163439719 | 15 | 1.5 kb downstream | G/A | 0.56 | 1.06 (1.04–1.08) |
SNPs are ordered by odds ratio (OR) for T2D. Reference for each SNP is from the original report, and OR for T2D is from that reference cited unless otherwise noted. Risk allele frequencies are from HapMap CEU unless otherwise noted. Chr, chromosome. CI, confidence interval. kb, kilobase. Adapted from Billings LK, Florez JC. Ann NY Acad Sci. 2010 Nov; 1212:59–77.
These SNPs are in low linkage disequilibrium (LD) in Europeans (r2 = 0.01) and likely represent independent association signals.
Risk allele frequency from HapMap-JPT.
These two SNPs are in strong LD in adjacent genes in Europeans (r2 = 0.87).
Traditional type 2 diabetes risk predictors
As an “exposure” in the epidemiologic sense, a T2D-associated genetic variant is fixed from conception, and this permanence should give it some predictive advantage over other risk predictors that vary over the life course, such as body-mass index (BMI) and even family history. Nonetheless, routine clinical T2D risk prediction has incorporated inexpensive and easily measured predictors, which, as a result, are the gold standard against which any new predictor has to be judged.
T2D risk increases with age throughout adulthood,20 and members of non-white race/ethnic groups have increased risk compared with whites.21,22 Lifestyle factors associated with an increased T2D risk include cigarette smoking,23 physical inactivity24 and diets with high glycemic load25 or trans-fat content.26 Elements of the medical history also predict T2D: a family history of diabetes20,27 or a personal history of gestational diabetes28 markedly increase an individual’s personal T2D risk. Obesity strongly increases an individual’s risk for T2D: a BMI ≥ 30 kg/m2 is associated with an odds ratio of 6.4 for T2D over 7 years in the Framingham Offspring Study, after adjustment for age and family history, compared with a BMI <25 kg/m2.20 This direct relationship between T2D risk and adiposity persists even in the normal range of BMI values. In the Nurses Health Study, the age-adjusted relative risk of T2D over 14 years was 5.0 in BMI range 24.0–24.9 ≥ 30 kg/m2 compared with BMI values <22 kg/m2.29 Elevated values of blood pressure, another common clinical measurement, are also associated with risk of future T2D.20,30
Not surprisingly, the strongest laboratory predictors of T2D measure glycemia. Higher fasting plasma glucose levels, even in the normal range, predict future T2D,31 while individuals with impaired fasting glucose (IFG, 100–125 mg/dL or 5.6–7.0 mmol/L) have an annualized relative risk of T2D of 4.7 compared to normoglycemic individuals. Impaired glucose tolerance (IGT) after an oral glucose tolerance test performs similarly as a predictor, although the combination of IFG and IGT carries much greater risk than either alone.32 In one Dutch study, 65% of people with IFG and IGT at baseline developed T2D over a mean follow-up of 6.4 years.33 Elevated values of hemoglobin A1c also predict future T2D, and a recent meta-analysis demonstrated that values of 6.0–6.5% were associated with an annual T2D incidence of 25–50% over 5 years.34 Measures of dyslipidemia, particularly elevated triglyceride and low high-density lipoprotein (HDL) levels, also predict future T2D independently of glycemic measures.20,31
Clinical type 2 diabetes prediction models
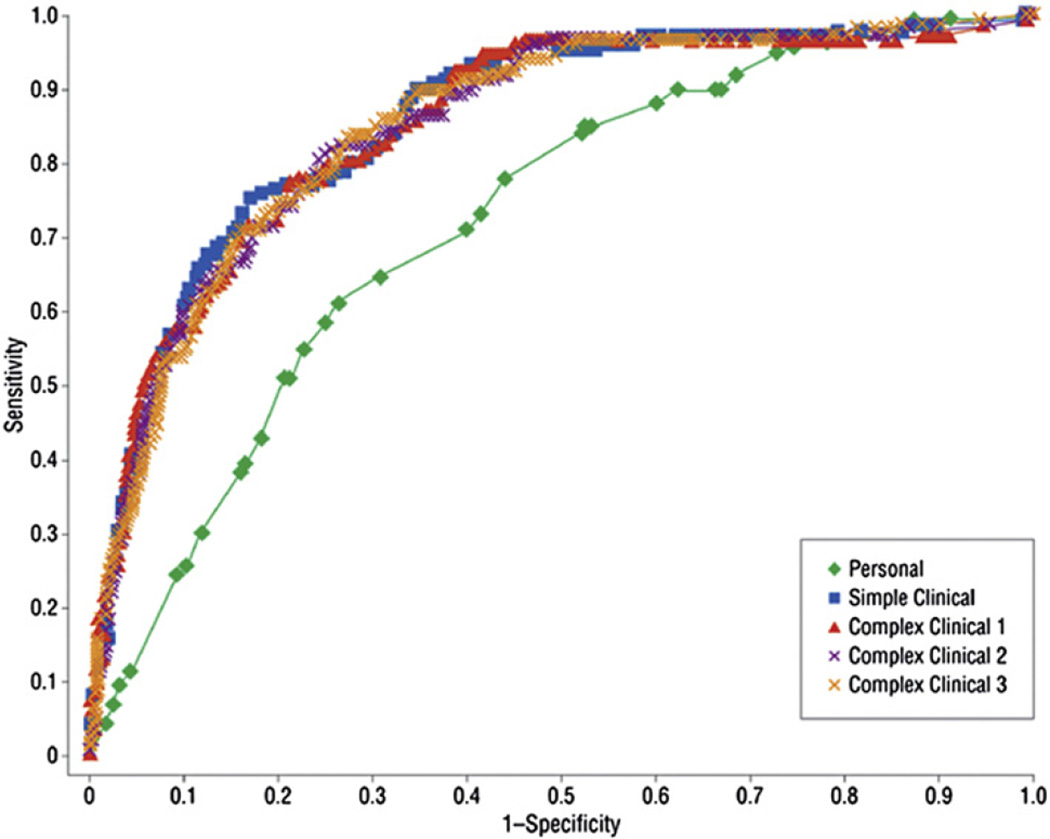
While some of these T2D risk predictors, particularly lifestyle and dietary factors, are important for public health policy, they are less easily measured in busy clinical settings. Groups from several large prospective cohort studies, such as the Framingham Offspring Study and the Atherosclerosis Risk in Communities Study (ARIC), have developed prediction models for T2D in middle-aged adults using common clinical measurements.20,35–37 These models have more similarities than differences and usually include measures of glycemia, adiposity, dyslipidemia, and blood pressure, in addition to age, family history of diabetes and, in some cases, race. The ability of these models to discriminate future cases from non-cases is usually very good, with C statistics (also called areas-under-the-receiver operating characteristic curve, see Glossary) ranging from 0.7 to 0.9 (Fig. 1). When these models are applied to racial groups other than those in the original validation studies, recalibration can preserve their discriminatory ability.38 Other clinical models have additionally included lifestyle predictors such as fruit and vegetable consumption and physical activity, with modest improvement in prediction.39,40
Fig. 1.
ROC curves for five type 2 diabetes prediction models from Framingham Offspring Study. The four clinical models have higher C statistics than the personal model. Reprinted with permission from Wilson, et al. Arch Intern Med. 2007;167(10):1068–1074. Copyright © 2007 American Medical Association. All rights reserved.
Genetic prediction of type 2 diabetes
Individual genetic variants, too, can predict incident T2D. In the Swedish Malmö Preventive Project, 11 SNPs individually predicted incident T2D in more than 15,000 participants over a median 24.8 years of follow-up, after adjustment for sex and baseline age. The odds ratio per risk allele ranged from 1.07 for the variant near HHEX to 1.30 for that in TCF7L2, compared with 1.67 for having a first-degree family member with diabetes and 1.84 for each standard deviation increase in BMI.41
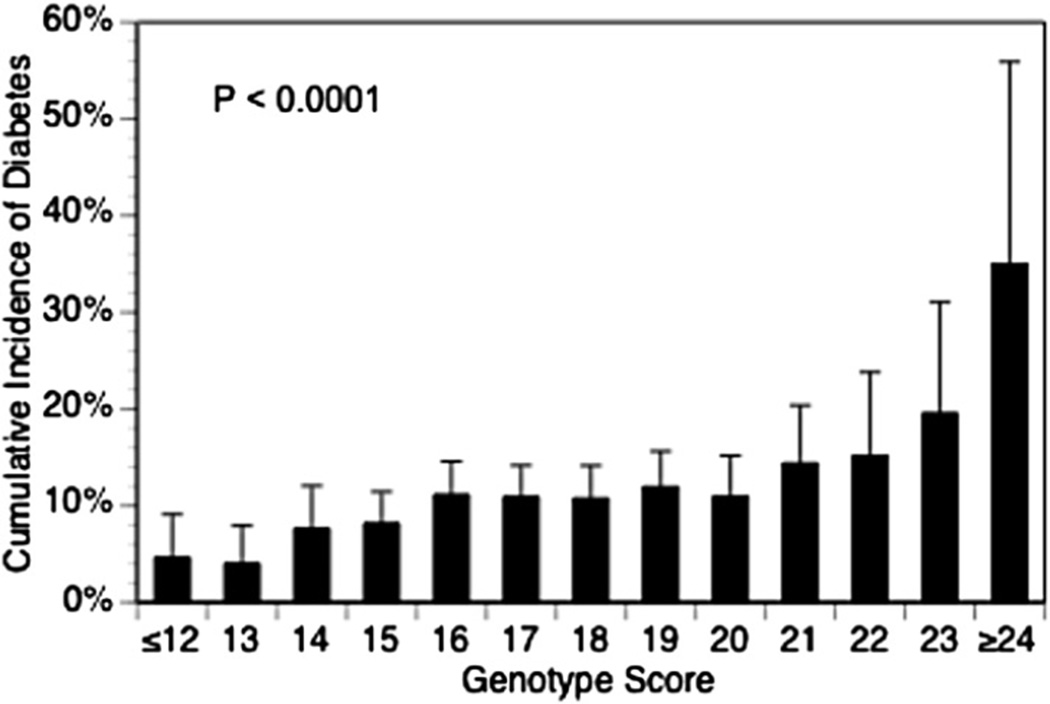
These relatively modest effect sizes of individual SNPs have prompted efforts to combine SNPs into composite genotype risk scores. To construct these scores, an individual’s risk alleles at each T2D-associated locus (ranging from 0 to 2 copies at each locus) are summed. Commonly, each locus is weighted according to its T2D effect size from the original GWAS. For instance, with this approach, each TCF7L2 risk allele carries a greater weight than each HHEX risk allele. A simulation study by Janssens et al., however, suggests that such weighting might not substantially improve the C statistic for alleles with small effects sizes (odds ratio 1.10–1.25).42 To date, several T2D genotype risk scores have been published in prospective studies of middle-aged adults (reviewed in43 and44), and the results have been similar. For population-level T2D prediction, prospective cohort studies (Table 2) are preferable to crosssectional case–control designs, since the latter do not reflect the true disease incidence in the population. Genetic risk scores alone, consisting of between 2 and 40 SNPs, have C statistics for T2D prediction ranging from 0.54 to 0.68. In the Framingham Offspring Study, cumulative T2D incidence over 28 years of follow-up increased significantly with genotype risk score (Fig. 2), and each 1-point increase in the score increased the odds of T2D over 8 years by 12%. The group with the highest genotype scores had an odds ratio for T2D of 2.6 compared to those with the lowest scores.45
Table 2.
Eight prospective studies comparing clinical and genetic type 2 diabetes prediction models.
| Study | n | Clinical model and AUC | Genetic model (no. of SNPs) and AUC |
Clinical + genetic model AUC |
p | ||
|---|---|---|---|---|---|---|---|
| Rotterdam46 | 5297 | Age, sex, BMI | 0.66 | 18 | 0.60 | 0.68 | <0.0001 |
| Malmö41 | 16,061 | Age, sex, BMI, FG, BP, TG, FH | 0.74 | 11 | 0.63 | 0.75 | 0.0001 |
| Botnia41 | 2770 | Age, sex, BMI, FG, BP, TG, FH | 0.79 | 11 | 0.68 | 0.80 | 0.23 |
| Framingham Offspring45 | 2377 | Age, sex, BMI, FG, BP, TG, FH | 0.900 | 18a | 0.581 | 0.901 | 0.49 |
| Framingham Offspring47 | 3471 | Age, sex, BMI, FG, BP, TG, FH | 0.903 | 40a | 0.606 | 0.906 | 0.04 |
| Whitehall II80 | 5135 | Age, sex, BMI, FG, BP, TG, FH | 0.78 | 20 | 0.54 | 0.78 | 0.10 |
| DESIR81 | 3442 | Age, sex, average BMI over 9 years | 0.82 | 3 | 0.56 | 0.83 | NR |
| DESIR37 | 3817 | WC, HTN, FG, GGT, FHD, smoking (men), BMI (women) | 0.850 (men) | 2 | NR | 0.851 (men) | NR |
| 0.917 (women) | 0.912 (women) | ||||||
P value compares areas under the receiver operating characteristic curves (AUCs) from clinical + genetic model to clinical model alone. BMI, body-mass index. BP, blood pressure. FG, fasting plasma glucose. FH, family history of diabetes. GGT, gamma-glutamyl transferase. HTN, hypertension. NR, not reported. TG, triglycerides. WC, waist circumference.
Genetic model also adjusted for sex.
Fig. 2.
The 28-year cumulative incidence of type 2 diabetes by genotype risk score in 2377 participants in the Framingham Offspring Study.45 Reprinted with permission from the Massachusetts Medical Society, Copyright© 2008.
However, the clinical T2D prediction models that consist of basic demographic, clinical, and laboratory predictors have C statistics ranging from 0.66 in the Rotterdam Study46 to 0.90 in the Framingham Offspring Study (Table 2),45,47 values superior to what genotype scores alone have yet achieved. Moreover, the addition of genotype risk scores to clinical prediction models only modestly improves the C statistic. For example, the C statistic improves from 0.903 to 0.906 with the addition of a 40-SNP score to the clinical model in the Framingham Offspring Study,47 and from 0.74 to 0.75 in the larger Malmö Preventive Project.41
Although the C statistic has relevance for population-level prediction, it is not the best measure of clinical utility. A higher C statistic corresponds to a higher likelihood that a test can distinguish an individual who will develop incident T2D from one who will not. But for T2D prevention, the clinician is asked to risk-stratify an individual patient, not to choose which one of a pair of patients is likelier to develop T2D. For this reason, Pencina, et al., introduced the net reclassification improvement (NRI) index,48 which compares the abilities of two prediction models to put individuals correctly into predicted risk categories (see Glossary). Using T2D risk categories of 0 to <2%, 2 to 9003C;8%, and ≥8% over 8 years in the Framingham Offspring Study, the addition of an 18-SNP genotype risk score to a model consisting only of sex and family history correctly reclassified only 2.6% of individuals (p = 0.22).45 Genetic risk prediction may work better in younger individuals. In Framingham, the addition of a 40-SNP score to a full clinical model, achieved an NRI of 10.2% (p = 0.001) among those younger than 50 years at baseline.47 It is important to note, however, that these categories of risk (0 to <2%, 2 to <8%, and ≥8% over 8 years) do not inform widely-accepted clinical prevention targets. This is in contrast to the Framingham Risk Score for 10-year risk of coronary disease, whose categories help tailor individual cholesterol level targets.49 Genetic testing will likely not have a role in clinical T2D prediction unless its addition to prediction models correctly reclassifies individuals as having lower or higher risk than previously thought based on patient phenotype, and unless the prevention strategies targeted for the individual change as a result of the marginal information value afforded by the genetic test. Recently, a large multi-disciplinary European consortium recommended against the use of genetic information in targeting T2D preventive efforts.50
Family history and heritability of type 2 diabetes
If genetic information does not improve T2D prediction compared with clinical prediction models, a next reasonable question is how genetic information compares with family history of T2D alone, itself a strong risk predictor. In the pre-genomic era, twin and family studies played an important role in separating the putative environmental and genetic components of T2D. Twin studies have estimated the genetic proportion of variance in T2D (or heritability, see Glossary) to be between 25% and 40%,51,52 which suggests that both genetic and non-genetic factors contribute substantially to an individual’s T2D risk. Having one parent with T2D doubles an individual’s risk, and having two affected parents can increase an individual’s risk up to six-fold.20,27 Family history does correlate with T2D genetic risk: data from the PPP-Botnia and Framingham Offspring Studies show that T2D genetic risk scores increase slightly with the degree of T2D family history (i.e. 0, 1, or 2 affected parents).53 However, the known T2D genetic variants do not account for the strong relationship between family history and T2D risk. In the Malmö Preventive Project, a self-reported first-degree family history of diabetes carried an odds ratio for incident T2D of 1.62 after adjustment for clinical predictors. When added to this multivariate model, the genetic risk score was an independent predictor of T2D (odds ratio 1.12 per 1-point increase in score), but the effect of family history was unchanged (odds ratio 1.65).41 In the Framingham Offspring Study, the addition of a genetic risk score to the clinical T2D prediction model modestly decreased the adjusted odds ratio for first-degree family history from 1.72 to 1.64.45 Indeed, the estimate of the degree of T2D heritability explained by known genetics variants is only 10%.54 Measurement error in both the numerator and denominator of this heritability proportion may account for the discrepancy between family history and genetic variants as T2D predictors. First, a greater percentage of heritability (numerator) may be explained with the future discovery of rare and private variants and the identification of the true causal variants for which the discovered T2D-associated variants are imperfect proxies (see below). Second, the total T2D heritability (denominator) may be overestimated, since its calculation ignores the potential contributions of intrauterine effects, gene–gene and gene-environment interactions, and the insufficient accounting for shared environmental, behavior and normative factors in family studies.55,56 However, the evidence above demonstrates that, compared with currently identified genetic variants, family history remains a more powerful T2D predictor that likely captures the genetic and environmental determinants of T2D risk.
Limitations of the current evidence base
The evidence to date does not justify the routine use of genetic testing for T2D prediction in clinical care. At present, certain limitations of the evidence base contribute to the modest performance of genotype information as a risk predictor. GWAS, including those that discovered the known T2D variants, rely on genotyping array platforms that may miss up to 20% of common variation in the human genome.57 These platforms fail to capture an even greater percentage of common variation in non-European ancestral populations, due to lower linkage disequilibrium (LD, see Glossary) rates in those groups.58 It is also worth noting that the majority of known T2D-associated variants (Table 1) are in non-coding intronic or even intergenic regions of the genome.13 This observation suggests that these variants may only be proxies in LD with the true causal variants. Fine-mapping and resequencing these regions should identify those causal variants, which should have stronger associations with T2D than their proxies and thus improve genetic prediction. This anticipated progress in defining the genetic architecture of T2D highlights the preliminary nature of the current state of genetic testing for T2D prediction. In an analysis of 5297 participants of the Rotterdam study, updating a T2D genetic risk test with the inclusion of more recently discovered T2D-associated variants changed the risk category (above or below average risk) for 34% of people.59 Clinicians may engender mistrust from their patients if the genetic risk predictions they deliver are overturned by the next wave of genetic discovery.
Future directions
Although GWAS have likely identified most common (risk allele frequency ≥5%) T2D-associated loci in populations of European ancestry, T2D genetics is still an active area of research. The nascent search for interactions between genes and between genes and environmental factors (including diet60 and physical activity61) has necessitated new analytic methods to preserve statistical power when looking for potentially small interaction effect sizes among trillions of combinations.62 It is not known how these future discoveries might be incorporated into risk prediction models.
Extending T2D genetic knowledge to non-European ancestral populations
The largest gap in the current evidence base for T2D genetic prediction is the racial homogeneity of studies to date. The largest T2D GWAS meta-analyses have taken advantage of the lesser degree of LD in European populations to identify genetic associations with T2D. Thus it is appropriate that T2D genetic prediction studies (Table 2) first applied these genetic discoveries to T2D prediction in populations of European ancestry similar to the ones in which they were discovered. Genetic prediction modeling in other racial groups, however, lags behind. It is well known that the risk allele frequencies for many cardiometabolic traits vary by race and ethnicity.63–65 However, genetic association studies are generally finding that T2D-associated variants from European GWAS have similar effects in non-European populations.66 Still, the European T2D-associated SNPs are likely much poorer proxies for the true causal variants in older ancestral populations such as Africans, which have more fragmented genomic LD. The Candidate gene Association Research Project (CARe), the largest T2D GWAS reported to date in African ancestral populations consisted of 8090 people,67 compared to more than 140,000 in the most recent DIAGRAM meta-analysis.19 Of all the known T2D association from European populations, this consortium replicated only that at TCF7L2. Larger GWAS in more diverse populations will bring at least two advantages. First, it will extend the knowledge of T2D genetic architecture to those racial and ethnic groups disproportionately affected by the disease.22 Second, any T2D-associated variant replicated or discovered in older populations such as Africans will likely be in an LD region smaller than the corresponding European region, narrowing the genome necessary to map and resequence for identification of the causal variant(s).68 The inclusion of these causal variants, instead of their originally identified proxy SNPs, have the potential improve T2D genetic prediction models for individual patients.
Specifying the time horizon of T2D genetic risk prediction
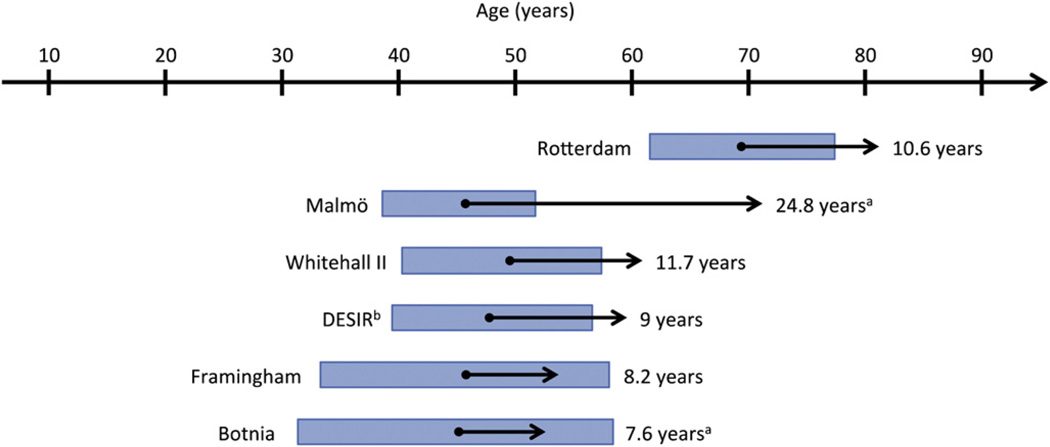
To address the limitations of the evidence base in genetic prediction, the U.S. Centers for Disease Control and Prevention sponsored the Genetic Risk Prediction Studies (GRIPS) Group. In 2011, GRIPS made several recommendations for standardizing the presentation of results from genetic prediction studies, including specifying the time horizon over which each risk prediction model applies.69 Applying this recommendation to T2D genetic risk prediction highlights another research gap in the field: the limited range of ages and durations of follow-up over which T2D genetic prediction has been modeled. Fig. 3 shows that the prospective studies of T2D genetic prediction include participants with baseline ages generally in middle adulthood and, with the exception of the Malmö study, have follow-up durations of around ten years. This time horizon is familiar to clinicians who use the Framingham Risk Score for 10-year coronary risk stratification.49 However, because obesity is increasingly affecting younger age groups70 and T2D can be prevented with lifestyle modification,3–5 a prediction model validated in younger age groups with a longer prediction time horizon may help target earlier prevention. A longer time horizon is also likely to improve the predictive value of genetic testing relative to other T2D risk predictors that vary with time, such as BMI and fasting glucose. Indeed, the Framingham Offspring Study showed that a 40-SNP genotype risk score significantly improved the T2D net reclassification improvement in participants younger than, but not older than, 50 years at baseline.47 T2D genetic risk testing may prove to have clinical utility only in populations younger than those in whom prediction models have been published to date.
Fig. 3.
Baseline ages and time horizons of six prospective studies reporting genetic prediction of type 2 diabetes.41,46,47,80,81 Arrow origins and bars correspond to mean age +/−1 standard deviation and arrow length corresponds to mean follow-up time, unless otherwise noted. aMedian baseline age and interquartile range are shown. bMedian follow-up time is shown.
Conclusions
Genetic testing has many potential future roles in the clinical management and prevention of T2D. It may encourage patients to engage with their health and risk factors and motivate them for lifestyle modification.71,72 Clinicians may be able to tailor T2D therapy to individual genotypes.73 Genotype information does significantly predict T2D, but it is outperformed in adults, at least, by measurements already routinely incorporated into clinical medicine. At present, this lack of utility does not support the clinical use of genetic testing for T2D prediction in adults.
Practice points.
About 40 genetic variants can significantly predict T2D, although, individually, most variants increase the odds of T2D only modestly (5–15%). “High” versus “low” genetic risk scores comprised of sums of variants are associated with a ~2-fold increased relative risk of incident T2D.
Prediction models consisting of routine clinical risk factors such as family history, BMI, and elevated fasting plasma glucose outperform genetic prediction models in middle-aged adults of European ancestry.
The addition of genotype information to clinical T2D prediction models does not meaningfully improve prediction for middle-aged adults of European ancestry.
Research agenda.
Expanded GWAS efforts in non-European populations will allow targeted sequencing of risk loci and the identification of true causal variants that are likely to improve predictive performance for individuals.
Studies with longer prediction time horizons may demonstrate that genotype information performs better than clinical risk predictors over a longer period of the life course.
Acknowledgments
Role of funding source
JLV is supported by NIH National Research Service Award grant T32 HP12706 from the Health Resources and Services Administration and the NIH Loan Repayment Program (NIDDK). JBM is supported by NIDDK K24 DK080140 and R01 DK078616. The sponsors had no role in the collection, analysis, or interpretation of data or in the writing of this manuscript.
Glossary
- Allele
A possible form that a particular region of DNA may take in an individual’s genome. For example, an individual may have a T allele at rs7903146 in his maternal copy of TCF7L2 and a C allele in his paternal copy. In addition to single-nucleotide polymorphisms (SNPs) such as this example, allele may also refer to other forms of genetic variation, including deletions and insertions.
- C statistic
Also known as the area under the receiver operating characteristic (ROC) curve (Fig. 1). For a given model that aims to predict an outcome (for example, incident type 2 diabetes), the ROC curve plots the sensitivity against 1 minus the specificity at all possible dichotomous cutpoints of the model. The area under that curve corresponds to the model’s discrimination, or the probability that it will distinguish a case from a non-case. The C statistic ranges from 0.5 (no discrimination, similar to a coin toss) to 1 (perfect discrimination).
- Candidate gene study
A hypothesis-driven method to identify genotype-phenotype associations by examining genetic variations only within genes which, because of their known function or protein product, are thought likely to be implicated in the phenotype of interest.
- Heritability
The proportion of the population variance in a phenotype (e.g. type 2 diabetes) due to genetic effects, often calculated from twin, family, and adoption studies. Underlying these studies is the assumption that siblings reared together share the same environment, at least in childhood, and thus any phenotypic variance between them is due to unshared genetic factors.
- Genome-wide association study
A hypothesis-free method of identifying genotype-phenotype associations. In a large population of cases and controls, the phenotype of interest is tested for statistical association with each of several hundred-thousands of single nucleotide polymorphisms (SNPs), which because of linkage disequilibrium (LD), capture a majority of human genetic variation. To minimize type I error, a genome-wide association is considered statistically significant at p<5×10−8.
- Linkage analysis study
A method to identify genotype-phenotype associations by looking at the inheritance patterns of certain genetic markers in families with high prevalence of the phenotype of interest.
- Linkage disequilibrium (LD)
The state when two alleles are inherited together more frequently than by chance because of their close proximity on a chromosome. LD is often measured by r2, the square of the correlation coefficient for the two allele frequencies, which ranges from 0 (linkage equilibrium) to 1 (perfect linkage disequilibrium). If LD is present between two loci, an allele at one genotyped locus can be a reasonable proxy for an allele at another, non-genotyped locus. Because recombination events degrade LD over generations, older ancestral groups such as Africans have lower LD values for neighboring alleles in a given chromosomal region than European ancestral populations.
- Net reclassification improvement (NRI) index
A measure of whether a new prediction model meaningfully improves upon a previous model by correctly reclassifying individuals into risk categories (e.g. low-, medium-,and high-risk). The NRI is higher as it correctly upgrades the risk category of future cases (for example, incident type 2 diabetes) and correctly downgrades the risk category for non-cases. The NRI is lower as it incorrectly downgrades the risk category of future cases and incorrectly upgrades the risk category of non-cases.
- Phenotype
An observed trait resulting from gene expression. Phenotypes can be discrete diseases like type 2 diabetes and coronary artery disease or quantitative traits such as fasting glucose or low-density lipoprotein (LDL) levels.
Footnotes
Conflict of interest
JLV has no conflict of interest to disclose. JBM has a consulting agreement with Interleukin Genetics, Inc.
References
- 1.U.S. Department of Energy Genome Programs. [cited 2011 June 20, 2011];Potential Benefits of Human Genome Project Research. Available from 2009, http://www.ornl.gov/sci/techresources/Human_Genome/project/benefits.shtml.
- 2.Centers for Disease Control and Prevention. [updated March 30, 2011; cited 2011 July 17, 2011];Crude and Age-Adjusted Percentage of Civilian, Noninstitutionalized Population with Diagnosed Diabetes, United States, 1980–2009. Atlanta. Available from 2011, http://www.cdc.gov/diabetes/statistics/prev/national/figage.htm.
- 3.Knowler WC, Barrett-Connor E, Fowler SE, et al. Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. The New England Journal of Medicine. 2002 Feb 7;346(6):393–403. doi: 10.1056/NEJMoa012512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuomilehto J, Lindstrom J, Eriksson JG, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. The New England Journal of Medicine. 2001 May 3;344(18):1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 5.Pan XR, Li GW, Hu YH, et al. Effects of diet and exercise in preventing NIDDM in people with impaired glucose tolerance. The Da Qing IGT and Diabetes Study. Diabetes Care. 1997 Apr;20(4):537–544. doi: 10.2337/diacare.20.4.537. [DOI] [PubMed] [Google Scholar]
- 6.Hill JO, Wyatt HR, Reed GW, et al. Obesity and the environment: where do we go from here? Science. 2003 Feb 7;299(5608):853–855. doi: 10.1126/science.1079857. [DOI] [PubMed] [Google Scholar]
- 7.Elinder LS, Jansson M. Obesogenic environments–aspects on measurement and indicators. Public Health Nutrition. 2009 Mar;12(3):307–315. doi: 10.1017/S1368980008002450. [DOI] [PubMed] [Google Scholar]
- 8.Reynisdottir I, Thorleifsson G, Benediktsson R, et al. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. American Journal of Human Genetics. 2003 Aug;73(2):323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grant SFA, Thorleifsson G, Reynisdottir I, et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nature Genetics. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 10.Helgason A, Palsson S, Thorleifsson G, et al. Refining the impact of TCF7L2 gene variants on type 2 diabetes and adaptive evolution. Nature Genetics. 2007;39(2):218–225. doi: 10.1038/ng1960. [DOI] [PubMed] [Google Scholar]
- 11.Altshuler D, Hirschhorn JN, Klannemark M, et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nature Genetics. 2000 Sep;26(1):76–80. doi: 10.1038/79216. [DOI] [PubMed] [Google Scholar]
- 12.Gloyn AL, Weedon MN, Owen KR, et al. Large-scale association studies of variants in genes encoding the pancreatic beta-cell KATP channel subunits Kir6.2 (KCNJ11) and SUR1 (ABCC8) confirm that the KCNJ11 E23K variant is associated with type 2 diabetes. Diabetes. 2003 Feb;52(2):568–572. doi: 10.2337/diabetes.52.2.568. [DOI] [PubMed] [Google Scholar]
- * 13.Billings LK, Florez JC. The genetics of type 2 diabetes: what have we learned from GWAS? Annals of the New York Academy of Sciences. 2010 Nov;1212:59–77. doi: 10.1111/j.1749-6632.2010.05838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sladek R, Rocheleau G, Rung J, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007 Feb 22;445(7130):881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 15.Zeggini E, Weedon MN, Lindgren CM, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007 Jun 1;316(5829):1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007 Jun 1;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research. Saxena R, Voight BF, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007 Jun 1;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 18.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nature Genetics. 2008 May;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 19.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nature Genetics. 2010 Jul;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 20.Wilson PWF, Meigs JB, Sullivan L, et al. Prediction of incident diabetes mellitus in middle-aged adults: the Framingham Offspring study. Archives of Internal Medicine. 2007 May 28;167(10):1068–1074. doi: 10.1001/archinte.167.10.1068. [DOI] [PubMed] [Google Scholar]
- 21.Shai I, Jiang R, Manson JE, et al. Ethnicity, obesity, and risk of type 2 diabetes in women: a 20-year follow-up study. Diabetes Care. 2006 Jul;29(7):1585–1590. doi: 10.2337/dc06-0057. [DOI] [PubMed] [Google Scholar]
- 22.U.S. Department of Health and Human Services. Healthy people 2010: objectives for improving health. Washington, D.C.: 2000. [Google Scholar]
- 23.Willi C, Bodenmann P, Ghali WA, et al. Active smoking and the risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2007 Dec 12;298(22):2654–2664. doi: 10.1001/jama.298.22.2654. [DOI] [PubMed] [Google Scholar]
- 24.Rana JS, Li TY, Manson JE, et al. Adiposity compared with physical inactivity and risk of type 2 diabetes in women. Diabetes Care. 2007 Jan;30(1):53–58. doi: 10.2337/dc06-1456. [DOI] [PubMed] [Google Scholar]
- 25.Barclay AW, Petocz P, McMillan-Price J, et al. Glycemic index, glycemic load, and chronic disease risk–a meta-analysis of observational studies. American Journal of Clinical Nutrition. 2008 Mar;87(3):627–637. doi: 10.1093/ajcn/87.3.627. [DOI] [PubMed] [Google Scholar]
- 26.Salmeron J, Hu FB, Manson JE, et al. Dietary fat intake and risk of type 2 diabetes in women. American Journal of Clinical Nutrition. 2001 Jun;73(6):1019–1026. doi: 10.1093/ajcn/73.6.1019. [DOI] [PubMed] [Google Scholar]
- * 27.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000 Dec 1;49(12):2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 28.Kim C, Newton KM, Knopp RH. Gestational diabetes and the incidence of type 2 diabetes: a systematic review. Diabetes Care. 2002 Oct;25(10):1862–1868. doi: 10.2337/diacare.25.10.1862. [DOI] [PubMed] [Google Scholar]
- 29.Colditz GA, Willett WC, Rotnitzky A, et al. Weight gain as a risk factor for clinical diabetes mellitus in women. Annals of Internal Medicine. 1995 Apr 1;122(7):481–486. doi: 10.7326/0003-4819-122-7-199504010-00001. [DOI] [PubMed] [Google Scholar]
- 30.Conen D, Ridker PM, Mora S, et al. Blood pressure and risk of developing type 2 diabetes mellitus: the Women’s Health Study. European Heart Journal. 2007 Dec;28(23):2937–2943. doi: 10.1093/eurheartj/ehm400. [DOI] [PubMed] [Google Scholar]
- 31.Tirosh A, Shai I, Tekes-Manova D, et al. Normal fasting plasma glucose levels and type 2 diabetes in young men. The New England Journal of Medicine. 2005 Oct 6;353(14):1454–1462. doi: 10.1056/NEJMoa050080. [DOI] [PubMed] [Google Scholar]
- 32.Gerstein HC, Santaguida P, Raina P, et al. Annual incidence and relative risk of diabetes in people with various categories of dysglycemia: a systematic overview and meta-analysis of prospective studies. Diabetes research and clinical practice. 2007 Dec;78(3):305–312. doi: 10.1016/j.diabres.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 33.de Vegt F, Dekker JM, Jager A, et al. Relation of impaired fasting and postload glucose with incident type 2 diabetes in a Dutch population: the Hoorn study. JAMA. 2001 Apr 25;285(16):2109–2113. doi: 10.1001/jama.285.16.2109. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Gregg EW, Williamson DF, et al. A1C level and future risk of diabetes: a systematic review. Diabetes Care. 2010 Jul;33(7):1665–1673. doi: 10.2337/dc09-1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidt MI, Duncan BB, Bang H, et al. Identifying individuals at high risk for diabetes: the Atherosclerosis Risk in Communities study. Diabetes Care. 2005 Aug;28(8):2013–2018. doi: 10.2337/diacare.28.8.2013. [DOI] [PubMed] [Google Scholar]
- 36.Stern MP, Williams K, Haffner SM. Identification of persons at high risk for type 2 diabetes mellitus: do we need the oral glucose tolerance test? Annals of Internal Medicine. 2002 Apr 16;136(8):575–581. doi: 10.7326/0003-4819-136-8-200204160-00006. [DOI] [PubMed] [Google Scholar]
- 37.Balkau B, Lange C, Fezeu L, et al. Predicting diabetes: clinical, biological, and genetic approaches: data from the Epidemiological Study on the Insulin Resistance Syndrome (DESIR) Diabetes Care. 2008 Oct;31(10):2056–2061. doi: 10.2337/dc08-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mann DM, Bertoni AG, Shimbo D, et al. Comparative validity of 3 diabetes mellitus risk prediction scoring models in a multiethnic US cohort: the Multi-Ethnic Study of Atherosclerosis. American Journal of Epidemiology. 2010 May 1;171(9):980–988. doi: 10.1093/aje/kwq030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lindström J, Tuomilehto J. The diabetes risk score. Diabetes Care. 2003 Mar 1;26(3):725–731. doi: 10.2337/diacare.26.3.725. [DOI] [PubMed] [Google Scholar]
- 40.Simmons RK, Harding AH, Wareham NJ, et al. Do simple questions about diet and physical activity help to identify those at risk of Type 2 diabetes? Diabetic Medicine. 2007 Aug;24(8):830–835. doi: 10.1111/j.1464-5491.2007.02173.x. [DOI] [PubMed] [Google Scholar]
- * 41.Lyssenko V, Jonsson A, Almgren P, et al. Clinical risk factors, DNA variants, and the development of type 2 diabetes. The New England Journal of Medicine. 2008 Nov 20;359(21):2220–2232. doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 42.Janssens AC, Moonesinghe R, Yang Q, et al. The impact of genotype frequencies on the clinical validity of genomic profiling for predicting common chronic diseases. Genetics in Medicine. 2007 Aug;9(8):528–535. doi: 10.1097/gim.0b013e31812eece0. [DOI] [PubMed] [Google Scholar]
- * 43.Herder C, Roden M. Genetics of type 2 diabetes: pathophysiologic and clinical relevance. European Journal of Clinical Investigation. 2010 doi: 10.1111/j.1365-2362.2010.02454.x. [DOI] [PubMed] [Google Scholar]
- 44.Mihaescu R, Meigs J, Sijbrands E, et al. Genetic risk profiling for prediction of type 2 diabetes. PLoS Currents. 2011;3:RRN1208. doi: 10.1371/currents.RRN1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 45.Meigs JB, Shrader P, Sullivan LM, et al. Genotype score in addition to common risk factors for prediction of type 2 diabetes. The New England Journal of Medicine. 2008 Nov 20;359(21):2208–2219. doi: 10.1056/NEJMoa0804742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Hoek M, Dehghan A, Witteman JC, et al. Predicting type 2 diabetes based on polymorphisms from genome-wide association studies: a population-based study. Diabetes. 2008 Nov;57(11):3122–3128. doi: 10.2337/db08-0425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Miguel-Yanes JM, Shrader P, Pencina MJ, et al. Genetic risk reclassification for type 2 diabetes by age below or above 50 Years using 40 type 2 diabetes risk single nucleotide polymorphisms. Diabetes Care. 2011 Jan 1;34(1):121–125. doi: 10.2337/dc10-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pencina MJ, D’Agostino RB, Sr, D’Agostino RB, Jr, et al. Evaluating the added predictive ability of a new marker: from area under the ROC curve to reclassification and beyond. Statistics in medicine. 2008 Jan 30;27(2):157–172. doi: 10.1002/sim.2929. [DOI] [PubMed] [Google Scholar]
- 49.National Cholesterol Education Program. [cited 2011 July 7, 2011];Risk assessment tool for estimating 10-year risk of developing hard CHD. Available from 2004, http://hp2010.nhlbihin.net/atpiii/calculator.asp?usertype=prof. [Google Scholar]
- 50.Paulweber B, Valensi P, Lindstrom J, et al. A European evidence-based guideline for the prevention of type 2 diabetes. Hormone and Metabolic Research. 2010 Apr;42(Suppl. 1):S3–S36. doi: 10.1055/s-0029-1240928. [DOI] [PubMed] [Google Scholar]
- 51.Poulsen P, Kyvik KO, Vaag A, et al. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance–a population-based twin study. Diabetologia. 1999 Feb;42(2):139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 52.Kaprio J, Tuomilehto J, Koskenvuo M, et al. Concordance for type 1 (insulin-dependent) and type 2 (non-insulin-dependent) diabetes mellitus in a population-based cohort of twins in Finland. Diabetologia. 1992 Nov;35(11):1060–1067. doi: 10.1007/BF02221682. [DOI] [PubMed] [Google Scholar]
- 53.Vassy JL, Shrader P, Jonsson A, et al. Association between parental history of diabetes and type 2 diabetes genetic risk scores in the PPP-Botnia and Framingham Offspring Studies. Diabetes Research and Clinical Practice. 2011 May 11; doi: 10.1016/j.diabres.2011.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy MI. Casting a wider net for diabetes susceptibility genes. Nature Genetics. 2008 Sep;40(9):1039–1040. doi: 10.1038/ng0908-1039. [DOI] [PubMed] [Google Scholar]
- 55.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 56.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era–concepts and misconceptions. Nature Reviews Genetics. 2008 Apr;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 57.Pe’er I, de Bakker PI, Maller J, et al. Evaluating and improving power in whole-genome association studies using fixed marker sets. Nature Genetics. 2006 Jun;38(6):663–667. doi: 10.1038/ng1816. [DOI] [PubMed] [Google Scholar]
- 58.Gabriel SB, Schaffner SF, Nguyen H, et al. The structure of haplotype blocks in the human genome. Science. 2002 Jun 21;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- * 59.Mihaescu R, van Hoek M, Sijbrands EJ, et al. Evaluation of risk prediction updates from commercial genome-wide scans. Genetics in Medicine. 2009 Aug;11(8):588–594. doi: 10.1097/GIM.0b013e3181b13a4f. [DOI] [PubMed] [Google Scholar]
- 60.Nettleton JA, McKeown NM, Kanoni S, et al. Interactions of dietary whole-grain intake with fasting glucose- and insulinrelated genetic loci in individuals of European descent: a meta-analysis of 14 cohort studies. Diabetes Care. 2010 Dec;33(12):2684–2691. doi: 10.2337/dc10-1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Brito EC, Lyssenko V, Renström F, et al. Previously associated type 2 diabetes variants may interact with physical activity to modify the risk of impaired glucose regulation and type 2 diabetes. Diabetes. 2009 Jun 1;58(6):1411–1418. doi: 10.2337/db08-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Thomas D. Gene–environment-wide association studies: emerging approaches. Nature Reviews Genetics. 2010 Apr;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chang MH, Yesupriya A, Ned RM, et al. Genetic variants associated with fasting blood lipids in the U.S. population: Third National Health and Nutrition Examination Survey. BMC Medical Genetics. 2010 Apr 20;11:62. doi: 10.1186/1471-2350-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang Q, Liu T, Shrader P, et al. Racial/Ethnic differences in association of fasting Glucose‘ÄìAssociated genomic loci with fasting glucose, HOMA-B, and impaired fasting glucose in the U.S. adult population. Diabetes Care. 2010 Nov 1;33(11):2370–2377. doi: 10.2337/dc10-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vassy JL, Shrader P, Yang Q, et al. Genetic associations with Metabolic Syndrome and its Quantitative traits by Race/ Ethnicity in the United States. Metabolic Syndrome and Related Disorders. 2011 Aug 17; doi: 10.1089/met.2011.0021. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waters KM, Stram DO, Hassanein MT, et al. Consistent association of type 2 diabetes risk variants found in europeans in diverse racial and ethnic groups. PLoS Genetics. 2010 Aug;6(8) doi: 10.1371/journal.pgen.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lettre G, Palmer CD, Young T, et al. Genome-wide association study of coronary heart disease and its risk factors in 8,090 African Americans: the NHLBI CARe Project. PLoS Genetics. 2011;7(2):e1001300. doi: 10.1371/journal.pgen.1001300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Palmer ND, Hester JM, An SS, et al. Resequencing and analysis of variation in the TCF7L2 gene in African Americans suggests that SNP rs7903146 is the causal diabetes susceptibility variant. Diabetes. 2011 Feb 1;60(2):662–668. doi: 10.2337/db10-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- * 69.Janssens AC, Ioannidis JP, van Duijn CM, et al. Strengthening the reporting of genetic risk prediction studies: the GRIPS Statement. Annals of Internal Medicine. 2011 Mar 15;154(6):421–425. doi: 10.7326/0003-4819-154-6-201103150-00008. [DOI] [PubMed] [Google Scholar]
- 70.Lee JM, Pilli S, Gebremariam A, et al. Getting heavier, younger: trajectories of obesity over the life course. International Journal of Obesity. 2010;34(4):614–623. doi: 10.1038/ijo.2009.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Loos RJ. Genetics: genome-wide risk profiles - will they change your life(style)? Nature Reviews Endocrinology. 2011 May 7;(5):252–254. doi: 10.1038/nrendo.2011.41. [DOI] [PubMed] [Google Scholar]
- 72.Markowitz SM, Park ER, Delahanty LM, et al. Perceived impact of diabetes genetic risk testing among patients at high Phenotypic risk for type 2 diabetes. Diabetes Care. 2011 Feb 1; doi: 10.2337/dc10-1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pearson ER. Pharmacogenetics in diabetes. Current Diabetes Reports. 2009 Apr;9(2):172–181. doi: 10.1007/s11892-009-0028-3. [DOI] [PubMed] [Google Scholar]
- 74.Yasuda K, Miyake K, Horikawa Y, et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nature Genetics. 2008 Sep;40(9):1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 75.Rung J, Cauchi S, Albrechtsen A, et al. Genetic variant near IRS1 is associated with type 2 diabetes, insulin resistance and hyperinsulinemia. Nature Genetics. 2009 Oct;41(10):1110–1115. doi: 10.1038/ng.443. [DOI] [PubMed] [Google Scholar]
- 76.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nature Genetics. 2010 Feb;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gudmundsson J, Sulem P, Steinthorsdottir V, et al. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nature Genetics. 2007 Aug;39(8):977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- 78.Winckler W, Weedon MN, Graham RR, et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007 Mar;56(3):685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 79.Qi L, Cornelis MC, Kraft P, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Human Molecular Genetics. 2010 Jul 1;19(13):2706–2715. doi: 10.1093/hmg/ddq156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Talmud PJ, Hingorani AD, Cooper JA, et al. Utility of genetic and non-genetic risk factors in prediction of type 2 diabetes: Whitehall II prospective cohort study. BMJ. 2010 Jan Thu, 14;340 doi: 10.1136/bmj.b4838. b4838. jan14_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vaxillaire M, Veslot J, Dina C, et al. Impact of common type 2 diabetes risk polymorphisms in the DESIR prospective study. Diabetes. 2008 Jan;57(1):244–254. doi: 10.2337/db07-0615. [DOI] [PubMed] [Google Scholar]