Abstract
VEGFR-2 tyrosine kinase receptor draws attention of the scientific fraternity in drug discovery for its important role in cancer, cardiopulmonary, cardiovascular diseases etc. Hence there is a need for novel VEGFR-2 inhibitors screening and testing for their biological activities. The 3D-structure was collected from PDB and stability was checked by using WHATIF and PROCHECK programs and subjected for virtual screening on Zinc database. We used virtual screening method to screen new VEGFR-2 blocker molecules based on their binding energies and then docked with active site on the receptor with the help of AUTODOCK software. Based on the results obtained top three molecules (VRB1-3) were selected and tested in Cardiomyocytes H9c2 cells for cell viability under hypoxic condition. The invitro studies showed VRB2 as the best molecule among the selected three molecules as well as with a standard commercial drug Sunitinib.
Keywords: VEGF, VEGFR-2, Cell viability, Virtual screening, Docking
Background
The vascular angiogenesis is an important process in the development and differentiation of the entire organ in the body. When this angiogenesis is affected e.g. in various pathological and environmental conditions it ultimately leading towards cancers, cardiopulmonary, cerebrovascular disorders etc., [1, 2]. In the above said diseases various pathologies lead to hypoxia condition in the cells. Vascular endothelial growth factor (VEGF) is known to be a potent hypoxia induced mitogen and activator of angiogenesis, cellular proliferation, migration etc. This VEGF molecule interacts with various tyrosine kinase receptors (RTKs) present on the cell surface. VEGFR-2 receptor gains importance among tyrosine receptors as this is the main receptor which regulates various downstream signal transduction pathways related to vascular growth in the cells.
VEGF is also known to be associated with normal cardiac morphogenesis and the causative agent of various cardiovascular disorders such as valvuloseptal defects under hypoxia, which may prove lethal, if untreated [3]. Biological activation of VEGEFR2 activates cascade of downstream reactions, which leads to enhanced cell damage and cardiovascular pathologies [4]. In the light of the above information, we select rat ventricular cardiomyocytes because these cells are sensitive to hypoxia and mimic all the in vivo molecular events.
VEGFR-2 receptor hence becomes the choice of the scientists working all over the world in the field of drug discovery for designing or screening new inhibitors [5].
In this connection the present study has been focused to develop novel inhibitors of VEGFR-2. For achieving this purpose firstly, on the 3D structure of VEGFR-2 virtual screening and docking approaches have been used to select novel drug molecules. Later some selected molecules, have been tested in vitro for their efficacy on cell viability in rat ventricular cardiomyocytes (H9c2) under hypoxia (0.5%) condition. commercially available VEGF receptor blocker Sunitinib is used in the study as a reference molecule for comparisons purpose.
Methodology
Protein Homology:
VEGFR-2 Protein 3D structure with ID 1VR2 was downloaded from RCSB Protein Data bank (PDB) [6]. Protein stability was analyzed through Ramachandran plot by using PROCHECK program [7].
Protein Prepration:
VEGFR-2 protein 3D structure (PDB ID: 1VR2) was subjected to various tools like CASTp [8] and DogSiteScorer [9]. Prediction of binding site in the protein structure was done by web server CASTp and DogSite Scorer. They analyzes the drugability by taking into account of its geometric and physiochemical properties [10]. While, CASTp is used for the study of protein and its surface shape to visualize, locates and measuring voids and pockets on 3D structure of protein [11]. The selected active site was further put in a box of dimensions 26.33Å × 22.67Å × 16.00Å and a total box volume of 9550 Å3 with the aid of “Make Receptor” module of the Open Eye software (http://www.eyesopen.com/) [12].
Ligand preparation:
Lipinski rule of five was instrumental in generating a set of ligands from ZINC database.The Structures of 5384 lead like molecules were downloded in mol2 format [13]. The molecular weight of the lead molecules range fixed between 35 to 350 and the value of xlogP fixed between -4 to 3.5 for downloading.
Virtual Screening:
The Virtual screening of ligands for pose prediction and scoring were done with high dock resolution against VEGFR-2 protein active site by utilizing the exhaustive search algorithm [14]. The protein target structure was then subjected to FRED 3.0, a docking module of Open Eye software, for pose prediction and scoring. The exhaustive search algorithm was used for scoring based on Chemgauss4 scoring functions. It recognizes the shape, hydrogen bond interactions and hydrogen bond geometry and also hydrogen bond networks for scoring. The results obtained with lowest score are considered to be the best screened molecules [15& 16]. Hence, the top 10 ranked molecules were taken into account as the best molecules.
Molecular Docking:
Auto Dock 4.0 [17] software tool was used for molecular docking of the top ten molecules obtained from virtual screening with VEGFR-2. The water molecules, co-factors and ligands were removed from the protein structure and then checked for polar hydrogen atom in the macromolecule. This was then followed by Atomic Kollman charges and atomic solvation parameters, which were assigned and the torsion bonds of the ligands were selected. The binding energy of the macromolecule coordinate was evaluated by a three dimensional grid box of 60 60 60 (num.grid points in xyz) and grid center 44.325 32.386 15.763 (xyz-coordinates) was created with a spacing of 0.375Å. The bound ligand and actual target docking site [18] was represented based on the calculation of the grid map.
Validation:
The docking energies of commercially available drug Sunitinib (marketed as Sutent by Pfizer, and previously known as SU11248) (Sunitinib malate, SIGMA, PZ0012) was compared along with the top ten ligands obtained after screening for the validation of the results. The docking energy for a given macromolecule-ligand pair comprised of the intermolecular interaction energies including internal steric energy, hydrogen bond interaction energy, van der Waals forces and columbic electrostatic energy of the ligand. The lowest binding energy of protein-ligand complex has been considered to be the best [19].
Maintenance of H9c2 cells:
Rodent H9c2 were procured from NCCS, Pune, India and maintained in DMEM (Sigma), using 10% fetal bovine serum at 37°C and 5% CO2 in the CO2 incubator (Galaxy 170R, New Burnswick).
Cells were collected by de-adherence from the culture flask by trypsinization (0.25%), counted using Neubauer haemocytometer and seeded in 48-well (Nunc, Denmark), with a cell count of ~105 viable cells/cm2 by trypan blue exclusion method. The culture plates were then incubated in the CO2 incubator (Galaxy 170R, New Burnswick), maintained at a temperature of 37°C and 5% CO2 overnight. The adhered cells were grown to 70-80% confluence.
Experimental Design of VRB treatment of H9c2 cells:
VRB 1, 2, 3 and Sunitinib were obtained. H9c2 cells seeded in 48-well plates were kept in the incubator (Galaxy 170R, New Burnswick) for 24 hours with 21% O2, i.e., normal conditions, with one set being kept in the CO2 incubator with 0.5% O2, i.e., hypoxic condition
Experimental groups were divided into 10 different sets of, i.e., Normoxia (N), Normoxia + ZINC04652104 (N+VRB1), Normoxia + ZINC00484682 (N+VRB2), Normoxia + ZINC00677022 (N+VRB3), Normoxia + Sunitinib (N+Sut), Hypoxia controls (H), Hypoxia + ZINC04652104 (H+VRB1), Hypoxia + ZINC00484682 (H+VRB2), Hypoxia + ZINC00677022 (H+VRB3).
Assessment of cellular viability by MTT reduction assay treated with different VRB molecules:
Cellular viability, in vitro, was assessed by MTT [3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide] assay as described previously [20]. Various concentrations of VRB (VRB1-3) were used for experiments (12.5 nM, 25 nM, 50 nM, 100 nM, 200 nM and 400 nM). Twenty-four hours after hypoxic treatment, 0.5 mg/mL MTT was added to each well and cells were incubated upto 4 hours at 37°C. Absorbance was measured at 550 nm in Spectrophotometer (FLUOstar Omega, BMG LABTECH).
Statistical analysis:
Data was expressed as Mean±S.D. for each experimental group. The results were analyzed for statistical significance using Student׳s t-test, p<0.05 was considered to be significant.
Results
Stability analysis of 3D protein structure:
VEGFR-2 protein 3D structure was obtained ID 1VR2 from Protein Data bank (PDB). It was subjected for validation of structure by Ramachandran plot through PROCHECK program. According to the Ramachandran plot 87.3% residues are in the most favorable region, 12.2% residues are in allowed region and 0.0% residues were found in disallowed region. Based on the values obtained in the favorable and allowed regions of the plot it can be deduced that the protein structure got the most stable structure.
Active Site Prediction:
The active site of a protein directly participates in making and breaking of chemical bonds between the substrate and residues present in it. CASTp and DogSiteScorer were used in the present study for the active site prediction. The results obtained are shown in the Figure 1. Some of the Active site residues were already experimentally determined in the earlier studies [20].
Figure 1.
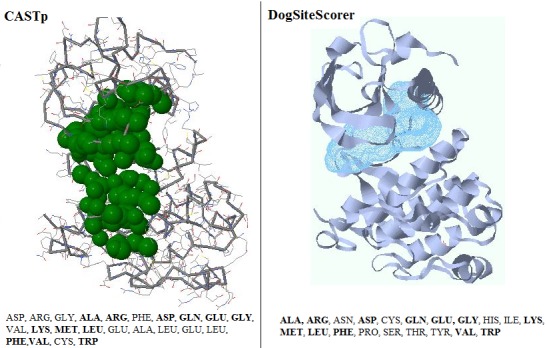
Comparing active site residues on VEGFR-2 through CASTp and DogSiteScorer. The residues highlighted are common in above two structures and are also earlier reported from the literature.
Virtual Screening:
The Open eye software module “FRED” was used for virtual screening of the ligands. The top 10 molecules obtained after screening is mentioned below in Table 1 (see supplementary material). Sorting of the ligands is based on their chemgauss4 score. The ligands scored with the lowest chemgauss4 score were placed at the top of the list and only top 10 ranked molecules represented in Table 1.
Molecular Docking:
The molecular docking of the individual top 10 molecules was done by using the Auto Dock docking tool (Figure 2). The validation of the results was performed based on the individual docking on the active site of the protein. The docking results obtained were then compared with the commercially available drug Sunitinib. The docking results are tabulated in Table 1.
Figure 2.
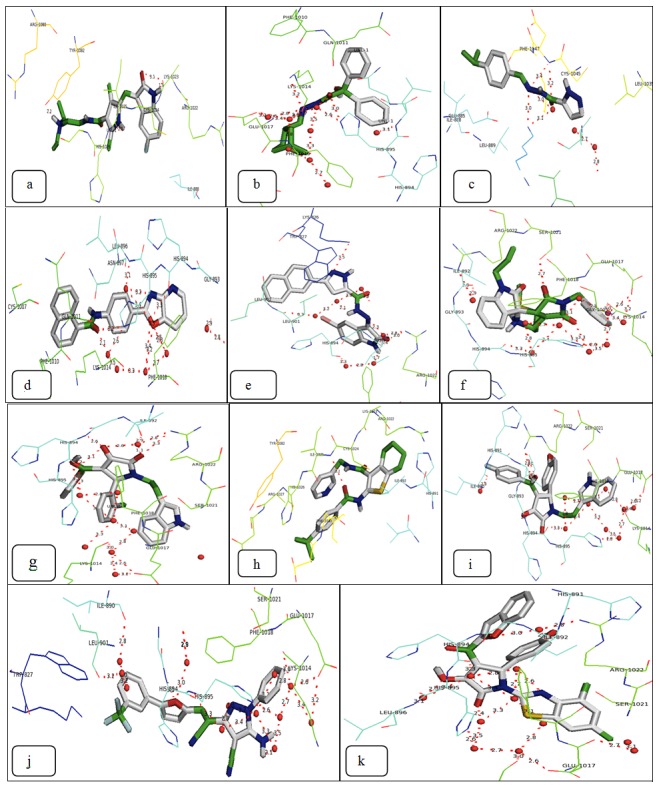
Docked structures of the Sunitinib and top ten molecules on the protein receptor: (a) Sunitinib (b) ZINC04652104 (c) ZINC00484682 (d) ZINC00677022 (e) ZINC09065134 (f) ZINC08439539 (g) ZINC00626508 (h) ZINC05944355 (i) ZINC01414763 (j) ZINC08424401 (k) ZINC00703128.
Effect of novel drug molecules (VRBs) on cellular viability under hypoxia:
Three candidate molecules, VRB-1 (ZINC04652104), VRB-2 (ZINC00484682) and VRB-3 (ZINC00677022) along with Sunitinib as control, were checked for improvement in cellular viability in H9c2 cells under hypoxia. Various concentrations of VRBs were used (12.5, 25, 50, 100, 200nM) and it was found that VRB2 at a concentration of 25nM showed maximum improved cellular viability. Among three candidate molecules and Sunitinib, both under normoxia and hypoxia compared to normoxia and hypoxia controls VRB2 was found as best in conferring cell survivability (Figure 3).
Figure 3.
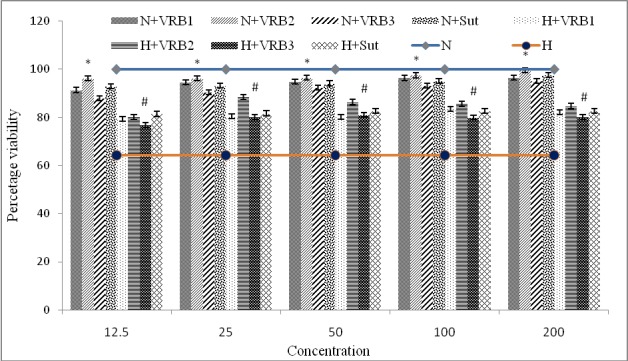
Figure showing effect of VRB1-3 on cellular viability in H9c2 cells by MTT assay at 24h hypoxia. Control cells were also assessed at same time-points. Maximum cellular viability was observed at 24h hypoxia exposure at a concentration of 25nM VRB2.Values are represented as mean±S.D. and significant values are represented at p≤0.05 as * when compared to normoxia control and # when compared to hypoxia control respectively.
Discussion
Vascular endothelial growth factor (VEGF) is an important vasogenic factor reported to be active in the early phase of the development and required for proper growth and vascular supply [1]. VEGF is activated through upstream regulator and primary hypoxia responsive gene, Hypoxia Inducible Factor (HIF1α). VEGF molecules interact with tyrosine kinase receptors (VEGFR-1 and VEGFR-2) and regulate various downstream pathways. The interaction of VEGF with KDR, i.e. VEGFR-2 is of particular importance not only during neonatal development of cardiac muscles but also to maintain homeostasis under stress [21]. VEGFR-2 thus provides an ideal target for designing novel candidate drug molecules by inhibiting the interaction of VEGF with VEGFR-2 [22& 23].
In the present study, in silico virtual screening approach was used to examine potential candidate molecule by using different bioinformatics tools, ten candidate molecules for VEGFR inhibitory molecules (VRB1-10) were screened [15& 16]. The top ten selected molecules were further checked for their active site prediction and docking performed between the active site and the protein molecules by using AutoDock 4.0. The scoring of the molecules was recorded by using chemgauss4 score [17& 18].
The top three in silico selected molecules were then tested in vitro by using the H9c2 cardiomyocytes cells for improving the cell viability under hypoxic stress (0.5% oxygen). The cardiomyocytes were chosen because under hypoxic conditions these cells tend to increase the VEGF levels so as to sustain in the hypoxic condition but later, overexpression of VEGF shifts the progression towards pathology of heart like hypertrophy and apoptosis [24, 25& 26].
Hence in the present study the new molecules screened through in silico virtual screening have been tested for checking their efficiency in improving the cell viability under hypoxic condition. All the results were compared with the commercially available Sunitinib drug which clearly suggested that VRB2 (ZINC00484682, N'-(4-isopropylbenzylidene)-1H-pyrazole-5- carbohydrazide) gave better cell survival compared to Sunitinib [18]. In the above case better cell viability is may be due to suppression of VEGFR-2 mediated signaling cascade.
These results thus opened a new window for the selected best molecules to study further in detail on the animal model for their clinical acceptability.
Conclusion
VEGF-receptor inhibitor appears to be an impressive strategy to restore normal cellular functions under hypoxic stress conditions. We virtually screened novel VEGF-receptor inhibitor molecules (VRB 1 to 10). Top three molecules were further tested in rat ventricular cardiomyocytes H9c2 (in vitro system) under hypoxia and VRB2 was found to be the best molecule for improving cellular viability compared to Sunitinib. However, this needs further validation in animal model to be used as a potential drug molecule.
Supplementary material
Footnotes
Citation:Saraswat et al, Bioinformation 10(5): 273-280 (2014)
References
- 1.Gaudue Y, Duvelleroy M. J Mol cell Cardiol. 1984;16:459. doi: 10.1016/s0022-2828(84)80617-3. [DOI] [PubMed] [Google Scholar]
- 2.Weis SM, Cheresh DA. Nature. 2005;437:497. doi: 10.1038/nature03987. [DOI] [PubMed] [Google Scholar]
- 3.Conway EM, et al. Cardiovascu Res. 2001;49:507. doi: 10.1016/s0008-6363(00)00281-9. [DOI] [PubMed] [Google Scholar]
- 4.Nicosia RF, et al. Am J Pathol. 1998;153:11. doi: 10.1016/S0002-9440(10)65539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bertl E, et al. Mol Cancer Ther. 2006;5:575. doi: 10.1158/1535-7163.MCT-05-0324. [DOI] [PubMed] [Google Scholar]
- 6.Wiesmann C, et al. Cell. 1997;91:695. doi: 10.1016/s0092-8674(00)80456-0. [DOI] [PubMed] [Google Scholar]
- 7.Laskowski RA, et al. J Appl Cryst. 1993;26:283. [Google Scholar]
- 8. http://sts-fw.bioengr.uic.edu/castp/calculation.php.
- 9. http://dogsite.zbh.uni-hamburg.de/
- 10.Volkamer A, et al. Bioinformatics. 2012;28:2074. doi: 10.1093/bioinformatics/bts310. [DOI] [PubMed] [Google Scholar]
- 11.Dundas J, et al. Nucleic acids research. 2006;34:116. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wlodek S, et al. Acta Cryst. 2006;62:741. doi: 10.1107/S0907444906016076. [DOI] [PubMed] [Google Scholar]
- 13.Irwin JJ, Shoichet BK. Journal of chemical information and modeling. 2005;45:177. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGann M. Journal of computer-aided molecular design. 2012;26:897. doi: 10.1007/s10822-012-9584-8. [DOI] [PubMed] [Google Scholar]
- 15.McGann M. Journal of chemical information and modeling. 2012;51:578. doi: 10.1021/ci100436p. [DOI] [PubMed] [Google Scholar]
- 16.Sanner MF. J Mol Graphics Mod. 2009;17:57. [PubMed] [Google Scholar]
- 17.Garrett M, et al. Journal of Computational Chemistry. 1998;19:1639. [Google Scholar]
- 18.Laura Q, et al. Journal of clinical oncology. 2007;25:884. [Google Scholar]
- 19.Denizot F, Lang R. J Immunol Method. 1986;89:271. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 20.Gamacy EL, et al. Bioinformation. 2011;7:52. [Google Scholar]
- 21.Izumiya Yasuhiro, et al. Hypertension. 2006;47:887. doi: 10.1161/01.HYP.0000215207.54689.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Olsson Anna-Karin, et al. Nature Reviews/Molecular Cell Biology. 2006;7:359. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 23.Madonna R, Caterina R. Cardiovas Res. 2009;84:4. doi: 10.1093/cvr/cvp270. [DOI] [PubMed] [Google Scholar]
- 24.Levy AP, et al. Cardiovas Res. 1995;76:758. [Google Scholar]
- 25.Bruggen N, et al. J Clin Invest. 1999;104:1613. [Google Scholar]
- 26.Izumiya Y, et al. Hypertension. 2006;47:887. doi: 10.1161/01.HYP.0000215207.54689.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


