Abstract
Periodontitis have been referred to as the sixth complication of diabetes found in high prevalence among diabetic patients than among healthy controls. The aim of the present study was to examine the periodontal disease status among collected dental plaque samples. Chromosomal DNA was isolated and amplified by universal primers. The DNA was sequenced for bacterial confirmation and phylogenetic analysis performed for the evolutionary relationship with other known pathogens. No amplification products were observed in groups labeled non periodontal and non Diabetes (NP&ND) and non Periodontal and Diabetes (NP&D). But in the case of Periodontal and non Diabetes (P&ND) groups 22 amplified products were observed. In case of Periodontal and Diabetes (P&D), 32 amplified products were positive for microbes. Among the four microbial groups, Treponemadenticola, and Tannerella forsythia were found to be prevalent in P&D. The phylogenetic analysis of 16s rRNA of Treponemadenticola showed the relationship with other Treponema oral pathogen species and with the Spirochaetazuelaera. Tannerella forsythia shows its evolutionary relationship only with four oral pathogens (Macellibacteroidesfermentans, Porphyromadaceae bacterium, Parabacteroidesmeredae and Bacillus fosythus). Prevotellaintermedia also showed its evolutionary relationship only with Prevotella Spcs while Fusobacterium revealed close evolutionary relationship only with Porpiromonasgingivalis.
Keywords: Periodontal diseases, Diabetes, Microbial pathogens, PCR
Background
Periodontitis differs from many other types of infections since it is not caused by a single bacterium but by a group of bacteria. Although more than 500 different types of bacteria have been isolated from the oral cavity [1] only a small fraction of these bacteria has the potential to cause destruction of periodontal tissues [2]. Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis have considered the major pathogenic species in destructive periodontal disease [3]. However, other subgingival species have also been related to periodontal disease as described for Prevotella intermedia, Prevotella nigrescens (formerly P. intermedia), Bacteroides forsythus, Fusobacterium nucleatum, Campylobacter rectus, Eikenell acorrodens, Treponemadenticola, Micromonas micros (formerly Peptostreptococcus micros) and some other species. Detection of these putative pathogens in periodontal health and disease strongly depends on the techniques used. Microbial diversity can be greatly under estimated by cultivation studies because many microorganisms cannot be cultivated by standard techniques. Human saliva is an attractive body fluid for disease diagnosis and monitoring because saliva testing is simple, safe, low-cost, and noninvasive. The use of saliva for diagnosis and monitoring of periodontal disease is highly desirable because periodontal pathogens, host antibacterial products, and other host-related proteins are readily detectable in saliva by new and highly sensitive technologies, such as real-time polymerase chain reaction (RT-PCR) analysis [4, 5]. It has been determined that RT-PCR provides a new rapid diagnostic tool and opens the opportunity to detect small numbers of oral pathogens in clinical specimens, that are under the detection limit by the culture technique [6, 7]. Therefore, a novel rapid method of RT-PCR can be used for identification and quantification of periodontopathic bacteria in saliva samples [6, 7, 8]. PCR has become an important tool for the rapid, sensitive, and specific detection of bacterial pathogens. Recent advances in molecular biology have made it possible to study microbial communities, including uncultivable species. Direct amplification by PCR of housekeeping genes from mixed culture biomass followed by purification and sequencing has allowed the analysis of complex communities [9]. The gene encoding the small-subunit rRNA has been used particularly for this purpose, and large databases of 16S rRNA sequences, such as that made available by the Ribosomal Database Project [10], now exist. These techniques have been applied to the micro flora associated with dento alveolar abscesses [11, 12]. A number of novel sequences which do not correspond to known, culturable organisms have been identified. Novel taxa identified by phylogenetic analysis in this way are designated “phylotypes”. Three of these, which each made up a substantial proportion of the flora in the abscesses in which they were detected, were selected for further study in five major periodontal pathogens present in periodontal, periodontal with diabetes, diabetes, non periodontal and non diabetic cases by PCR analysis, sequencing and assess the evolutionary relationship between the other microbial pathogens and estimated these five major microbial pathogen levels by the instate and the status of the periodontal condition. On the basis of their phylogenetic positions, phylotype PUS3.42 represents a new genus related to the genera Bacteroides and Prevotella, PUS9.170 represents a new genus related to the oral asaccharolytic Eubacterium species, and PUS9.180 represents a new species in the genus Prevotella [11, 12].
Methodlogy
Study of clinical Indices:
These four groups were paired with age; in addition to health conditions, and teeth number. The study procedure was approved by the Ethics Committee of Sri Venkateswara Medical College, Tirupati, Andhra Pradesh, India on 29th Dec, 2011 and an informed consent was signed by the study participants. To examine the periodontal disease status in patients by observing the destruction of tooth surrounding tissue and Clinical indices for the collection of microbial plaque sample with the two expert periodontologists. The initial clinical exam was performed by a single, previously trained examiner, who used a periodontal probe of the Williams type No. 23, and determined. Probing depth: distance from the bottom of the sulcus up to gingival margin in six points: mesio-vestibular, vestibular, disto-vestibular, disto-lingual/palatine, lingual/palatine and mesio-lingual/palatine of each tooth examined. Clinical attachment level: also determined in the same six points as the probing depth.
Collection of subgingival plaque samples:
A total of 120 subgingival plaque samples were collected from patients and divided them into four groups, each group consisting 30 patients such as Non-Periodontal and Non-Diabetes (np&nd), Periodontal and Non-diabetes (p&nd), Non-Periodontal and Diabetes (np&d) and Diabetes and Periodontal (d&p) groups. Both normal and diseased cases (age range 30-75 years) with probing depth (PD) values >= than 6 mm in the experimental sites. The sites were isolated with cotton rolls; removal of supra gingival plaque by a sterile curette (Asadental, Bozzano,Italy); gingival surface was dried and plaque samples were obtained by insertion of three standardized #30 sterile paper points (Inline, Turin, Italy) at the deepest part of each periodontal pocket and left in situ for 15 s. The paper points were transferred to a test tube containing 1 ml of the rout transport medium under anaerobic conditions.
DNA-extraction:
Chromosomal DNA was extracted from specimen collection by SDS lysis method. Concentration of DNA was caluculated by spectrophotometric analysis by using the formula Abs at 260x 50x dilution factor purity by using formula Abs at260/abs at 280. Samples having ratio close to 1.8 were taken for study. Integrity was checked by Electrophoretic analysis.
PCR amplification:
Multiplex PCR was performed using specific primers for the 16S rRNA gene of each bacterium (CalinAlexandruCristea., 2005). PCR amplification reactions were carried out in a reaction mixture in a final volume of 50µl consisting of 5µl of DNA sample, 25µl of master mix, 4µl of primers (both F.P and R.P) and autoclaved millique water16µl. The PCR protocol was as follows:98°C for 15 min followed by 40 cycles of 95°C for30 s, 60°C for 1 min, 72°C for 1 min, and a finalstep of 72°C for 10 min.PCR amplification was performed in an i Cycler System (Animal biotechnology laboratory, S.V. University, Tirupati, India). Amplicons were detected by electrophoresis of 10µl of samples from each PCR tube in a 1% agarose gel in TAE (Tris-Acetate-EDTA buffer) for 2 h at 80 V. The amplification products were visualized and photographed under a UV light trans illuminator. A positive or negative identification was based on the presence of clear bands of the expected molecular size using a commercial DNA molecular weight marker (number VIII; Roche DiagnosticsS.p.A., Milan, Italy). Amplified products were sequenced and phylogenetic analysis for the evolutionary relationship with other pathogens.
Phylogenetic analysis:
All the periodontal pathogens are subjected to similarity search for their 16s rRNA to find out their homologs. Phylogenetic analysis has been carried out to find out the evolutionary relationship among the group of homologs for each pathogen. The homologous rRNA sequences were obtained from the BLAST search and a sequence alignment was carried out using Clustal X tool along with the construction of Phylogenetic trees. A phylogenetic tree (or) evolutionary tree can shows the inferred evolutionary relationships among the group of biological species based upon similarities and differences in their genetic characteristics.
Results
The frequency of detection of the four periodonto pathogens as identified by PCR and sequencing. An important relationship between the presence of periodonto pathogenic bacteria and the serious form of periodontal disease was analyzed. In particular, in the 120 samples of subgingival plaque analysed, the prevalence of the 5 species monitored varied as regards the species itself and the method of detection. Among the 30 sample taken in each group DNA was obtained. No amplification product were observed in groups NP&ND and NP&D which clearly indicate that the patients were free of periodontal microbial pathogens belongs to the selected microbial pathogens. But in case of NDP group 22 amplified products were observed (Figure 1: NDP A, B, C, D). Which clearly indicate patients were positive for those microbes. Among the four microbial groups Trepanemadenticola was found to be prevalent in NDP. In case of P&D 32 amplified products which were positive for microbes (Figure 2: P&D A, B, C, D). Among the five microbial groups Treponemadenticola, Prevotellaintermedia and Tannerella forsythia were found to be prevalent in P&D. The Phylogenetic analysis of 16s rRNA of Treponemadenticola showed the relationship with other Treponema oral pathogen Spcs to a major extent and also with the Spirochaetazuelaera (Figure 3B). Tannerella forsythia shows it evolutionary relationship only with four oral pathogens i.e Macellibacteroides fermentans, Porphyromadaceae bacterium, Parabacteroidesmeredae and Bacillus fosythus (Figure 3C). Prevotellaintermedia also showed its evolutionary relationship only with Prevotella Spcs (Figure 3D). The Phylogenetic analysis of Fusobacterium (Figure 3A) revealed that it has a close evolutionary relationship only with Porpiromonasgingivalis.
Figure 1.
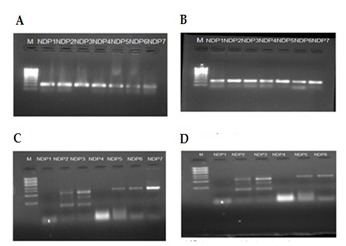
Non diabetes and periodontal disease group (ND&P) A, B, C, D Showing amplified products from different samples of ND&P group.
Figure 2.
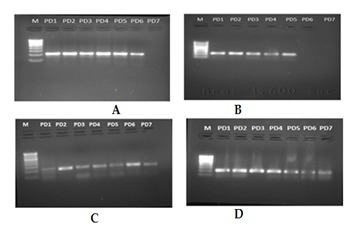
Diabetes and Periodontal disease group (D&P). A, B, C, D Showing amplified products from different samples of D&P group.
Figure 3.
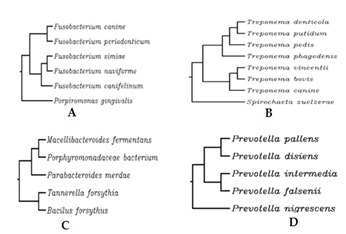
Phylogenetic analysis of different oral pathogen groups. A, B, C, D Showing selected 4 Periodontal microbial pathogens.
Discussion
The present study aimed at investigating the occurrence of some principal periodontal pathogens in combined periodontities and diabetic lesions. It is known that most periodontal pathogens are also endodontic pathogens [13]. Because of lack of dental history it was not possible to subgroup the lesions according to classification [14]. For practical reasons, the study focused only on five selected periodontal pathogens and single periodontal lesions were included as a reference group. The aim of the present study was to screening and molecular identification of periodontal microbial pathogens and their role in diabetic patients of south Indian population. In this study, to determine the detection of selected five periodontal microbial pathogens and these pathogens were analysis evolutionary relationship with other pathogens using the PCR and phylogenitic analysis and estimate the microbial population by statistic analysis. My results showed percentages in groups among the four microbial groups Fusobacterium and Trepanema denticola were found to be prevalent in P&ND. In case of P&D 32 amplified products which were positive for microbes. Among the four microbial groups Treponemadenticola, Prevotellaintermedia and Tannerella forsythia were found to be prevalent in P&D. Real-time PCR using universal 16S rRNA primer and specific Fusobacterium, primers allowed quantification of the total number of bacteria as well as of P.g. and A.a. My results are not accordance but some differ with the previous reports of real-time quantification for total bacteria [15], for P.g. [15, 16] and for A.a. [16]. Absolute quantification for P.g. and A.a. made possible the calculation of their proportions in the total bacteria. My results showed percentages in the similar range for Prevotella intermedia and Tannerella forsythia but fusobacterium and Treponem denticola showed higher percentage which is different as published elsewhere [16] and higher percentage values for A.a. than previously reported [17]. To my knowledge such data have not been presented before especial in south Indian population. The higher number could be explained by bacterial migration between the diabetes and the periodontal disease. In this study clearly indicate that people suffering periodontal disease from the Fusobacterium and Treponema denticola pathogens and Prevotella intermedia, Tannerella forsythia also causing periodontal disease but its low amount when compared to Fusobacterium and Treponema denticola. The elucidation of more specific disease associations are presently hampered by the lack of a reliable method for strain identification and a very poor understanding of how strains differ at the genetic level. Phylogenetic analysis has been carried out to find out the evolutionary relationship among the group of homologs for each pathogen. The homologous rRNA sequences were obtained form the BLAST search and a sequence alignment was carried out using ClustalX tool along with the construction of Phylogenetic trees. Our results revealed that there are relatively high levels of genetic diversity amongst Tannerella forsythia and Treponemadenticola, strains; with gene sequence similarities ranging between ca. 84 − 100% between the strains. These levels are considerably higher than in T. pallidum; where strains of the pallidum and pertenue subspecies share ca. 100-99.6% genome sequence identity, and genetic differences are largely confined to recombination “hotspots” or other areas of acquired DNA sequence [18]. 16S rRNA gene sequences, revealed that the A. actinomycetemcomitans serotype strains formed a separate branch. Previously [18, 19, 20], already noticed serotype e strains with an unusual 16S rRNA gene sequence (Type V). The oral spirochete bacterium Treponema denticola has postulated to play an important role in the pathogenesis of periodontal disease; in particular chronic periodontitis, which is estimated to affect ca. 10-15% of the global population [21, 22, 23, 24, 25, 26]. It is also implicated in the etiology of acute necrotizing ulcerative gingivitis (ANUG) [27], and orofacial noma [28], two other tissue-destructive diseases of the orofacial region. However, T. denticola has commonly detected in the oral microbiota in dentulous adults; albeit at relatively low levels, and its precise etiopathogenic mechanisms remain to be established. The elucidation of more specific disease associations are presently hampered by the lack of a reliable method for strain identification, and a very poor understanding of how strains differ at the genetic level [29], did a comprehensive study to determine the phylogenetic relationships within the family Bacteroidaceae, using sequence analysis of the 16S rRNA. They found Porphyromonas gingivalis in the Porphyromonas cluster, and Prevotella intermedia and Prevotella nigrescens closely related but clearly separated within the Prevotella cluster, showing a clear difference between the two genera as well as between the two species Prevotella intermedia and Prevotella nigrescens.
Conclusion
In this study no amplification product were observed in groups NP&ND and NP&D which clearly indicate that the patients were free of periodontal microbial pathogens. Among the five microbial groups Phorphyromonas gingivalis and Trepanema denticola were found to be prevalent in ND&P but other microbes like Treponema denticola, Prevotella intermedia and Tannerella forsythia were found to be prevalent in P&D. These results may be useful for the dental field of research to understand the disease and in which organism involving more in periodontal disease, which in turn can help for treatment. In addition this study may be helpful for the clinicians to get detailed knowledge and easy to select newer drug against bacterial pathogens.
Footnotes
Citation:Chiranjeevi et al, Bioinformation 10(4): 241-245 (2014)
References
- 1.Paster BJ, et al. J Bacteriol. 2001;183:3770. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, et al. Periodontol. 2000;20:341. doi: 10.1111/j.1600-0757.1999.tb00165.x. [DOI] [PubMed] [Google Scholar]
- 3.Slots J, Ting M. Periodontol. 2000;20:82. doi: 10.1111/j.1600-0757.1999.tb00159.x. [DOI] [PubMed] [Google Scholar]
- 4.Kononen E, et al. Oral Microbiol Immunol. 1994;9:126. doi: 10.1111/j.1399-302x.1994.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 5.Umeda M, et al. J Periodontol. 1998;69:828. doi: 10.1902/jop.1998.69.7.828. [DOI] [PubMed] [Google Scholar]
- 6.Boutaga K, et al. J Clin Periodontol. 2006;33:427. doi: 10.1111/j.1600-051X.2006.00925.x. [DOI] [PubMed] [Google Scholar]
- 7.Boutaga K, et al. J Periodontol. 2007;78:79. doi: 10.1902/jop.2007.060078. [DOI] [PubMed] [Google Scholar]
- 8.Morillo JM, et al. J Periodontal Res. 2003;38:518. doi: 10.1034/j.1600-0765.2003.00684.x. [DOI] [PubMed] [Google Scholar]
- 9.Wilson MJ, et al. Rev Med Microbiol. 1997;8:91. [Google Scholar]
- 10.Maidak BL, et al. Nucleic Acids Res. 1994;22:3485. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dymock D, et al. J Clin Microbiol. 1996;34:537. doi: 10.1128/jcm.34.3.537-542.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wade WG, et al. Clin Infect Dis. 1997;25:S235. doi: 10.1086/516215. [DOI] [PubMed] [Google Scholar]
- 13.Siqueira JF Jr, et al. J Endod. 2003;29:619. doi: 10.1097/00004770-200310000-00001. [DOI] [PubMed] [Google Scholar]
- 14.Simon JH, et al. J Periodontol. 1972;43:202. doi: 10.1902/jop.1972.43.4.202. [DOI] [PubMed] [Google Scholar]
- 15.Lyons SR, et al. J Clin Microbiol. 2000;38:2362. doi: 10.1128/jcm.38.6.2362-2365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakamoto M, et al. Microbiol Immunol. 2001;45:39. doi: 10.1111/j.1348-0421.2001.tb01272.x. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida A, et al. J Clin Microbiol. 2003;41:863. doi: 10.1128/JCM.41.2.863-866.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smajs D, et al. Infect Genet Evol. 2012;12:191. doi: 10.1016/j.meegid.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan JB, et al. J Clin Microbiol. 2002;40:1181. doi: 10.1128/JCM.40.4.1181-1187.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Norskov-Lauritsen N, Kilian M. Int J Syst Evol Microbiol. 200;56:2135. doi: 10.1099/ijs.0.64207-0. [DOI] [PubMed] [Google Scholar]
- 21.Pihlstrom BL, et al. Lancet. 2005;366:1809. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- 22.Petersen PE, Ogawa H. J Periodontol. 2005;76:2187. doi: 10.1902/jop.2005.76.12.2187. [DOI] [PubMed] [Google Scholar]
- 23.Ellen RP, Galimanas VB. Periodontol. 2005;38:13. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 24.Sela MN, et al. Crit Rev Oral Biol Med. 2001;12:399. doi: 10.1177/10454411010120050301. [DOI] [PubMed] [Google Scholar]
- 25.Dashper SG, et al. J Dent Res. 2011;90:691. doi: 10.1177/0022034510385242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishihara K, et al. Periodontol. 2010;54:117. doi: 10.1111/j.1600-0757.2009.00345.x. [DOI] [PubMed] [Google Scholar]
- 27.Gmur R, et al. Eur J Oral Sci. 2004;112:33. doi: 10.1111/j.0909-8836.2004.00103.x. [DOI] [PubMed] [Google Scholar]
- 28.Paster BJ, et al. J Bacteriol. 1994;176:725. doi: 10.1128/jb.176.3.725-732.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paster BJ, et al. J Clin Microbiol. 2002;40:2187. doi: 10.1128/JCM.40.6.2187-2191.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]


