Abstract
Molecular chaperones triage misfolded proteins via action as substrate selectors for quality control (QC) machines that fold or degrade clients. Herein, the endoplasmic reticulum (ER) associated Hsp40 JB12 is reported to participate in partitioning mutant conformers of GnRHR, a G-protein coupled receptor, between ER-associated degradation (ERAD) and a novel ERQC-autophagy pathway for membrane proteins. ERQC-autophagy degrades E90K-GnRHR because pools of its partially folded and detergent soluble degradation intermediates are resistant to ERAD. S168R-GnRHR is globally misfolded and disposed of via ERAD, but inhibition of p97, the protein retrotranslocation motor, shunts S168R-GnRHR from ERAD to ERQC autophagy. Partially folded and grossly misfolded forms of GnRHR associate with JB12 and Hsp70. Elevation of JB12 promotes ERAD of S168R-GnRHR, with E90K-GnRHR being resistant. E90K-GnRHR elicits association of the Vps34 autophagy initiation complex with JB12. Interaction between ERassociated Hsp40s and the Vps34 complex permits the selective degradation of ERAD-resistant membrane proteins via ERQC-autophagy.
Introduction
Biogenesis of membrane proteins initiates in the endoplasmic reticulum (ER) and is a complex process, as domains on both sides of the ER membrane, and within the membrane bilayer, must fold and assemble (Buchberger et al., 2010; Houck and Cyr, 2012). Loss of protein function diseases, including cystic fibrosis and hypogonadotropic hypogonadism, arise from missense mutations in membrane proteins that lead to misfolding and premature degradation. Misfolded polytopic membrane proteins are targeted for ER associated degradation (ERAD) by E3 ubiquitin ligase complexes that utilize the membrane inserted Hsp40 DNAJB12 (JB12) and cytosolic Hsp70 for substrate selection (Grove et al., 2011; Hirsch et al., 2009; Meacham et al., 2001; Younger et al., 2006). A key reaction in ERAD is the p97 dependent extraction, or retrotranslocation, of ubiquitinated membrane proteins from the membrane to the cytosol, which permits proteasomal degradation to occur (Brodsky, 2012).
Misfolded proteins with structural restraints that prevent entrance into the proteolytic chamber of the proteasome, such as aggregates, are subject to autophagic degradation (Webb et al., 2003). In some cases, mutant membrane and ER luminal proteins form aggregates, so they are not optimal ERAD clients, and are partitioned to the lysosome for degradation via autophagy (Houck and Cyr, 2012). How misfolded transmembrane and ER luminal proteins are partitioned between pathways for folding and proteasomal or lysosomal degradation is unclear. Seminal studies on ERQC of mutant α1-antitrypsin (α1AT) show that aggregation-prone α1AT mutants fail to retrotranslocate from the ER lumen, and consequentially are degraded by autophagy (Teckman and Perlmutter, 2000). Pools of mutant membrane proteins such as dysferlin, rhodopsin, and mOR-EG are believed to aggregate in the ER membrane, so a fraction of these mutant proteins are degraded by autophagic mechanisms (Fujita et al., 2007; Kaushal, 2006; Lu et al., 2003). Yet, how ERAD-resistant forms of mutant proteins that accumulate in misfolded states in the ER membrane, but don’t aggregate, are disposed of is not clear?
Molecular chaperones, such as Hsp70, function in the selection of misfolded proteins for proteasomal degradation (Meacham et al., 2001), and Hsp70 also functions in selection of cytosolic aggregates for autophagy (Gamerdinger et al., 2011). Thus, components of the Hsp70 system could cooperate with ER-associated forms of the Vps34/beclin-1 autophagosome initiation complex (Axe et al., 2008; Koyama-Honda et al., 2013; Yang et al., 2013) to select misfolded membrane proteins for autophagic degradation. Yet, whether bulk autophagy or a putative ERQC-autophagy pathway degrades ERAD resistant proteins is a mystery (Bernales et al., 2007).
To uncover new routes for ERQC of membrane proteins, the biogenesis of wild-type and mutant forms of the G-protein coupled receptor (GPCR) gonadotropin-releasing hormone receptor (GnRHR) was evaluated. The proper folding of human GnRHR requires action of the molecular chaperone calnexin (Ayala Yanez and Conn, 2010) and contacts formed between its seven transmembrane helices are stabilized by two disulfide bonds (Janovick et al., 2006; Jardon-Valadez et al., 2008). Folding of rat and mouse GnRHR is very efficient, but folding of human GnRHR is made inefficient by insertion of amino acid K191, which hinders assembly steps that involve disulfide bond formation (Janovick et al., 2006). The folding of human GnRHR therefore involves multiple steps with pools of on- and off-pathway intermediates being triaged for folding and degradation.
Hypogonadotropic hypogonadism is associated with over 20 mutations that further decrease the intrinsically low efficiency of nascent GnRHR folding and cause nearly all of the mutant forms to be retained in the ER (Conn and Ulloa-Aguirre, 2011). Disease-causing mutations in GnRHR include E90K and S168R, with E90K causing folding defects that are correctable with chemical folding correctors (pharmacoperones), and S168R leading to non-correctable global misfolding (Conn and Ulloa-Aguirre, 2011). Wild-type and mutant forms of GnRHR are referred to here as WT, E90K and S168R. Models of GPCRs show the location of residue E90 to be within transmembrane domain (TM) 2, where it forms a salt bridge with K121 in adjacent TM3 (Jardon-Valadez et al., 2008). E90K folding appears to arrest after it adopts a near native conformation because it can complete folding and escape the ER upon post-translational application of a pharmacoperone that interacts with the ligand binding site (Conn and Ulloa-Aguirre, 2011). Yet, rescued E90K has an abnormal conformation because its cell surface forms are locked in a constitutively active state (Conn and Ulloa-Aguirre, 2011).
Here, we identify an ERQC-autophagy pathway for selective degradation of detergent soluble and non-aggregated forms of partially folded E90K. Globally misfolded S168R is primarily degraded by the proteasome. In contrast, a pool of E90K is resistant to ERAD and accumulates in a complex with Derlin-1, JB12 and the Hsp70, but is degraded by autophagy. E90K folds to a protease resistant conformation and disruption of GnRHR folding through impairment of calnexin function or mutation of cysteines involved in disulfide bond formation permits its disposal by ERAD. Accumulation of partially folded E90K elicits association of the Vps34/beclin-1 autophagy initiation machinery with ER-associated complexes that contain E90K, JB12, and Hsp70. E90K dependent association of Vps34/beclin-1 with JB12 suggests a mechanism for targeted degradation of ERAD-resistant forms of misfolded GnRHR via ERQC-autophagy.
Results
GnRHR mutants are differentially sensitive to proteasome and lysosome inhibitors
Mechanisms for triage of misfolded membrane proteins and the relative contributions of ERAD and autophagy in maintenance of ER homeostasis are unclear. Herein, we identify differences in the conformation of Wt, E90K and S168R that dictate whether GnRHR degradation intermediates are selected for proteasomal or lysosomal degradation. In these studies GnRHR-GFP is employed and it migrates as a doublet on SDS-PAGE (Figure 1B). C-terminally GFP- tagged membrane proteins commonly migrate as a doublet (Geertsma et al., 2008) and doublets represent two distinct conformational states of the GFP tag. The lower band retains fluorescence on an SDS-PAGE gel, but the upper band does not (Geertsma et al., 2008). Both GnRHR-GFP bands in the doublet are equally sensitive to modulators of protein homeostasis, so they are discussed as a single entity and quantitation of this signal represents the sum of both bands.
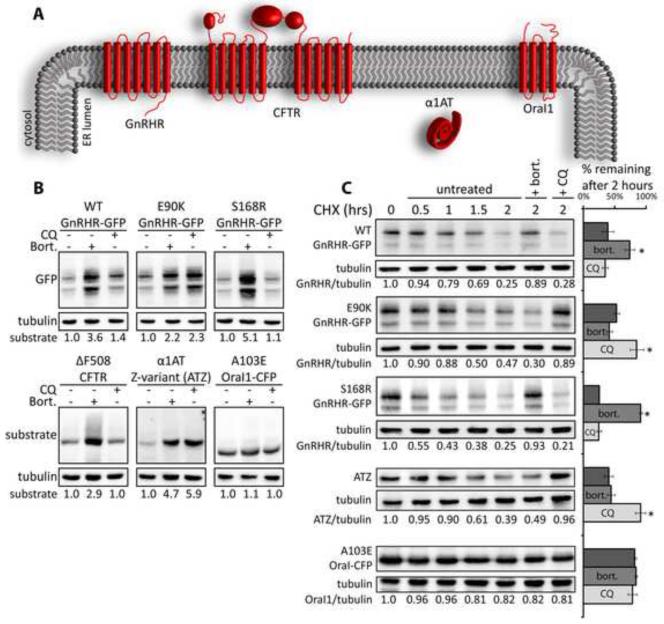
Figure 1. Transmembrane and secretory proteins exhibit differential sensitivity to proteasome and lysosome inhibitors.
(A) Topology of nascent disease-associated proteins. (B) Western blot analysis shows sensitivity of disease protein accumulation to inhibitors of the proteasome (bort; 10μM) or lysosomal proteases (CQ; 15μM). Transiently transfected Cos-7 cells were treated for 5 hours. (C) CHX chase analysis shows the impact of bort and CQ on the kinetics of disease protein degradation; 10μg/ml CHX, bort., or CQ were added at t=0. Relative amounts of respective disease proteins remaining at indicated time points was quantified and the average quantity −/+ standard error of mean (SEM) for the different disease protein remaining after 2 hours is depicted in bar graphs. Asterisks (*) indicate a p<0.05 as determined by the students t-test; n=3. Half-life curves are shown in Figure S1D. See also Figure S1.
Study of GnRHR degradation is aided by companion studies with model ERAD substrates that have different domain structures, which invoke use of different sets QC factors for disposal (Figure 1A). Cystic Fibrosis Transmembrane Conductance Regulator (CFTR) differs from GnRHR since CFTR has several large cytoplasmic domains, which make CFTR a substrate for a ERQC complexes that contain JB12 and cytosolic Hsp70 (Grove et al., 2011; Meacham et al., 2001; Younger et al., 2006). ATZ is a mutant variant of α1AT that misfolds and aggregates in the ER lumen, with its soluble and aggregated conformers being degraded by either ERAD or autophagy, respectively (Teckman and Perlmutter, 2000). Orai1 is a plasma membrane Ca2+ channel, which has four transmembrane spans and functions as homohexamer. A103E Orai1 has a poorly defined biogenic defect that leads to its retention in the ER and causes immunodeficiency (McCarl et al., 2009). The contribution of the proteasome and lysosome to degradation of ERQC substrates is estimated via comparing changes in their steady-state levels caused by the proteasome inhibitor bortezomib (bort) or the lysosome inhibitor chloroquine (CQ) (Figure 1B). CQ increases the pH of lysosomes and thereby functions in Cos-7 cells like the vacuolar-type H+-ATPase inhibitor Bafilomycin A1 to inhibit activity of lysosomal proteases (Figures S1A-C). This in turn drives the accumulation of misfolded proteins in autophagosomes and lysosomes (Klionsky et al., 2008).
During 5 hr incubations with Bort, levels of WT, E90K, and S168R increase differentially; S168R levels increase 5.5 fold versus 3.6 and 2.2 fold for WT and E90K respectively (Figure 1B). Likewise, WT, E90K, and S168R levels exhibit differential sensitivity to CQ; WT, E90K, and S168R increased 1.4, 2.2, and 1.1 fold respectively. ΔF508 CFTR increases in the presence of bort, but not CQ. In contrast, ATZ increases in the presence of both bort and CQ. Surprisingly, A103E Orai1-CFP is insensitive to both drugs. WT, E90K, and S168R exhibited half-lives in cycloheximide (CHX) chase experiments of 1.6, 1.9, and 1.0 hr, respectively (Figures 1C and S1D). Degradation of WT and S168R over a 2 hr CHX chase period is inhibited by bort, but not CQ. Whereas E90K degradation during this short-chase is inhibited by CQ, but not bort. A103E Orai1-CFP, is degraded slowly, such that 82% of the protein still remains after 2 hr of incubation with CHX.
These data suggest that sub-populations of WT and mutant GnRHR conformers -are degraded by different PQC pathways, with a large pool of E90K being degraded by autophagy. WT shows greatest sensitivity to bort, but levels do increase 40% with CQ, suggesting that non-native conformers of WT are also degraded by autophagy. E90K and ATZ have similar half-lives and CQ sensitivity, which supports the notion that E90K degradation involves autophagy. In contrast, S168R and ΔF508 CFTR have half-lives that are primarily sensitive to bort, suggesting that they are ERAD substrates.
Examination of the sensitivity of E90K and ATZ to bort and CQ show that steady-state accumulation of these proteins is sensitive to long-term inhibition of the proteasome and lysosome. Yet, short-term degradation rates of newly synthesized E90K and ATZ are most sensitive to CQ, suggesting that their autophagic degradation can occur at a rapid rate. In contrast, A103E Orai1-CFP appears resistant to the degradation pathways that clear E90K and ATZ. E90K appears to be partially folded, and S168R is globally misfolded (Conn and Ulloa-Aguirre, 2011), so data presented suggest a conformation specific mechanism for autophagic degradation of membrane proteins.
E90K is soluble in non-ionic detergents, but accumulates in puncta
Protein aggregates are degraded by autophagy and a portion of E90K is degraded by the lysosome, so the aggregation state of E90K was evaluated (Figure 2A). As expected, ATZ, but not ATM (a non-disease causing variant of α1AT), forms aggregates because it fractionates to a Triton X-100 insoluble fraction of cell extracts. A small portion of WT Orai1-CFP accumulates in Triton X-100 insoluble aggregates, with most A103E Orai1-CFP being detergent insoluble. Surprisingly, even though pools of E90K appear to be degraded by autophagy, it is soluble in Triton X-100, and therefore does not form detergent insoluble aggregates.
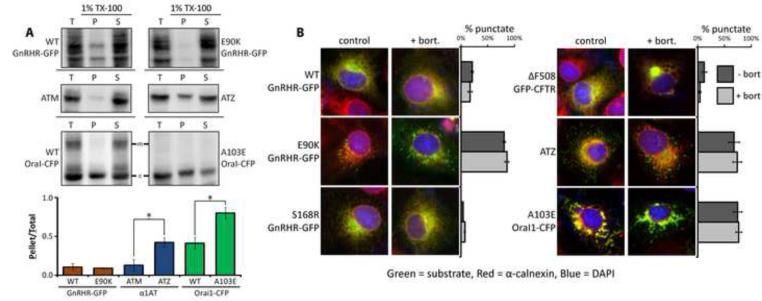
Figure 2. Analysis of the E90K aggregation state.
(A) Analysis of disease protein solubility in the non-ionic detergent Triton X-100. Cos-7 cell extracts were prepared in lysis buffer supplemented with 1% Triton X-100 and spun at 4°C at 20,000xg for 15 min: total (T), pellet (P), and supernatant (S). Mature glycosylated Orai1-CFP is indicated (#). Proteins were detected by western blot and the ratio of pellet to total was quantified for 3 experiments and graphed +/− SEM. Asterisks (*) indicate p<0.05 via a students t-test. (B) Fluorescence analysis of disease proteins localization in fixed Cos-7 cells. GFP fusion proteins were detected in the green channel. ATZ (green) was detected by immunofluorescence, and so was the ER marker calnexin (red). One hundred cells per slide were scored for the presence of puncta and cells that contained >10 puncta were judged as punctate. Asterisks (*) indicate a p<0.05 in a t-test, N=3. Green and red channels are shown unmerged in Figure S2.
Microscopic analysis shows WT and S168R to colocalize with the ER membrane marker calnexin in a pattern similar to GFP-ΔF508 CFTR (Figures 2B and S2). Interestingly, some E90K is found in a diffuse pattern that co-localizes with calnexin, and 80% of cells also contain E90K puncta that are not enriched in calnexin. E90K puncta are similar in size and distribution to puncta containing ATZ. A103E Orai1-CFP formed puncta that were enriched with calnexin and much larger than ATZ and E90K puncta, suggesting that A103E Orai1-CFP aggregates within the ER membrane. Proteasome inhibition caused GFP-ΔF508 CFTR to localize in perinuclear aggresomes, but bort does not lead GnRHR-GFP to enter aggresomes or impact E90K puncta. Thus, the cell utilizes specific QC mechanisms for handing different conformers of GnRHR, CFTR and Oral1. E90K appears to be the substrate of a novel ERQC system that targets misfolded and non-aggregated transmembrane proteins to autophagic puncta for lysosomal degradation.
E90K puncta contain autophagy markers
The mechanism for CQ sensitive E90K degradation was explored through definition of the requirements for E90K puncta formation. If puncta that contain E90K are autophagic then modulation of discrete steps of autophagy should impact E90K puncta accumulation (Figure 3A). Rapamycin induces autophagy by inhibiting the kinase mTOR, while LiCl-stimulates autophagy through inhibition inositol-monophosphatase (IMPase) and the resultant downstream effects on ER-dependent calcium signaling (Cardenas et al., 2010; Sarkar et al., 2005). LiCl strongly reduced E90K levels (Figures 3B and S3A), but rapamycin caused an increase (Figure S3A). Reduction of E90K by LiCl was suppressed by CQ, which suggests that LiCl stimulates autophagy of E90K (Figure S3B). In parallel, experiments with ATZ, the same differential sensitivity to rapamycin and LiCl was observed. Similar, results were observed when the effects of the carbamazepine (an IMPase inhibitor) and rapamycin (an mTOR inhibitor) on ATZ levels were tested (Hidvegi et al., 2010). LiCl at the high dose of 10mM partially reduced WT and S168R, and inhibition of autophagy with CQ prevented this effect(Figures S3A-B). ΔF508 CFTR and A103E Orai1-CFP levels were unchanged by LiCl, suggesting that effects seen on GnRHR-GFP and ATZ are not due to bulk turnover of the ER (Figure S3A).
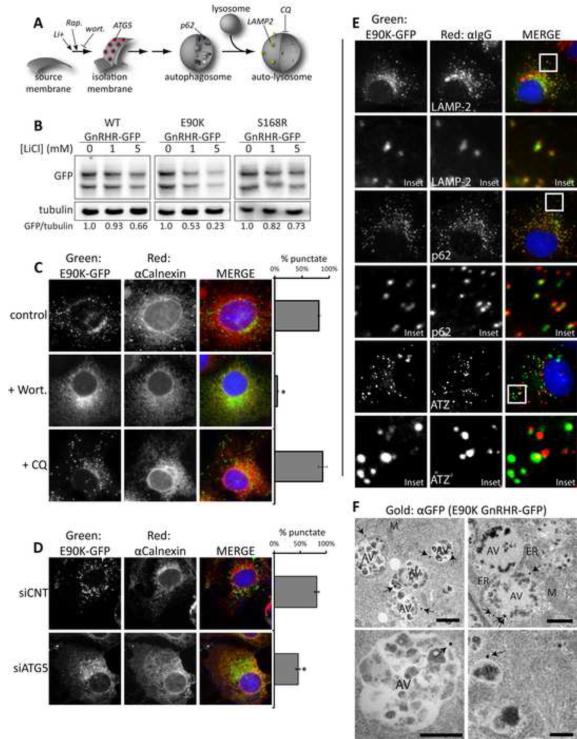
Figure 3. E90K puncta colocalize with autophagic markers and localization is sensitive to chemical and genetic inhibitors of autophagy.
(A) Model pathway for autophagosome formation. Sites of action for CQ, Wortmannin (wort), rapamycin (Rap), and lithium chloride (LiCl) are shown. (B) Impact of LiCl on steady-levels of indicated disease proteins as determined by western blot. Cells were treated with for 18 hr prior to lysis of Cos-7 cells. C-E show fluorescence micrographs of Cos-7 cells treated as indicated. (C) The impact of Wort and CQ on the accumulation of E90K in puncta. (D) The impact of siRNA KD of ATG5 on the accumulation of E90K in puncta. One hundred cells per slide were scored for the presence of puncta and cells that contained >10 puncta were judged as punctate. Data are represented as mean +/−SEM. Asterisks (*) indicates a p<0.05 in a t-test ; n=3. (E) Localization of E90K in relation to autophagic markers. Cells immunostained for LAMP-2 (lysosome marker), or p62 (autophagy marker) are shown. Additionally, cells co-expressing E90K and ATZ were immunostained with α1AT. Insets show colocalization of LAMP-2 and p62 with E90K puncta. (F) Immunogold transmission electron microscopy (TEM) of Cos-7 cells expressing E90K GnRHR-GFP. Immunogold staining was performed against the GFP tag and gold particles are indicated with arrows. Mitochondria (M), endoplasmic reticulum (ER), nuclei (N), and autophagic vesicles (AV) are labeled. Scale bar = 1 μM. See also Figures S2 and S3.
LiCl induces degradation of a broad spectrum of autophagic substrates, so we carried out titrations with LiCl to determine if WT, E90K, and S168R exhibited differential sensitively to the drug. Indeed, 1mM LiCl selectively reduced WT, E90K, and S168R levels to 93, 53, and 82% of control (Figure 3B). At 5mM LiCl the levels of WT, E90K, and S168R were 66, 23, and 73% of control. A significant portion of E90K is degraded by autophagy, whereas the conformation of S168R appears to favor degradation by ERAD. Folding of WT appears to go off-pathway at different stages with most degradation intermediates being destroyed by the proteasome and a smaller fraction by autophagy.
Initiation of autophagy requires the type-III phosphatidylinositol 3-phosphate kinase (PI3K) VPS34, so entry of E90K into puncta should be blocked by the PI3K inhibitor wortmannin (Klionsky et al., 2008). Indeed, formation of E90K puncta is disrupted by wortmannin and insensitive to the late stage autophagy inhibitor CQ (Figure 3A and C). Autophagosome formation is also dependent upon ATG5 (Figure 3A), so ATG5 knockdown should inhibit entrance of E90K into puncta. ATG5 levels are reduced 3-fold by siRNA, and this reduces E90K puncta formation and increases E90K co-localization in the ER with calnexin (Figures 3D and S3C). In wortmannin treated cells, and when ATG5 is depleted, most E90K co-localizes with calnexin, so E90K accumulates in the ER prior to wortmannin sensitive and ATG5 dependent passage to puncta.
Authentic autophagic puncta contain marker proteins such as LAMP-2 and p62, and E90K colocalizes with these autophagic markers (Figure 3E). In contrast, E90K puncta do not contain markers of the secretory pathway, ERAD, or the proteasome (Figures S3D-F). Additionally, E90K puncta do not colocalize with SEL1L, a marker of the autophagosome-like ERAD tuning vesicles (TVs) (Bernasconi et al., 2012). E90K puncta formation is insensitive to Sel1 knockdown (Figures S3G-H), so E90K puncta are autophagic in nature, but distinct from ERAD TVs. Misfolded E90K and ATZ are both degraded by LiCl stimulated autophagic mechanisms. However, ATZ that is co-expressed with E90K does not colocalize in puncta with E90K (Figure 3E). E90K accumulates in autophagic puncta, as does ATZ, but ATZ aggregates and detergent soluble E90K are independently selected for autophagy.
ER-Golgi trafficking is not required for autophagy of E90K
ER-associated E90K may enter autophagy as a passenger of ER membranes that serve as a source of autophagosomal membranes (Axe et al., 2008; Koyama-Honda et al., 2013). Alternatively, regions of the ER in which misfolded E90K accumulates could be engulfed by autophagosomes (Bernales et al., 2007). To address this question we employed transmission electron microscopy (TEM) coupled with GFP immunogold staining to show that E90K is on the surface of large 0.5-2μm vesicles (Figures 3F and S4A). The size of the immunogold positive vesicles and the fact that they contain cellular debris fit criteria of autophagic vesicles (AVs) (Klionsky et al., 2008). Very few gold particles are seen in cells expressing untagged E90K GnRHR (Figure S4B). As expected, GFP immunogold staining of Cos-7 cells expressing GFP-LC3 show localization to the cytosol and surface of AVs (Figure S4C). These data infer the existence of a pathway for movement of E90K degradation intermediates from the ER membrane into the autophagosomal membrane system. Therefore, the engulfment of bulk ER by autophagy is not a likely mechanism for E90K entry into puncta.
There are multiple potential sources of autophagosomal membrane (Mizushima et al., 2011). Therefore, support for E90K moving with ER membrane into autophagosomes was obtained by demonstrating that ER localized E90K moves from the ER to puncta (Figure 4A). A 4 hr treatment with the reversible wortmannin analog LY-294002 resulted in accumulation of a large ER pool of E90K that colocalizes with calnexin (Figure 4A). Upon LY-294002 wash-out and CHX addition, the ER localized E90K moved into puncta.
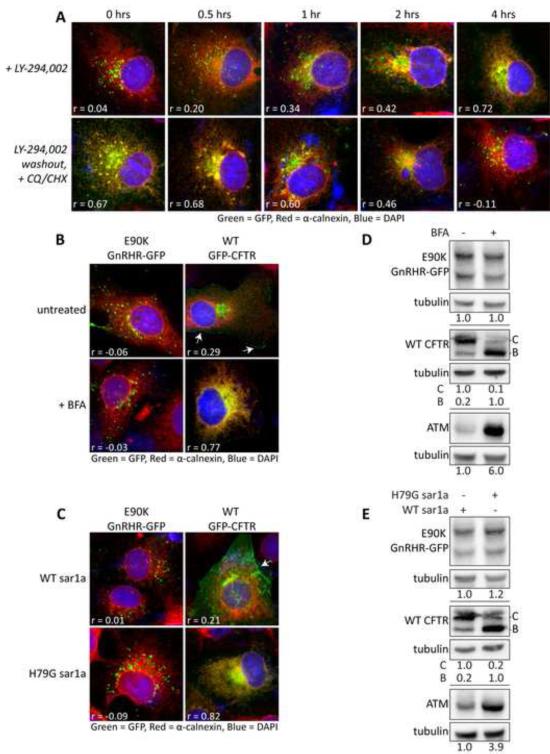
Figure 4. E90K puncta formation and protein levels are not affected by ER-golgi trafficking inhibition.
(A) Effects of LY-294002 treatment and washout on E90K localization in Cos-7 cells. After 4 hours of treatment with LY-294002, media was removed and replaced with media containing 10μg/ml CHX and 15μM CQ. (B) Effects of the ER-golgi transport inhibitor Brefeldin-A (BFA) on levels of E90K, WT CFTR, or α1AT M-variant (ATM). (C) Effects of overexpression of a dominate negative mutant (H79G) of Sar1a on levels of E90K, WT CFTR, or α1AT M-variant (ATM). Panels B and C show western blots of Cos-7 cells transiently transfected on E90K GnRHR-GFP, WT CFTR, or α1AT M-variant (ATM). CFTR blots show core-glycosylated (B-band) and mature-glycosylated (C-band). (D) Effects of the ER-golgi transport inhibitor Brefeldin-A (BFA) on localization of E90K and GFP-CFTR. (E) Effects of overexpression of a dominate negative mutant (H79G) of Sar1a on localization of E90K and GFP-CFTR. Fluorescence micrographs of Cos-7 cells transiently transfected with substrates are shown in A, D and E. Cell surface localization of CFTR is shown with arrows. Pearson's correlation coefficients (r) between calnexin (Red) and GFP (Green) signal are shown. DAPI staining is shown in blue. Data are represented as mean +/−SEM.
To further demonstrate that E90K enters puncta from the ER, we utilized two approaches to inhibit ER-golgi trafficking and assessed their impact on puncta formation and E90K levels: (1) Brefeldin A (BFA) and (2) overexpression of H79G Sar1, a dominant negative form of a small GTPase that inhibits ER-Golgi trafficking (Aridor et al., 1995). BFA and H79G Sar1a caused intracellular accumulation of the secretory protein ATM and blocked ER-Golgi trafficking of CFTR as it drove accumulation of the ER localized and core glycosylated B-band at the expense of more heavily glycosylated C-band (Figure 4B-E). Under these conditions, BFA and H79G Sar1a, had no effect on E90K levels and accumulation in puncta. Taken together all these data support the claim that E90K enters autophagic puncta from the ER membrane.
Conformation Dependent Triage of Misfolded GnRHR for Autophagy or ERAD
E90K folding arrests at a late stage (Conn and Ulloa-Aguirre, 2011), so thermodynamically stable, but misfolded, forms of E90K could be poor substrates for retrotranslocation and therefore ERAD-resistant. To test this supposition, we modulated or inhibited specific steps in GnRHR folding and evaluated changes in E90K’s accumulation and sensitivity to CQ or bort (Figure 5). GnRHR is a glycoprotein and contains two disulfide bonds (Figure 5A-B), so its proper folding and QC is facilitated by the glycochaperone calnexin and its associated protein disulfide isomerase ERp57 (Ayala Yanez and Conn, 2010). When the GnRHR structure is perturbed by the E90K mutation, the disulfides bonds may be inaccessible to ERp57 and this could make E90K resistant to ERAD. Indeed, overexpression of ERp57 and calnexin had no effect on E90K levels (Figure S5C).
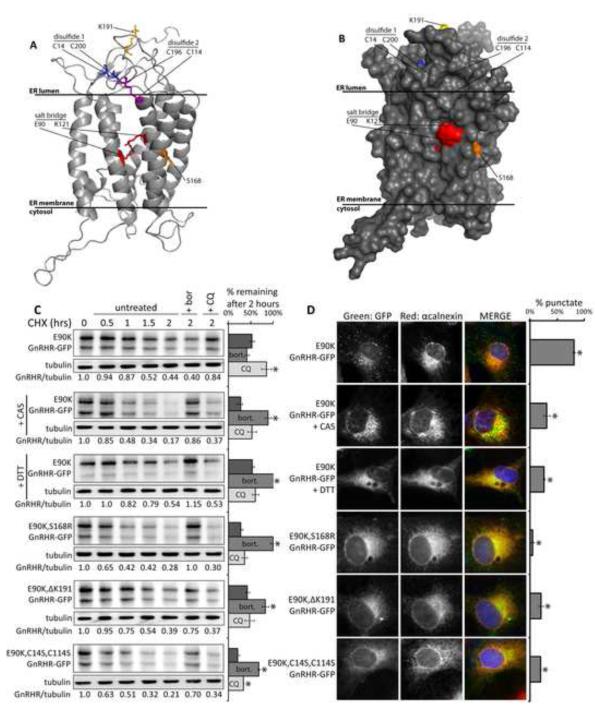
Figure 5. Interference with GnRHR folding impacts triage of E90K.
(A) Ribbon diagram of a GnRHR homology model (Jardon-Valadez et al., 2008). (B) Space filling model of the GnRHR structure. (C-D) Effects of 5mM castanospermine (CAS), 2mM DTT (4 hour pre-treatment), and second-site mutations in GnRHR on the half-life and localization of E90K. Cos-7 cells were treated with 10μg/ml CHX, 10μM bort., or 15μM CQ at t=0. Bar graphs show the relative amounts of protein remaining at 2 hours. Asterisks (*) indicate a p<0.05 in a t-test. Half-life curves are shown in Figure S5D. E90K localization was determined by fluorescence microscopy of cells treated as described. 100 cells/slide were scored for the presence of puncta. Asterisks (*) indicates p<0.05 in a t-test; N=3. See also Figure S5. Data are represented as mean +/−SEM.
The glucosidase inhibitor castanospermine (CAS), which blocks calnexin binding to glycoproteins, and inhibition of disulfide bond formation with the reducing agent DTT both caused E90K degradation to become sensitive to bort and insensitive to CQ (Figures 5C and S5D). Likewise, DTT treatment also accelerated the rate of WT degradation (Figure S5E-F). These same treatments also prevented E90K accumulation in puncta (Figure 5D). CAS has no negative effect on global autophagy as seen by monitoring LC3-RFP puncta (Figure S5B). Therefore, inhibition of GnRHR folding favors proteasomal degradation, which suggests that abnormally folded forms of E90K are degraded by autophagy.
To directly modulate E90K folding, the destabilizing mutation S168R was introduced into E90K to form the double mutant, E90K-S168R GnRHR-GFP (E90K-S168R). Additionally, folding of human GnRHR is made inefficient by the insertion of a lysine (K191) in a position that appears to destabilize the conformation of the disulfide bond containing loops, which are located near E90. Deletion of K191 from GnRHR partially suppresses folding defects caused by E90K, but the majority of E90K K191 GnRHR remains misfolded (Janovick et al., 2006). Changes in GnRHR caused by these second site mutations prevented detection of E90K-S168R and E90K-ΔK191 in puncta, and their degradation was sensitive to bort instead of CQ (Figures 5C-D and S5D). Similarly, a form of E90K, (E90K,C14S, C114S), in which one cysteine required to form each disulfide bond in GnRHR was mutated, was not detected in puncta, and its degradation was sensitive to bort instead of CQ.
Changes in the conformation of GnRHR caused by the aforementioned manipulations were confirmed by limited proteolysis (Figure S5A). WT and E90K exhibited similar resistance to digestion with trypsin, which is consistent with E90K accumulating in a partially folded state. CAS and mutations that cause misfolding of GnRHR caused E90K to accumulate in conformation that is as sensitive to trypsin as globally misfolded S168R. Partially folded GnRHR conformers, but not globally misfolded forms, are degraded by autophagy.
Inefficient ERAD leads misfolded GnRHR to be degraded by autophagy
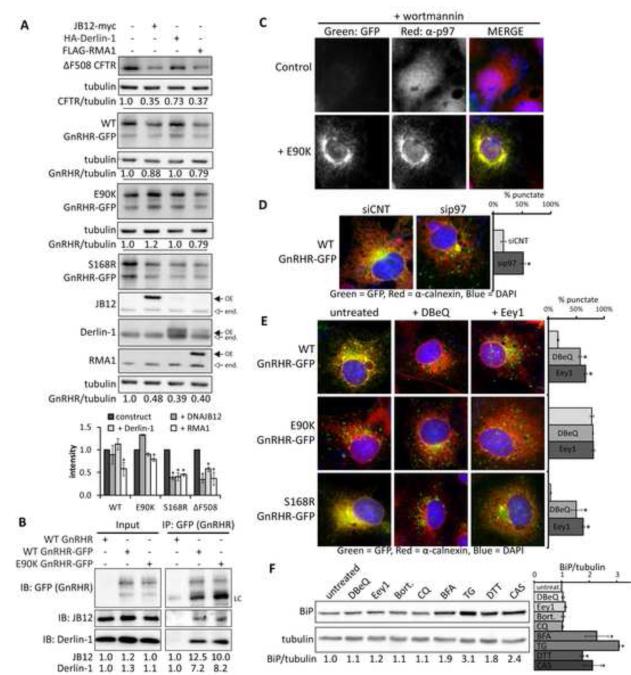
E90K may be degraded by autophagy because its partially folded and detergent soluble forms are ERAD-resistant. Thus, we compared the sensitivity of WT, E90K, or S168R to overexpression of components of the RMA1/RNF5 E3 complex. The Hsp40 JB12 cooperates with Hsp70 and Derlin-1 to select membrane proteins for RMA1 dependent ubiquitination (Grove et al., 2011). Activity of these ERQC factors was elevated to levels that decrease ΔF508 CFTR accumulation around 70% (Figure 6A). WT and E90K are largely insensitive to JB12, where as S168R is reduced 60% (Figure 6A). When compared to S168R and ΔF508 CFTR, E90K is also relatively insensitive to RMA1 and Derlin-1.
Figure 6. Interference with ERAD causes misfolded GnRHR to be partitioned to autophagy.
(A) The effect of ER quality control factors overexpression on steady-state levels of WT, E90K, and S168R GnRHR-GFP in Cos-7 cells. (B) Co-immunoprecipitation of endogenous derlin-1 and JB12 with WT and E90K GnRHR-GFP from Cos-7 cell lysates. IgG light chains (LC) are indicated. (C)Colocalization of E90K with immunostained p97 in wort-treated cells. (D) Inactivation of p97 by siRNA KD leads to detection of WT GnRHR-GFP in puncta. (E) The impact of the p97 inhibitors DBeQ and Eeyarestatin 1(Eey1) on accumulation GnRHR-GFP in puncta. (F) Changes in BiP levels in Cos-7 cell treated for 4 hours with indicated chemicals. Western blots were probed for the ER stress marker BiP and tubulin. Average band density and SEM, N=3, is shown in the bar graph. In panels C-F, cells were immunostained for Calnexin, JB12, or p97 (Red) and nuclei were stained with DAPI (Blue). 100 cells/slide were scored for the presence of puncta and quantification shown the average +/− SEM for N=3. Asterisks (*) indicate a p<0.05 in a t-test. See also Figure S6.
If E90K is initially recognized by ERQC factors, but resistant to ERAD its degradation intermediates should accumulate in association with ERAD machinery. Indeed, E90K coimmunoprecipitates (co-IPs) with JB12 and Derlin-1 (Figure 6B). The retrotranslocation factor p97 associates Derlin-1 and RMA1/RNF5 (Younger et al., 2006), and upon inhibition of early-stages of autophagy with wortmannin, E90K colocalizes in a perinuclear location with p97 (Figure 6C). These data support the concept that E90K associates with components of RMA1/RNF5 E3 complex that include p97, but a portion of E90K adopts an ERAD-resistant conformation and therefore is degraded by autophagy.
To test this concept, we asked if inhibition of p97 function in retrotranslocation caused WT and S168R to accumulate in puncta. This was accomplished via p97 siRNA knockdown and chemical inhibition of p97 with DBeQ and Eeyarestatin-1 (Eey1)(Chou et al., 2011; Wang et al., 2008). SiRNA-mediated p97 knockdown (3-fold KD) promotes accumulation of WT in puncta (Figures 6D and S6A). Treatment of cells with DBeQ and Eey1 for 4 hours also caused WT and S168R to localize to E90K-like puncta (Figure 6E). Importantly, DBeQ and Eey1 did not lead GFP-ΔF508 CFTR to re-localize in puncta, so effects on WT and S168R are not from gross changes in ER morphology (Figure S6C). Inhibition of autophagy with wortmannin blocks the DBeQ or Eey1 induced accumulation of WT in puncta (Figure S6D). Additionally, dual inhibition of the proteasome and lysosome with bort and CQ does not cause WT to partition to puncta (Figure S6E).
The short exposure of cells to Eey1 and DBeQ used to induce GnRHR accumulation in puncta does not cause an increase in levels of the ER-stress marker BiP (Figures 6F). Indicating that autophagic partitioning of WT and S168R GnRHR in response to these drugs is not due to ER stress. In addition to facilitating ER-protein extraction, p97 also has a late stage role in autophagy (Chou et al., 2011). Thus, examination of the time course for degradation of WT and S168R in p97 inhibitor-treated cells is difficult. Nevertheless, inefficiencies in p97-mediated protein extraction can impact protein triage decisions and lead forms of GnRHR that are normally degraded by ERAD to accumulate in autophagic puncta.
Selective autophagy is mediated by ubiquitin binding proteins (Kirkin et al., 2009; Pankiv et al., 2007), so we explored the role of ubiquitin in autophagic degradation of GnRHR. Interestingly, Eey1, but not DBeQ, caused WT to accumulate as a ubiquitinated species (Figure S6F-G). Thus, DBeQ doesn't drive accumulation of ubiquitinated GnRHR, but still causes WT and S168R to be partitioned to autophagic puncta. This is consistent with the observation that Eey1 blocks retrotranslocation, via a mechanism that involves inhibition of p97-associated deubiquitinases (Wang et al., 2008). Additionally, siRNA knockdown of the ubiquitin-mediated autophagy cargo selectors p62 and NBR1 did not cause E90K to accumulate in the ER or disrupt puncta formation (Figure S6H). These data are consistent with the conformation of GnRHR degradation intermediates dictating whether pools of misfolded forms are destroyed via ERAD or ERQC-autophagy.
E90K drives association the Vps34 autophagy initiation complex with JB12
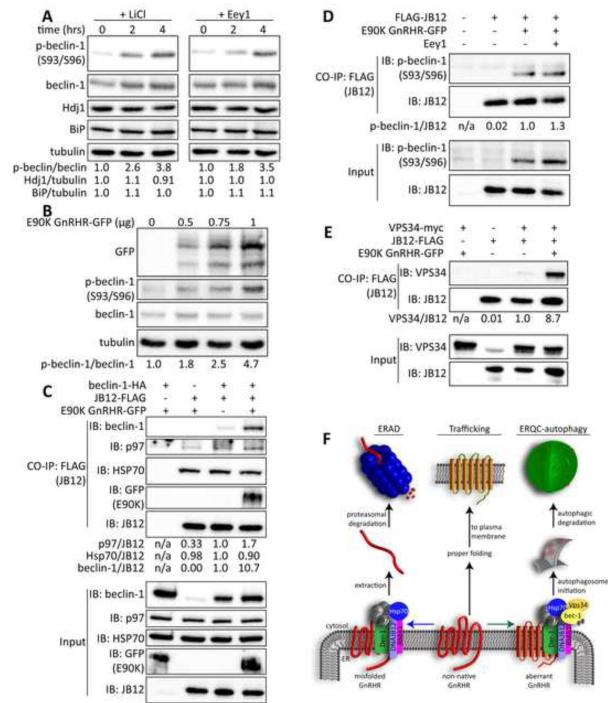
To understand how the cell senses accumulation of ERAD-resistant membrane proteins we asked if E90K impacts activity of the beclin-1/Vps34 PI3K-complex (Figure 7). Beclin-1 is a scaffold protein that directs Vps34 to drive autophagosome formation and associates with the ER membrane (Matsunaga et al., 2010). Beclin-1 function is regulated by several kinases and its activity in autophagy is increased by phosphorylation on residues that include S93 and S96 (Kim et al., 2013). We therefore determined if stimulation of ERQC-autophagy by LiCl or Eey1 is associated with accumulation of beclin-1 that is phosphorylated on S93 and S96 (p-beclin-1 S93/96)(Figure 7A). Indeed, phosphorylation of S93/S96 on beclin-1 rapidly increased by almost 4 fold in response to LiCl or Eey1. At the same time, no increase occurred in expression of the ER stress marker BiP or the heat shock marker Hdj1 (Figure 7A). Notably, E90K causes a dose dependent increase in accumulation of p-beclin-1 S93/96 (Figure 7B), and does not induce expression of BiP (Figure S7D). The cells respond to inhibition of p97 and E90K accumulation by activating beclin-1 to function in autophagy.
Figure 7. E90K promotes association of the ERAD factor JB12 with the beclin-1/Vps34 autophagy initiation complex.
(A) Phosphorylation of beclin-1-HA on S93/96 in response to treatment with 10mM LiCl and 15μM Eey1. (B) Phosphorylation of beclin-1-HA on S93/96 in response to E90K expression. (C) Native IP of JB12-FLAG in the presence and absence of beclin-1-HA and E90K. (D) Native IP of JB12-FLAG in the presence and absence of beclin-1-HA and E90K to look at interactions of JB12 with phosphorylated beclin-1. (E) Native IP of JB12-FLAG in the presence and absence of beclin-1-HA, ATZ, or E90K. (F) Native IP of JB12-FLAG in the presence of E90K and Vps34-myc. Western blots of samples from Cos-7 cells are depicted in all panels, see the supplemental experimental procedures for details. See also Figure S7.
P-beclin-1 S93/S96 functions in bulk autophagy (Kim et al., 2013), so we sought to explore the mechanism for specification of its action in ERQC-autophagy. Hsp70 and Hsp90 function in the formation of complexes that activate autophagy (Joo et al., 2011; Yang et al., 2013) and Hsp40 proteins play a major role in specification of Hsp70 function. Since JB12 is an Hsp40 that is present in complexes with ERAD-resistant forms of E90K, we hypothesized that interactions between JB12 and beclin-1 are associated ERQC-autophagy of E90K. Indeed, Hsp70, beclin-1, and p97 are present in co-immunoprecipitates with JB12, and association of beclin-1 with JB12 is dramatically increased by the presence of E90K (Figure 7C). Substrate dependent interaction between JB12 and beclin-1 is dependent upon the conformation of the misfolded proteins as S168R, ΔF508-CFTR, and A103E Ora1 has little detectable effect (Figures S7A-B). Wortmannin prevents E90K from increasing the association of JB12 and beclin-1, so active autophagy is required for substrate dependent association of JB12 and beclin-1 (Figure S7B). Consistently, the form of beclin-1 that associated with JB12 in response to E90K is p-beclin-1 S93/96 (Figure 7D). Additionally, Vps34 also co-immunoprecipitates with JB12, and E90K dramatically increases detection of Vps34/JB12 complexes (Figure 7E).
To evaluate the requirement of JB12 in ERQC of E90K, it was depleted via shRNA by over 90% (Figure S7E). Yet, interpretation of results is complicated because JB12 depletion decreased cell viability by 40% and caused apoptosis as high level of caspase-3 cleavage was now detected (Figure S7E-F). Under these conditions, accumulation of E90K, ATZ and Orai1 was dramatically reduced (Figure S7G), and E90K accumulation was relatively insensitive to CQ. Autophagy and apoptosis are intimately linked (Eisenberg-Lerner et al., 2009), so induction of apoptosis in response to loss of JB12 is consistent with it playing an important role in ERQC autophagy. Data presented place the Vps34/beclin-1 complex in association with E90K and ERQC complexes that help select globally misfolded GnRHR for ERAD. The Vps34/beclin-1/ E90K/JB12 complex suggests that E90K is selected for ERQC-autophagy through interaction between autophagy initiation factors and ERAD-complexes.
Discussion
An ERQC-autophagy pathway for membrane proteins is revealed via demonstration that different GnRHR mutants are selectively targeted for degradation via either ERAD or autophagy. Partially folded, detergent soluble, and ERAD-resistant forms of E90K are degraded by autophagy. Globally misfolded GnRHR, which accumulates resultant from the S168R mutation, inhibition of calnexin, or blockade of disulfide bonds, is degraded by ERAD. ERQC-autophagy disposes of E90K at rates similar to ERAD of WT and S168R, so flux through the pathway is rapid. ERQC-autophagy involves E90K dependent association of Vps34/beclin-1 with ERQC complexes that contain JB12 and Hsp70. We suggest that substrate driven association of Vps34/beclin-1 with JB12/Hsp70 initiates ERQC-autophagy at locations where ERAD-resistant membrane proteins accumulate (Figure 7F).
Cytosolic protein aggregates are degraded by autophagy because they can't be unfolded and threaded through the narrow proteolytic chamber of the proteasome (Webb et al., 2003). ER luminal ATZ aggregates are degraded by autophagy due to failed unfolding and retrotranslocation to cytosol (Hidvegi et al., 2010). Remarkably, nascent E90K is not aggregation prone and is partially folded. The folding defect in E90K occurs at a late stage and manifests itself after adoption of a protease resistant conformation, which is similar WT and requires conserved cysteines. Inefficiencies in disulfide bond formation limit human GnRHR folding efficiency (Janovick et al., 2006), and blockage of disulfide bond formation promotes proteasomal degradation of E90K. Thus, we propose that aspects of E90K's partially folded structure limit ERADs ability to retrotranslocate it to the cytosol, and this invokes the use of ERQC-autophagy. The disulfide bonds in GnRHR are highly conserved, so a portion of the misfolded GPCRs rhodopsin and mOR-EG may contain covalent bonds that limit sensitivity to ERAD and cause their degradation by autophagy (Kaushal, 2006; Lu et al., 2003). A general role for ERQC-autophagy in protein homeostasis will be established through identification of additional clients. Demonstration that ERQC-autophagy of E90K occurs in pituitary tissue where symptoms of hypogonadotropic hypogonadism originate will aid in demonstration of the physiologic significance of ERQC-autophagy.
Several pieces of data suggest that autophagy of E90K initiates soon after its biosynthesis and that E90K enters the autophagosomal membranes from ER membranes. E90K accumulates in the ER when reversible PI3K inhibitors inactivate Vps34, and E90K moves from the ER to autophagic puncta upon Vps34 reactivation. This is consistent with the Vps34/beclin-1 complex being associated with ER, and the human ER serving as a source of autophagic membranes (Axe et al., 2008). In addition, defects in retrotranslatoction of misfolded proteins from the ER caused by chemical inhibition of p97 leads ER localized forms of WT and S168R to be shunted away from ERAD into autophagic puncta. Finally, the entrance of E90K into autophagic puncta does not require ER to Golgi trafficking.
ERQC-autophagy is selective for partially folded E90K, so it is distinct from bulk ER-autophagy. In bulk ER-phagy, fragments of ER membranes are detected by EM in the autophagosome lumen (Bernales et al., 2007), but EM data indicates E90K is located within autophagosomal membranes. Interestingly, levels of ER lumenal chaperones are regulated by the membrane inserted ERQC factor Seǀ1ǀ via an autophagic-like pathway known as ERAD tuning (Bernasconi et al., 2012). Seǀ1ǀ could therefore participate in selection of E90K for autophagy, but Seǀ1ǀ is not required for E90K puncta formation and Sel1 does not co-localize with E90K puncta. Therefore, there are distinct mechanisms for tuning Seǀ1ǀ activity and autophagic degradation of E90K.
Inhibition of basal autophagy blocks E90K degradation, and ER-stress markers are not induced by E90K, so flux through ERQC-autophagy occurs under normal growth conditions. Thus, initiation E90K degradation by ERQC-autophagy is not necessarily dependent on ER-stress. E90K degradation intermediates accumulate in ERQC complexes that contain JB12, Hsp70 and derlin-1. A pool of beclin-1 is detected in complexes with JB12, and E90K dramatically increases the interactions of beclin-1 and Vps34 with JB12. Association of beclin-1 with JB12 is sensitive to the conformation of GnRHR, as an increase is observed with E90K, but not globally misfolded S168R or ΔF508-CFTR, or aggregated A103E OraI1. JB12 and Hsp70 select globally misfolded membrane proteins for ERAD (Grove et al., 2011), so association of Vps34/beclin-1 with JB12 may initiate targeted clearance of ERAD-resistant E90K from the ER via ERQC-autophagy.
Flux through autophagy is regulated by kinases and other factors that promote formation of distinct proautophagy Vps34/beclin-1 complexes (Itakura et al., 2008). Lithium chloride promotes the initiation of autophagy by antagonizing the ER localized IP3-receptor (Sarkar et al., 2005). Antagonism of the IP3-receptor induces autophagy by regulating activity of factors that modulate Vps34/beclin-1 activity (Cardenas et al., 2010). Consistently, lithium chloride induces ERQC-autophagy in a manner associated with increased levels of proautophagic p-beclin-1-S93/S96 (Kim et al., 2013). However, LiCl has effects on global autophagy and promotes the degradation of cytosolic protein aggregates as well as E90K (Sarkar et al., 2005). Thus, effects of LiCl on E90K may occur as a result of the general induction of autophagy that initiates in the ER. Yet, rapamycin does not induce degradation of E90K, so there must be a specific mechanism for entrance of ERAD-resistant membrane proteins into autophagic degradation pathways. Since E90K increases the association of JB12 and Vps34/beclin-1, we suggest that the specificity of ERQC-autophagy occurs at the substrate level. .E90K dependent association of beclin-1/Vps34 with JB12 requires PI3K activity, which is consistent with the participation of JB12/Vps34/beclin-1 in initiation of ERQC-autophagy.
The interaction of JB12 with the Vps34 complex provides a new link between ERAD and autophagy pathways that suggests a mechanism for conformation specific selection of membrane proteins for ERQC-autophagy. However, details on the mechanism for JB12/Hsp70 action in regulation of Vps34 in ERQC-autophagy requires study. Hsp70 selects misfolded proteins for degradation and also facilitates the association of beclin-1 with Vps34 (Yang et al., 2013). Thus, accumulation of E90K in association with JB12 and Hsp70 on the ER surface could trigger the targeted assembly of proautophagy Vps34 complexes for function in ERQC-autophagy.
Experimental Procedures
Plasmids, antibodies, and reagents
Reagents were obtained from commercial or academic sources as noted in the supplemental experimental procedures.
Cell culture , transfections, and chemical treatments
Cos-7 cells were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and maintained in accordance with ATCC recommendations. Plasmid transfections were performed using the using the Effectene transfection reagent (Qiagen, Alameda, CA) and siRNA transfections were performed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA). For experiments where siRNA and plasmid DNA were both transfected, siRNA transfection was performed prior to reseeding and subsequent plasmid transfection. Chemical treatments were performed on cells post-transfection. See supplemental experimental procedures for more details.
Cycloheximide chases
Transfected Cos-7 cells were incubated with 5μg/ml cycloheximide (CHX; Sigma-Aldrich, St. Louis, MO) in the presence and absence of 15μM CQ (Sigma-Aldrich, St. Louis, MO) or 10μM bortezomib (LC Laboratories, Woburn, MA). Where indicated, 2mM DTT (Thermo Fisher, Waltham, MA) or 5mM castanospermine (Sigma-Aldrich, St. Louis, MO) were added 4 hours prior to CHX addition. Cells were harvested at indicated time points. See supplemental experimental procedures for more details.
Immunoprecipitation (IP)
Transfected Cos-7 cells were lysed in 200μl of RIPA buffer + N-ethyl-malemide or 50mM Tris-Cl, 150mM NaCl, 1% Triton X-100, pH 7.6 (TBSt) prior to immunoprecipitation analysis. See supplemental experimental procedures for more details.
Triton X-100 solubility assay
Harvested cell pellets were lysed in TBSt(1%) and separated into soluble and insoluble fractions by centrifugation at 20,000xg for 15minutes at 4°C. See supplemental experimental procedures for more details.
Cell lysis and western blotting
For all non-immunoprecipitation or TX-100 solubility experiments, harvested cell pellets were lysed in 200μl of 50mM Tris-Cl pH 7.7, 150mM NaCl, 0.8% SDS, 10mM EDTA with 1× Complete Protease Inhibitor cocktail (Roche, Basel, Switzerland). Lysate was then subjected to two 30-second sonication prior to Western blotting and data analysis. See supplemental experimental procedures for more details.
Fluorescence Microscopy
Cos-7 cells were grown on sterile coverslips and fixed prior to immunostaining. In all images nuclei are stained with DAPI and shown in blue channel. In each experiment 100 cells were scored for a punctate staining pattern. A punctate cell was defined as a cell that contained more than 10 discrete puncta, of any size, in the indicated channel. See supplemental experimental procedures for more details.
Immunogold Electron Microscopy
Transfected Cos-7 cells were fixed with 2% paraformaldehyde/0.5% glutaraldehyde prior to immunostaining and sectioning. Sections were observed using a LEO EM-910 transmission electron microscope (LEO Electron Microscopy, Thornwood, NY). See supplemental experimental procedures for more details.
Supplementary Material
Highlights.
- A route for ERQC autophagy of soluble ERAD-resistant GnRHR mutants is discovered.
- Partitioning of E90K GnRHR between ERAD and ERQC autophagy is conformation dependent.
- ERQC autophagy initiates via E90K GnRHR driven association of Vps34 with DNAJB12.
- Rates of GnRHR degradation via ERAD or ERQC autophagy are similar.
Acknowledgements
This work was supported by grants from the National Institutes of Health to D.M.C (R01GM056981) and S.A.H. (F31AG042257). Thanks to KJ Liang for critical review of the manuscript and DH Perlmutter for α1AT plasmids.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aridor M, Bannykh SI, Rowe T, Balch WE. Sequential coupling between COPII and COPI vesicle coats in endoplasmic reticulum to Golgi transport. J Cell Biol. 1995;131:875–893. doi: 10.1083/jcb.131.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayala Yanez R, Conn PM. Protein disulfide isomerase chaperone ERP-57 decreases plasma membrane expression of the human GnRH receptor. Cell Biochem Funct. 2010;28:66–73. doi: 10.1002/cbf.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Noack J, Bianchi S, de Haan CA, Reggiori F, Molinari M. Role of the SEL1L:LC3-I complex as an ERAD tuning receptor in the mammalian ER. Mol Cell. 2012;46:809–819. doi: 10.1016/j.molcel.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Brodsky JL. Cleaning up: ER-associated degradation to the rescue. Cell. 2012;151:1163–1167. doi: 10.1016/j.cell.2012.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas C, Miller RA, Smith I, Bui T, Molgo J, Muller M, Vais H, Cheung KH, Yang J, Parker I, et al. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, et al. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proc Natl Acad Sci U S A. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conn PM, Ulloa-Aguirre A. Pharmacological chaperones for misfolded gonadotropin-releasing hormone receptors. Adv Pharmacol. 2011;62:109–141. doi: 10.1016/B978-0-12-385952-5.00008-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A. Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009;16:966–975. doi: 10.1038/cdd.2009.33. [DOI] [PubMed] [Google Scholar]
- Fujita E, Kouroku Y, Isoai A, Kumagai H, Misutani A, Matsuda C, Hayashi YK, Momoi T. Two endoplasmic reticulum-associated degradation (ERAD) systems for the novel variant of the mutant dysferlin: ubiquitin/proteasome ERAD(I) and autophagy/lysosome ERAD(II) Hum Mol Genet. 2007;16:618–629. doi: 10.1093/hmg/ddm002. [DOI] [PubMed] [Google Scholar]
- Gamerdinger M, Kaya AM, Wolfrum U, Clement AM, Behl C. BAG3 mediates chaperone-based aggresome-targeting and selective autophagy of misfolded proteins. EMBO Rep. 2011;12:149–156. doi: 10.1038/embor.2010.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geertsma ER, Groeneveld M, Slotboom DJ, Poolman B. Quality control of overexpressed membrane proteins. Proc Natl Acad Sci U S A. 2008;105:5722–5727. doi: 10.1073/pnas.0802190105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grove DE, Fan CY, Ren HY, Cyr DM. The endoplasmic reticulum-associated Hsp40 DNAJB12 and Hsc70 cooperate to facilitate RMA1 E3-dependent degradation of nascent CFTRDeltaF508. Mol Biol Cell. 2011;22:301–314. doi: 10.1091/mbc.E10-09-0760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hidvegi T, Ewing M, Hale P, Dippold C, Beckett C, Kemp C, Maurice N, Mukherjee A, Goldbach C, Watkins S, et al. An autophagy-enhancing drug promotes degradation of mutant alpha1-antitrypsin Z and reduces hepatic fibrosis. Science. 2010;329:229–232. doi: 10.1126/science.1190354. [DOI] [PubMed] [Google Scholar]
- Hirsch C, Gauss R, Horn SC, Neuber O, Sommer T. The ubiquitylation machinery of the endoplasmic reticulum. Nature. 2009;458:453–460. doi: 10.1038/nature07962. [DOI] [PubMed] [Google Scholar]
- Houck SA, Cyr DM. Mechanisms for quality control of misfolded transmembrane proteins. Biochim Biophys Acta. 2012;1818:1108–1114. doi: 10.1016/j.bbamem.2011.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janovick JA, Knollman PE, Brothers SP, Ayala-Yanez R, Aziz AS, Conn PM. Regulation of G protein-coupled receptor trafficking by inefficient plasma membrane expression - Molecular basis of an evolved strategy. Journal of Biological Chemistry. 2006;281:8417–8425. doi: 10.1074/jbc.M510601200. [DOI] [PubMed] [Google Scholar]
- Jardon-Valadez E, Ulloa-Aguirre A, Pineiro A. Modeling and molecular dynamics simulation of the human gonadotropin-releasing hormone receptor in a lipid bilayer. J Phys Chem B. 2008;112:10704–10713. doi: 10.1021/jp800544x. [DOI] [PubMed] [Google Scholar]
- Joo JH, Dorsey FC, Joshi A, Hennessy-Walters KM, Rose KL, McCastlain K, Zhang J, Iyengar R, Jung CH, Suen DF, et al. Hsp90-Cdc37 chaperone complex regulates Ulk1- and Atg13-mediated mitophagy. Mol Cell. 2011;43:572–585. doi: 10.1016/j.molcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaushal S. Effect of rapamycin on the fate of P23H opsin associated with retinitis pigmentosa (an American Ophthalmological Society thesis) Trans Am Ophthalmol Soc. 2006;104:517–529. [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL. Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell. 2013;152:290–303. doi: 10.1016/j.cell.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, et al. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abeliovich H, Agostinis P, Agrawal DK, Aliev G, Askew DS, Baba M, Baehrecke EH, Bahr BA, Ballabio A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Honda I, Itakura E, Fujiwara TK, Mizushima N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy. 2013;9 doi: 10.4161/auto.25529. [DOI] [PubMed] [Google Scholar]
- Lu M, Echeverri F, Moyer BD. Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors. Traffic. 2003;4:416–433. doi: 10.1034/j.1600-0854.2003.00097.x. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Morita E, Saitoh T, Akira S, Ktistakis NT, Izumi T, Noda T, Yoshimori T. Autophagy requires endoplasmic reticulum targeting of the PI3-kinase complex via Atg14L. J Cell Biol. 2010;190:511–521. doi: 10.1083/jcb.200911141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarl CA, Picard C, Khalil S, Kawasaki T, Rother J, Papolos A, Kutok J, Hivroz C, Ledeist F, Plogmann K, et al. ORAI1 deficiency and lack of store-operated Ca2+ entry cause immunodeficiency, myopathy, and ectodermal dysplasia. J Allergy Clin Immunol. 2009;124:1311–1318. doi: 10.1016/j.jaci.2009.10.007. e1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacham GC, Patterson C, Zhang W, Younger JM, Cyr DM. The Hsc70 co-chaperone CHIP targets immature CFTR for proteasomal degradation. Nat Cell Biol. 2001;3:100–105. doi: 10.1038/35050509. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Ohsumi Y. The role of Atg proteins in autophagosome formation. Annu Rev Cell Dev Biol. 2011;27:107–132. doi: 10.1146/annurev-cellbio-092910-154005. [DOI] [PubMed] [Google Scholar]
- Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Floto RA, Berger Z, Imarisio S, Cordenier A, Pasco M, Cook LJ, Rubinsztein DC. Lithium induces autophagy by inhibiting inositol monophosphatase. J Cell Biol. 2005;170:1101–1111. doi: 10.1083/jcb.200504035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teckman JH, Perlmutter DH. Retention of mutant alpha(1)-antitrypsin Z in endoplasmic reticulum is associated with an autophagic response. Am J Physiol Gastrointest Liver Physiol. 2000;279:G961–974. doi: 10.1152/ajpgi.2000.279.5.G961. [DOI] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. J Biol Chem. 2008;283:7445–7454. doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. Alpha-Synuclein is degraded by both autophagy and the proteasome. J Biol Chem. 2003;278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- Yang Y, Fiskus W, Yong B, Atadja P, Takahashi Y, Pandita TK, Wang HG, Bhalla KN. Acetylated hsp70 and KAP1-mediated Vps34 SUMOylation is required for autophagosome creation in autophagy. Proc Natl Acad Sci U S A. 2013;110:6841–6846. doi: 10.1073/pnas.1217692110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younger JM, Chen L, Ren HY, Rosser MF, Turnbull EL, Fan CY, Patterson C, Cyr DM. Sequential quality-control checkpoints triage misfolded cystic fibrosis transmembrane conductance regulator. Cell. 2006;126:571–582. doi: 10.1016/j.cell.2006.06.041. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.