Abstract
Objectives: In contrast to coronary heart disease (CHD), genetic variants that influence susceptibility to peripheral arterial disease (PAD) remain largely unknown.
Background: We performed a two-stage genomic association study leveraging an electronic medical record (EMR) linked-biorepository to identify genetic variants that mediate susceptibility to PAD.
Methods: PAD was defined as a resting/post-exercise ankle-brachial index (ABI) ≤0.9 or ≥1.4 and/or history of lower extremity revascularization. Controls were patients without history of PAD. In Stage I we performed a genome-wide association analysis adjusting for age and sex, of 537, 872 SNPs in 1641 PAD cases (66 ± 11 years, 64% men) and 1604 control subjects (61 ± 7 year, 60% men) of European ancestry. In Stage II we genotyped the top 48 SNPs that were associated with PAD in Stage I, in a replication cohort of 740 PAD cases (70 ± 11 year, 63% men) and 1051 controls (70 ± 12 year, 61% men).
Results: The SNP rs653178 in the ATXN2-SH2B3 locus was significantly associated with PAD in the discovery cohort (OR = 1.23; P = 5.59 × 10−5), in the replication cohort (OR = 1.22; 8.9 × 10−4) and in the combined cohort (OR = 1.22; P = 6.46 × 10−7). In the combined cohort this SNP remained associated with PAD after additional adjustment for cardiovascular risk factors including smoking (OR = 1.22; P = 2.15 × 10−6) and after excluding patients with ABI > 1.4 (OR = 1.24; P = 3.98 × 10−7). The SNP is in near-complete linkage disequilibrium (LD) (r2 = 0.99) with a missense SNP (rs3184504) in SH2B3, a gene encoding an adapter protein that plays a key role in immune and inflammatory response pathways and vascular homeostasis. The SNP has pleiotropic effects and has been previously associated with multiple phenotypes including myocardial infarction.
Conclusions: Our findings suggest that the ATXN2-SH2B3 locus influences susceptibility to PAD.
Keywords: genome-wide association study, peripheral arterial disease, ankle-brachial index, electronic medical records, biorepository
Introduction
Peripheral arterial disease (PAD) affects nearly 10 million people in the US and more than 200 million people worldwide (Hirsch et al., 2001; Fowkes et al., 2013). PAD is associated with significant mortality and morbidity, underscoring the need to discover genetic variants that mediate susceptibility to this disease (Leeper et al., 2012). In contrast to coronary heart disease (CHD), genetic variants that influence susceptibility to PAD remain unknown. A genome-wide association study (GWAS) of smoking quantity revealed a variant in CHRNA3 that was associated with PAD and lung cancer (Thorgeirsson et al., 2008).
Repositories of DNA from patients seen in the clinical setting and linked to the electronic medical record (EMR) systems can be leveraged to conduct genotyping or sequencing studies to identify genetic variants associated with human diseases and related quantitative traits. Extensive clinical data residing in the EMR can be leveraged for high-throughput phenotyping of medically relevant traits (Kullo et al., 2010). Such an approach may reduce the time, effort, and cost involved in conducting genomic studies to identify disease susceptibility loci.
The Electronic Medical Records and Genomics (eMERGE) consortium (McCarty et al., 2011) was created to develop and implement approaches for leveraging biorepositories linked to the EMR for large-scale genomic research, including but not limited to GWAS, sequencing, and structural variation (Kho et al., 2011). We undertook a GWAS of PAD cases and controls identified from the EMR using a two-stage study design. In Stage I we performed a GWAS of 1641 PAD cases and 1604 controls, and in Stage II we attempted replication of the top significant SNPs in an independent sample of 740 PAD cases and 1051 controls.
Materials and methods
Study participants
All participants gave written informed consent for participation in the study and the use of their data for future research. The Institutional Review Board of the Mayo Clinic approved the study protocol.
Ascertainment of PAD cases and controls
The PAD patients were recruited from the non-invasive vascular laboratory at the Mayo Clinic Rochester, MN, based on the following criteria: (1) an ankle brachial index (ABI) of ≤0.9 at rest or 1 min after exercise, along with an abnormal continuous wave Doppler signal in one of the lower extremity arteries; (2) history of lower extremity revascularization if the ABI was normal; and (3) ABI ≥ 1.4 or ankle systolic BP > 250 mm Hg, representing poorly compressible arteries. Exclusion criteria included PAD secondary to vasculitis, radiation to the abdomen or lower extremities, trauma to a lower extremity artery, thrombophilia, and arterial thrombosis. Controls were identified from patients referred to the Cardiovascular Health Clinic for exercise ECG to screen for cardiovascular disease. We excluded patients who had a positive exercise ECG, were younger than age 50, or had an abnormal ABI or history of PAD. A proportion (60%) of the subjects who underwent exercise ECG also underwent measurement of ABI. The prevalence of an abnormal ABI in patients who had a negative stress ECG was <1%.
Patient-level data elements in the Mayo EMR included demographics, outpatient visits and hospitalizations, providers, diagnosis and procedure codes, and results of non-invasive lower extremity arterial evaluation. Birth date, race, sex, and ethnicity were obtained from the demographic database; the categories for race were “White,” “Black or African American,” “Hispanic,” “Asian/Pacific Islander,” “American Indian/Alaskan Native,” “Others,” and “Unknown.”
Stage-I: high-density genotyping of discovery cohort
Genotyping was performed using the Illumina 660W-Quad BeadChip at the Center for Genotyping and Analysis at the Broad Institute, Cambridge, MA. This platform consists of 561,490 SNPs and 95,876 intensity-only probes. In addition to 3347 patient DNA samples, 58 blind duplicates, and 37 Coriell controls were genotyped. The Coriell controls include 1 trio (3 unique samples) that was duplicated on each plate. Genotyping calls were made using BeadStudio version 3.3.7 (2010).
Analysis tools used for quality control (QC) procedures included Illumina BeadStudio (2010), PLINK (Purcell et al., 2007), R (The R Development Core Team, 2007), STRUCTURE (Pritchard et al., 2000), and Eigenstrat in the Eigensoft package (Price et al., 2006). Data were cleaned using the QC pipeline developed by the eMERGE Genomics Working Group (Turner et al., 2011). This process includes evaluation of sample and marker call rate, gender mismatch and anomalies, duplicate and HapMap concordance, batch effects, Hardy-Weinberg equilibrium, sample relatedness, and population stratification. The data from all the patients, in addition to the HapMap II populations, were evaluated for population structure/substructure using Eigenstrat (Price et al., 2006). Of the 3347 unique samples, 3336 passed genotyping QC (see Supplementary Data and Figures S1–S3).
Stage-II: genotyping of lead SNPs in the replication cohort
The replication cohort consisted of 744 (470 males and 274 females) patients who had PAD based on the criteria listed above and 1053 (645 males and 408 females) controls with no prior history of PAD. The top 48 SNPs associated with PAD in the discovery cohort were genotyped using an Illumina custom genotyping panel with primers and probes from Assay-by-Design (Applied Biosystems, Foster City, CA). Custom capture and genotyping was performed at Mayo Clinic's Genotyping Core lab/Genotyping Shared Resource Lab.
Standard QC procedures were applied including evaluation of sample and marker call rate, HapMap concordance, and Hardy-Weinberg in controls only. We excluded six patients with low call rates (<95%). Of the 48 SNPs selected for replication, one (rs7900716) had a low call rate. All the 47 remaining SNPs had call rates >99% and Hardy-Weinberg P-value > 0.05 in the controls.
Statistical analyses
Statistical analyses were conducted using SAS v. 9.3 {SAS Institute Inc., Cary, NC} and PLINK v1.07 (Purcell et al., 2007), and plots were created using R v2.11.0 (The R Development Core Team). Descriptive analyses were performed for the covariates and outcome variables using t-tests for continuous variables and chi-square tests for discrete variables. To adjust for population stratification, we used principal components to identify outliers in the study cohort (Price et al., 2006). Quantile-quantile (QQ) plots of observed –log10 P-values for PAD association versus the expected –log10 P-values under the null hypothesis of no association were generated to display the potential significant associations and to calculate the genomic inflation factor λ and to check for over dispersion of the test statistics. For each locus, we determined the set of HapMap SNPs in linkage disequilibrium (LD) (r2 > 0.5) with the most significantly associated SNP. We then bounded the associated interval by the flanking HapMap recombination hotspots. These windows are likely to contain the causal variants explaining the associations. We used logistic regression analyses that adjusted for age and sex to identify the SNPs associated with PAD case/control status in the discovery, replication, and combined sets. All analyses were forced to test the same allele as the original sample. We performed sensitivity analyses by including additional adjustment variables for smoking, CHD, statin use, diastolic and systolic blood pressure, and diabetes. Since the additional adjustment variables did not have a qualitative impact on the final inferences, the results are not shown.
Functional annotation of the lead SNP
Data for the SNP rs3184504 (c.784T>C), which is in nearly complete LD with the most significant SNP, were obtained from the Exome Variant Server. The impact of the variant was assessed using SIFT (Ng and Henikoff, 2003), PoplyPhen2 (Adzhubei et al., 2010), and conservation based measures such as PhastCons (Siepel et al., 2005), GRANTHAM (Grantham, 1974), and GERP (Cooper et al., 2005) scores. We performed Gene Ontology (GO) term enrichment analysis of SH2B3 using first-degree interacting partners that were obtained from the protein-protein interaction database “STRING” (http://www.string-db.org). To understand the impact of SNP rs3184504 on protein structure, we performed a molecular dynamics simulation using GROMACS v4.5.7 (http://www.gromacs.org/) of the pleckstrin homolog (PH) domain of the SH2B3 protein where the SNP is localized.
Results
Discovery
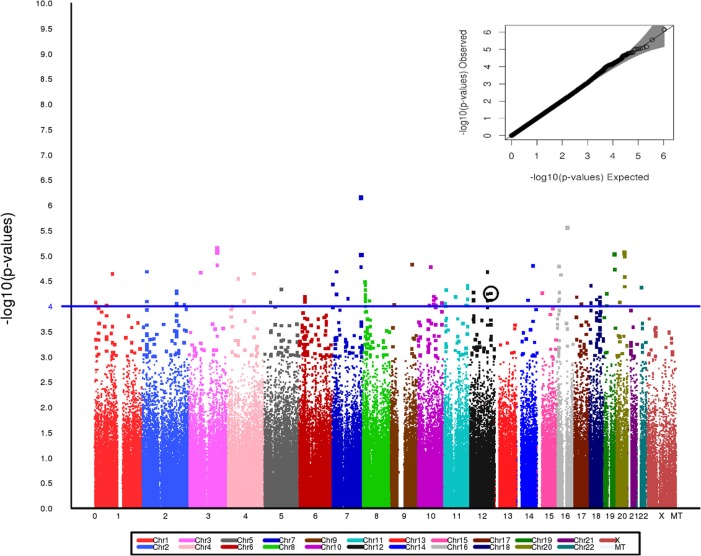
After exclusions based on QC, including removal of related individuals and those of non-European Ancestry, a total of 3245 individuals—1641 PAD subjects and 1604 controls—were included in the analyses. No evidence of population stratification was found and therefore correction for population stratification was not needed in the analyses. Since the estimate of λ was 1.0, the test statistics showed no significant over dispersion. The study population demographic and clinical characteristics by case-control status are presented in Table 1. Among PAD cases, 64.3% were men, while among the controls, 60.3% were men. The mean age of the PAD patients was higher than the mean age of the control patients (65.7 years vs. 60.8 years) (Table 1). Assuming an additive genetic model and adjusting for age and sex, 60 SNPs were associated with PAD at P < 1 × 10−4. Figure 1 presents a Manhattan plot of the P-values. Of these 60 SNPs, 48 were selected for replication based on Illumina designability score, LD, and minor allele frequency (MAF) in controls (see Supplementary Data for details).
Table 1.
Participant characteristics.
| Discovery cohort | Replication cohort | |||
|---|---|---|---|---|
| Cases (n = 1641) | Controls (n = 1604) | Cases (n = 740) | Controls (n = 1051) | |
| Men, n (%) | 1055 (64.3) | 968 (60.3) | 468 (63.2) | 643 (61.2) |
| Age, years | 65.7 ± 10.68 | 60.8 ± 7.41‡ | 70.6 ± 11.60 | 70.2 ± 12.42 |
| European ancestry, n (%) | 1547 (94.3) | 1512 (94.3) | 721 (97.4) | 1023 (97.3) |
| “Ever” smoker, n (%) | 1322 (80.5) | 963 (60.1) | 632 (85.4) | 641 (61.0) |
| ABI (pre-exercise) | 0.72 ± 0.25 | 1.1 ± 0.07‡ | 0.79 ± 0.30 | 1.07 ± 0.16‡ |
| ABI (post-exercise) | 0.54 ± 0.25 | 1.1 ± 0.12‡ | 0.56 ± 0.28 | 1.03 ± 0.19‡ |
| Hypertension, n (%) | 1358 (82.8) | 843 (52.6)‡ | 583 (78.8) | 634 (60.3)‡ |
| Type 2 diabetes, n (%) | 507 (30.9) | 141 (8.8)‡ | 225 (30.4) | 126 (12.0)‡ |
| Statin use, n (%) | 774 (49.2) | 398 (24.8)‡ | 532 (72.0) | 326 (61.1)‡ |
| CHD, n (%) | 903 (55) | 251 (15.6)‡ | 483 (65.3) | 235 (22.4)‡ |
Continuous traits are depicted as mean ± standard deviation and categorical traits as count (percent); ABI, ankle-brachial index; CHD, coronary heart disease.
P < 0.001 for differences between PAD cases and controls.
Figure 1.
Manhattan plot for the Stage I discovery cohort showing genome wide P-value distribution and the corresponding Q-Q plot (inset). SNP rs653178 is circled. A line at p = 1 × 10−4 is included on the Manhattan plot to provide a visual reference for p-values across the chromosomes.
Replication
Characteristics of participants in the discovery and replication cohorts are presented in Table 1. The allele C of the intronic SNP rs653178 at the ATXN2-SH2B3 locus on chromosome 12 was present more frequently in PAD cases (52%) than in controls (47%) with a resulting odds ratio (OR) of 1.23 (95% CI, 1.11–1.36, P = 5.59 × 10−5) in the discovery cohort (Table 2). In the replication cohort, the OR was 1.25 (95% CI, 1.10–1.40, P = 8.94 × 10−4) and in the combined sample, the OR was 1.22 (95% CI, 1.13–1.32, P = 6.46 × 10−7) (Table 2). The lead SNP rs653178 is in strong LD (r2 = 0.99) with a missense SNP (rs3184504) in SH2B3, an adapter protein that plays a key role in immune and inflammatory response pathways and vascular homeostasis (Devalliere and Charreau, 2011; Devalliere et al., 2012). A locus specific visualization of lead variants associated with PAD is provided in the Supplementary Data (Figure S4).
Table 2.
Association of rs653178 with PAD in discovery, replication, and combined cohorts, after adjustment for age and sex in logistic regression models.
| Cohort | PAD (n) | Controls (n) | Risk allele (frequency*) | OR (95% CI) | P-value |
|---|---|---|---|---|---|
| Discovery | 1641 | 1604 | C (0.469) | 1.23 (1.11,1.36) | 5.59 × 10−5 |
| Replication | 740 | 1051 | C (0.475) | 1.25 (1.10,1.40) | 8.90 × 10−4 |
| Combined | 2381 | 2655 | C (0.469) | 1.22 (1.13,1.32) | 6.46 × 10−7 |
CI, confidence interval; OR, odds ratio; PAD, peripheral disease.
Controls.
Two additional SNPs rs11726269 (intronic region of MAPK10) and rs131408 (intergenic region between LOC388882 and IGLL1) were significant at P < 0.05 in the replication cohort with similar direction of effect. However the P values exceeded the Bonferroni threshold for testing 48 SNPs (see Supplementary Table S1). The two most significant SNPs in the discovery cohort, rs7795096 in PRKAG2 on chromosome 7 and rs2587888 in GNAO1 on chromosome 16, did not replicate.
Structural and functional implications of the W262R variant in the SH2B3 protein
Our analyses indicate that SH2B3 encodes a multi-functional protein involved in diverse molecular pathways. Comparative protein sequence analyses using wild type and mutant sequences indicated that that rs3184504 leads to substitution of tryptophan with arginine (W262R) thereby introducing a new cAMP phosphorylation site in the PH domain of SH2B3 (see Supplementary Data and Figure S5). The PH domain in SH2B3 is important for lipid binding, membrane tethering and protein-protein interactions. GO terms (Ashburner et al., 2000) that are enriched among proteins interacting with SH2B3 include blood coagulation; wound healing, and cell signaling events (see Supplementary Data, Table S2 and Figure S6). Conservation measures like Genomic Evolutionary Rate Profiling (GERP: 2.97) and phastCons (posterior probability: 0.159) suggest the variant is marginally conserved. Effect prediction analysis using Variant Effect Predicator (McLaren et al., 2010) indicate the variant as tolerant (SIFT: score = 1) benign (PolyPhen-2; score = 0.0), and moderately radical (GRANTHAM; score = 101). Molecular dynamic simulation suggested that the mutation in the PH domain of the SH2B3 results in structural perturbations and conformational changes (see Supplementary Data; Figures S7 and S8).
Discussion
A better understanding of the genetic basis of PAD is required to improve risk stratification and identify new pathophysiologic pathways and drug targets. Conventional linkage and association approaches have failed to identify replicable susceptibility loci for PAD (Leeper et al., 2012) and the genome-wide association approach is currently the most promising design to uncover such loci. Heritable factors contribute to the risk of developing PAD. In the large population-based Swedish Twin Registry (Wahlgren and Magnusson, 2011), the odds ratio of having PAD in persons whose twin had PAD compared with persons whose twin did not have PAD was 17.7 (95% CI, 11.7–26.6) for monozygotic twins and 5.7 (95% CI, 4.1–7.9) for dizygotic twins. In a large case control study we found that family history of PAD was associated with doubling the odds of the presence of PAD (Khaleghi et al., 2014). Heritability estimates for ABI have varied from 0.21 (Kullo et al., 2006; Murabito et al., 2006) to 0.48 (Carmelli et al., 2000). In spite of evidence supporting the presence of heritable contribution to PAD, little is known about the genetic determinants of PAD.
In the present study, the SNP most strongly associated with PAD was an intronic SNP rs653178 in ATXN2 on chromosome 12q24-12q24.1. This SNP is in near-complete LD with a missense SNP in SH2B adaptor protein 3 gene (SH2B3) (rs3184504; r2 = 0.99) that is likely the causal SNP. The SNP in SH2B3 results in a substitution of tryptophan (large size and aromatic side chain) by arginine (large size and basic side chain) that induces changes in the structure and hydrophilic properties of the pleckstrin homology domain. This may result in altered lipid binding and protein–protein interactions as indicated by our molecular dynamics analyses. The variant also introduces a new phosphorylation site in the pleckstrin homology domain which may influence signaling pathways mediated by SH2B3. The SNP rs3184504 exhibits significant pleiotropic effects and has been implicated in immunological disorders, cardiovascular diseases (Gudbjartsson et al., 2009) and hematologic traits such as platelet count, mean-platelet volume (Gieger et al., 2011) and eosinophil count (Barrett et al., 2009). A summary of disease/trait associations of rs3184504 and rs653178 in the ATXN2-SH2B3 locus is provided in the Supplement (Table S3).
The pleiotropic nature of SH2B3 may be due to its role in immune and inflammatory signaling pathways including erythropoietin, cytokine receptor-mediated and integrin signaling (20). The protein also regulates hematopoietic cell lineage and endothelial cells, and influences adhesion and migration of platelets by modulating actin cytoskeleton organization (Takizawa et al., 2010; Gieger et al., 2011; Devalliere et al., 2012; Shameer et al., 2014). SH2B3 is also involved in platelet production via megakaryocyte development; mice lacking SH2B3 (Lnk/SH2B3−/−) (Kwon et al., 2009) have altered platelet function and thrombus development (Tong et al., 2005). The relatively high frequency of this SNP in the general population is speculated to be due to a protective effect against bacterial infection (Zhernakova et al., 2010). We (Ding and Kullo, 2011) and others (Pickrell et al., 2009) have previously demonstrated that the SNP may have been subject to natural selection.
Two GWAS in European ancestry cohorts have reported variants associated with PAD. Thorgeirsson et al (Thorgeirsson et al., 2008) found a common variant in the nicotinic acetylcholine receptor gene cluster on chromosome 15q24 to affect nicotine dependence, smoking quantity, and the risk of PAD and lung cancer. A synonymous SNP (rs1051730) within the cholinergic receptor nicotinic alpha 3 gene (CHRNA3) was significantly associated with PAD (OR = 1.19). In a meta-analysis (Murabito et al., 2012) of GWAS for ABI consisting of 21 population-based cohort studies and 41,692 participants of European ancestry among whom 3409 participants had PAD (ABI < 0.90), six SNPs were associated (P = 1 × 10−6) with PAD, but none at a genome-wide significance level. The ATXN2-SH2B3 locus was not associated with PAD in this study. One possible explanation may be the differences in case ascertainment, the present study including symptomatic PAD patients from the clinical setting whereas in the meta-analyses by Murabito et al, most individuals had undergone ABI measurement as part of prospective cohort studies. Koriyama et al. (2010) found the OSBPL10 locus to be associated with PAD in a Japanese cohort. We assessed the strength of association of these SNPs in our dataset and found that the 9p21 variant and the OSBPL10 variants were not associated, whereas the CHRNA3 variant was weakly (P = 1 × 10−3) associated with PAD case status.
In conclusion, our findings suggest that SNP rs653178 in the ATXN2-SH2B3 locus is associated with clinically defined PAD. The SNP is in near complete LD with rs3184504, a non-synonymous SNP in SH2B3, a gene implicated in immune, inflammatory, and hematopoietic pathways. This SNP is emerging as a key pleiotropic genetic variant influencing multiple cardiovascular traits. Our findings motivate additional investigation of this locus including sequencing, gene expression and drug targeting studies as well as studies in experimental animals.
Sources of funding
This work was supported by grants HG-04599 and HG-06379 from the National Human Genome Research Institute (NHGRI), Bethesda, MD. The eMERGE Network was initiated and funded by NHGRI, with additional funding from National Institute of General Medical Sciences (NIGMS), Bethesda, MD.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Genotyping Core Lab/Genotyping Shared Resource Lab at Mayo Clinic for technical assistance.
Supplementary material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fgene.2014.00166/abstract
References
- Adzhubei I. A., Schmidt S., Peshkin L., Ramensky V. E., Gerasimova A., Bork P., et al. (2010). A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., et al. (2000). Gene ontology: tool for the unification of biology. Gene Ontol. Consort. Nat. Genet. 25, 25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett J. C., Clayton D. G., Concannon P., Akolkar B., Cooper J. D., Erlich H. A., et al. (2009). Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 41, 703–707 10.1038/ng.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmelli D., Fabsitz R. R., Swan G. E., Reed T., Miller B., Wolf P. A. (2000). Contribution of genetic and environmental influences to ankle-brachial blood pressure index in the NHLBI Twin Study. National Heart, Lung, and Blood Institute. Am. J. Epidemiol. 151, 452–458 10.1093/oxfordjournals.aje.a010230 [DOI] [PubMed] [Google Scholar]
- Cooper G. M., Stone E. A., Asimenos G., Green E. D., Batzoglou S., Sidow A. (2005). Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 15, 901–913 10.1101/gr.3577405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devalliere J., Charreau B. (2011). The adaptor Lnk (SH2B3): an emerging regulator in vascular cells and a link between immune and inflammatory signaling. Biochem. Pharmacol. 82, 1391–1402 10.1016/j.bcp.2011.06.023 [DOI] [PubMed] [Google Scholar]
- Devalliere J., Chatelais M., Fitau J., Gerard N., Hulin P., Velazquez L., et al. (2012). LNK (SH2B3) is a key regulator of integrin signaling in endothelial cells and targets alpha-parvin to control cell adhesion and migration. FASEB J. 26, 2592–2606 10.1096/fj.11-193383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding K., Kullo I. J. (2011). Geographic differences in allele frequencies of susceptibility SNPs for cardiovascular disease. BMC Med. Genet. 12:55 10.1186/1471-2350-12-55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowkes F. G., Rudan D., Rudan I., Aboyans V., Denenberg J. O., McDermott M. M., et al. (2013). Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet 382, 1329–1340 10.1016/S0140-6736(13)61249-0 [DOI] [PubMed] [Google Scholar]
- Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., et al. (2011). New gene functions in megakaryopoiesis and platelet formation. Nature 480, 201–208 10.1038/nature10659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R. (1974). Amino acid difference formula to help explain protein evolution. Science 185, 862–864 10.1126/science.185.4154.862 [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D. F., Bjornsdottir U. S., Halapi E., Helgadottir A., Sulem P., Jonsdottir G. M., et al. (2009). Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 41, 342–347 10.1038/ng.323 [DOI] [PubMed] [Google Scholar]
- Hirsch A. T., Criqui M. H., Treat-Jacobson D., Regensteiner J. G., Creager M. A., Olin J. W., et al. (2001). Peripheral arterial disease detection, awareness, and treatment in primary care. JAMA 286, 1317–1324 10.1001/jama.286.11.1317 [DOI] [PubMed] [Google Scholar]
- Illumina. (2010). Illumina BeadStudio Data Analysis Software Modules. Available online at: http://www.illumina.com/Documents/products/datasheets/datasheet_beadstudio.pdf
- Khaleghi M., Isseh I. N., Jouni H., Bailey K. R., Kullo I. J. (2014). Family history as a risk factor for peripheral arterial disease. Am. J. Cardiol. (in press). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kho A. N., Pacheco J. A., Peissig P. L., Rasmussen L., Newton K. M., Weston N., et al. (2011). Electronic medical records for genetic research: results of the eMERGE consortium. Sci. Transl. Med. 3, 79re71 10.1126/scitranslmed.3001807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koriyama H., Nakagami H., Katsuya T., Sugimoto K., Yamashita H., Takami Y., et al. (2010). Identification of evidence suggestive of an association with peripheral arterial disease at the OSBPL10 locus by genome-wide investigation in the Japanese population. J. Atheroscler. Thromb. 17, 1054–1062 10.5551/jat.4291 [DOI] [PubMed] [Google Scholar]
- Kullo I. J., Fan J., Pathak J., Savova G. K., Ali Z., Chute C. G. (2010). Leveraging informatics for genetic studies: use of the electronic medical record to enable a genome-wide association study of peripheral arterial disease. J. Am. Med. Inform. Assoc. 17, 568–574 10.1136/jamia.2010.004366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kullo I. J., Turner S. T., Kardia S. L., Mosley T. H., Jr., Boerwinkle E., de Andrade M. (2006). A genome-wide linkage scan for ankle-brachial index in African American and non-Hispanic white subjects participating in the GENOA study. Atherosclerosis 187, 433–438 10.1016/j.atherosclerosis.2005.10.003 [DOI] [PubMed] [Google Scholar]
- Kwon S. M., Suzuki T., Kawamoto A., Ii M., Eguchi M., Akimaru H., et al. (2009). Pivotal role of lnk adaptor protein in endothelial progenitor cell biology for vascular regeneration. Circ. Res. 104, 969–977 10.1161/CIRCRESAHA.108.192856 [DOI] [PubMed] [Google Scholar]
- Leeper N. J., Kullo I. J., Cooke J. P. (2012). Genetics of peripheral artery disease. Circulation 125, 3220–3228 10.1161/CIRCULATIONAHA.111.033878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty C. A., Chisholm R. L., Chute C. G., Kullo I. J., Jarvik G., Larson E. B., et al. (2011). The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med. Genomics 4:13 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. (2010). Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26, 2069–2070 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murabito J. M., Guo C. Y., Fox C. S., D'Agostino R. B. (2006). Heritability of the ankle-brachial index: the Framingham Offspring study. Am. J. Epidemiol. 164, 963–968 10.1093/aje/kwj295 [DOI] [PubMed] [Google Scholar]
- Murabito J. M., White C. C., Kavousi M., Sun Y. V., Feitosa M. F., Nambi V., et al. (2012). Association between chromosome 9p21 variants and the ankle-brachial index identified by a meta-analysis of 21 genome-wide association studies. Circ. Cardiovasc. Genet. 5, 100–112 10.1161/CIRCGENETICS.111.961292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P. C., Henikoff S. (2003). SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 10.1093/nar/gkg509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell J. K., Coop G., Novembre J., Kudaravalli S., Li J. Z., Absher D., et al. (2009). Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 19, 826–837 10.1101/gr.087577.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Patterson N. J., Plenge R. M., Weinblatt M. E., Shadick N. A., Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38, 904–909 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Pritchard J. K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155, 945–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M. A., Bender D., et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer K., Denny J. C., Ding K., Jouni H., Crosslin D. R., de Andrade M., et al. (2014). A genome- and phenome-wide association study to identify genetic variants influencing platelet count and volume and their pleiotropic effects. Hum. Genet. 133, 95–109 10.1007/s00439-013-1355-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepel A., Bejerano G., Pedersen J. S., Hinrichs A. S., Hou M., Rosenbloom K., et al. (2005). Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 10.1101/gr.3715005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takizawa H., Nishimura S., Takayama N., Oda A., Nishikii H., Morita Y., et al. (2010). Lnk regulates integrin alphaIIbbeta3 outside-in signaling in mouse platelets, leading to stabilization of thrombus development in vivo. J. Clin. Invest. 120, 179–190 10.1172/JCI39503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The R Development Core Team. (2007). R: A Language and Environment for Statistical Computing. Available online at: http://www.r-project.org/
- Thorgeirsson T. E., Geller F., Sulem P., Rafnar T., Wiste A., Magnusson K. P., et al. (2008). A variant associated with nicotine dependence, lung cancer and peripheral arterial disease. Nature 452, 638–642 10.1038/nature06846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W., Zhang J., Lodish H. F. (2005). Lnk inhibits erythropoiesis and Epo-dependent JAK2 activation and downstream signaling pathways. Blood 105, 4604–4612 10.1182/blood-2004-10-4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner S., Armstrong L. L., Bradford Y., Carlson C. S., Crawford D. C., Crenshaw A. T., et al. (2011). Quality control procedures for genome wide association studies. Curr. Protoc. Hum. Genet. Chapter 1; Unit 1: 19 10.1002/0471142905.hg0119s68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahlgren C. M., Magnusson P. K. (2011). Genetic influences on peripheral arterial disease in a twin population. Arterioscler. Thromb. Vasc. Biol. 31, 678–682 10.1161/ATVBAHA.110.210385 [DOI] [PubMed] [Google Scholar]
- Zhernakova A., Elbers C. C., Ferwerda B., Romanos J., Trynka G., Dubois P. C., et al. (2010). Evolutionary and functional analysis of celiac risk loci reveals SH2B3 as a protective factor against bacterial infection. Am. J. Hum. Genet. 86, 970–977 10.1016/j.ajhg.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.