Abstract
Variations in a locus at chromosome 10q26 are strongly associated with the risk of age-related macular degeneration (AMD). The most significantly associated haplotype includes a nonsynonymous SNP rs10490924 in the exon 1 of ARMS2 and rs11200638 in the promoter region of HTRA1. It is under debate which gene(s), ARMS2, HTRA1 or some other genes are functionally responsible for the genetic association. To verify whether the associated variants correlate with a higher HTRA1 expression level as previously reported, HTRA1 mRNA and protein were measured in a larger human retina-RPE-choroid samples (n = 82). Results show there is no significant change of HTRA1 mRNA level among genotypes at rs11200638, rs10490924 or an indel variant of ARMS2. Furthermore, two AMD-associated synonymous SNPs rs1049331 and rs2293870 in HTRA1 exon 1 do not change its protein level either. These results suggest that the AMD-associated variants in the chromosome 10q26 locus do not significantly affect the expression of HTRA1.
Identifying and verifying functional consequences of disease-associated genetic variants remains a challenge in the post-genome wide association study (GWAS) era. Two loci at chromosome 10q26 and 1q31 have been strongly associated with the risk of developing age-related macular degeneration (AMD), the principal cause for irreversible visual loss in developed countries. In contrast to the successful identification of complement factor H (CFH) at chromosome 1q31 as the first major susceptibility gene for AMD, there is a debate in the scientific community about functional variants and susceptibility genes in the chromosome 10q26 locus. Three genes are located within the bounds of the chromosome 10q26 locus, pleckstrin homology domain containing family A member 1 (PLEKHA1), age-related maculopathy susceptibility 2 (ARMS2, formerly LOC387715), and HtrA serine peptidase 1 (HTRA1), which are all associated with AMD. The most significantly associated haplotype includes single nucleotide polymorphisms (SNPs) rs10490924 (nonsynonymous substitution A69S) in ARMS2 and rs11200638 in the promoter region of HTRA1. Some studies propose ARMS2 as the AMD susceptibility gene, whereas others propose HTRA1. One major reason for the inconclusive findings is that variants in genes ARMS2 and HTRA1 are in such strong linkage disequilibrium (LD) that their effects are indistinguishable using statistical analysis.
HTRA1 was proposed as the susceptibility gene primarily based on the assumption that the expression of HTRA1 is up-regulated by AMD-associated genotypes/haplotypes at the locus. SNP rs11200638 is located within a conserved CpG island and 497bp upstream from the transcription start site of HTRA1. Compared to the major allele G of rs11200638, the AMD-associated minor allele A may disrupt the CG pattern in the region, which is suggested to alter the transcription of HTRA1. One initial study showed that the risk allele of rs11200638 was correlated with a higher level of HTRA1 mRNA in blood lymphocytes by quantitative RT-PCR and a higher level of HTRA1 protein in human retinal pigment epithelium (RPE) by immunoblot (Yang et al., 2006). Subsequently, this correlation was replicated in archived retinas as well as fresh placenta tissues by immunohistochemistry and quantitative RT-PCR (Chan et al., 2007; Tuo et al., 2008; Yang et al., 2010). In contrast, several studies from independent groups, applying ex vivo and in vitro methods, have shown that the genotypes at rs11200638 or other AMD-associated variants in the chromosome 10q26 locus are not correlated with HTRA1 expression at either mRNA or protein level in human retinas or other tissues (Kanda et al., 2007; Chowers et al., 2008; Wang et al., 2010a; Kanda et al., 2010; Friedrich et al., 2011). The discordant results call into question the proposed HTRA1-AMD functional association and call for more experiments to clarify whether variants at the chromosome 10q26 locus affect the expression of HTRA1.
One major reason for the discordant results is a lack of simultaneous quantification of HTRA1 mRNA and protein in fresh human retinal tissues. To determine the relationship between variants at the chromosome 10q26 locus and HTRA1 expression, we examined HTRA1 in a larger sample of retina-RPE-choroids. A total of 82 human retina-RPE-choroids from 82 unrelated Caucasian subjects (age 75.0 ± 14.6 years old) without any known eye diseases were provided by the Florida Lions Eye Bank. Procedures for recruitment, requests for medical records, and consent forms were approved by the University of Miami, Miller School of Medicine Institutional Review Board. Eye tissues were retrieved and frozen in −80°C within 24 hrs post-mortem. Retinal tissues (including neuroretina, RPE and choroid) were punched from the macular region of frozen eyes for RNA and protein extractions. RNA was extracted by RNeasy lipid tissue kit (Qiagen) and protein was extracted by RIPA buffer (Thermo) according to the manufacturer's instructions. The quality and quantity of RNA were monitored by a Bioanalyzer 2100 (Agilent) and a NanoDrop 8000 Spectrophotometer (Thermo). The concentration of total protein was measured using a BCA protein assay kit (Thermo).
Three variants, including rs10490924, rs11200638 and ARMS2 3′UTR indel (a combination polymorphism of insertion and deletion) at the chromosome 10q26 locus have previously been strongly associated with AMD. All samples were genotyped for these variants. One other SNP rs2736911 (R38X in ARMS2) was reported to be associated inversely with AMD (Yang et al., 2010). We recently showed that this inverse association is insignificant after adjustment for sex and age. Additional analyses further suggested that the trending inverse association of rs2736911 (without adjustment for sex and age) appears to be due to strong LD with the non-risk wild-type allele at rs10490924 (Wang et al., 2012). For this reason, rs2736911 was not included in this study. Genotypes at SNPs rs10490924 and rs11200638 were assessed by Taqman assays (Life Technologies). Genotypes at the ARMS2 3′UTR indel were evaluated by PCR and gel assay as described previously (Fritsche et al., 2008; Wang et al., 2010b). The three variants are located in a strong LD region. Overall, samples include 54 individuals homozygous for protective haplotype (G-WT-G), 24 heterozygotes and 4 individuals homozygous for risk haplotype (T-indel-A).
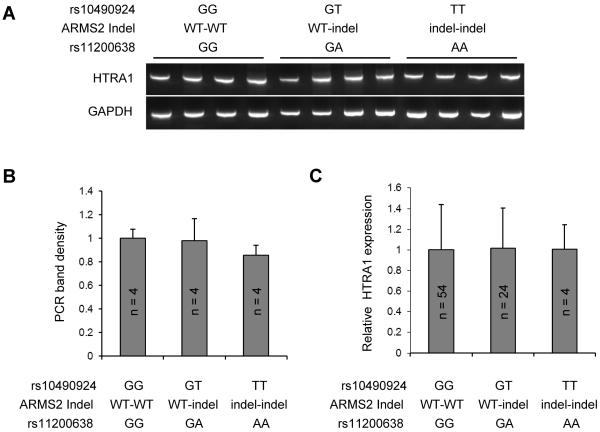
To test whether the risk haplotype is associated with a higher expression of HTRA1 as previous reported (Yang et al., 2010), primers (forward: 5′-CGGAAGATGGACTGATCGTGAC-3′; reverse: 5′-GGTGATGGCTTTTCCTTTGGC-3′) were applied for RT-PCR analysis. Twelve retina-RPE-choroid samples included 4 individuals homozygous for protective haplotype (G-WT-G), 4 heterozygotes, and 4 individuals homozygous for risk haplotype (T-indel-A). There was no significant difference in age among the three groups (data not shown). RT-PCR of housekeeper gene GAPDH was used for internal controls. PCR products were displayed at 2% agarose gel (Figure 1A) and the band density was quantified by AlphaImager software. After normalization with GAPDH band densities, no statistically significant difference was identified (P > 0.05) in HTRA1 RT-PCR bands among haplotypes of the three chromosome 10q26 locus variants using Student t test (Figure 1B).
Figure 1.
Variants at the chromosome 10q26 locus and the level of HTRA1 mRNA. A. Agarose gel images of the RT-PCR of HTRA1 and GAPDH in 12 human retina samples including 4 individuals homozygous for protective haplotype (G-WT-G), 4 heterozygotes and 4 individuals homozygous for risk haplotype (T-indel-A). B. Quantification of RT-PCR band densities. No statistically significant difference was found (P > 0.05) in HTRA1 RT-PCR bands among haplotypes of the three chromosome 10q26 locus variants. C. Analysis of quantitative RT-PCR of HTRA1 and GAPDH in 82 human retina samples including 54 individuals homozygous for protective haplotype, 24 heterozygotes and 4 individuals homozygous for risk haplotype. HTRA1 mRNA levels are not significantly different (P > 0.05) among haplotypes of the three chromosome 10q26 locus variants.
To more accurately quantify HTRA1 mRNA level, we performed quantitative RT-PCR using a Taqman gene expression assay (Life Technologies) for all 82 retina-RPE-choroid samples. There was no significant difference in age among the three haplotype groups (data not shown). The quantitative RT-PCR reaction mix contains 20× TaqMan Gene Expression Assay (1μl), 2× master mix (10μl), cDNA (1μl corresponding to the cDNA reverse transcribed from approximately 25 ng RNA) and nuclease free water (8μl). Each sample was repeated three times at different locations in the plate. The 384-well plate was then run on the 7900 HT (Life Technologies) at 50°C for 2 min, 95°C for 10 min, then 95°C for 15 s and 60°C for 1 min (for 45 cycles). The HTRA1 expression level was calculated using the 2−ΔΔCt method and normalized by GAPDH as internal controls. We did not find statistically significant differences (P > 0.05) in HTRA1 expression level among haplotypes of the three chromosome 10q26 locus variants using the Student t test (Figure 1C).
Previous studies examined HTRA1 mRNA in smaller samples with only a few samples homozygous for the risk haplotype. From 10 retinas including one sample homozygous for AA genotype rs11200638, Chan et al. observed a trending increase of HTRA1 mRNA with risk genotype at rs11200638 (Chan et al., 2007). By examining 35 retinas including six samples (5 controls and 1 AMD affected) homozygous for risk haplotype, Kanda et al found no association between AMD susceptibility variants at chromosome 10q26 and the expression level of HTRA1 (Kanda et al., 2010). Friedrich et al. quantified HTRA1 mRNA in 45 human retinas including 2 samples homozygous for risk haplotype and stated that the risk haplotype does not affect HTRA1 expression (Friedrich et al., 2011). Our group previously reported that there were no effects of rs11200638 genotype on HTRA1 expression by examining 24 retina-RPE-choroids (no homozygous risk allele sample included) (Wang et al., 2010a). In this updated sample set of 82 retina-RPE-choroids including 4 samples homozygous for risk haplotype, we found no statistically significant correlation between AMD-associated haplotype at chromosome 10q26 and the HTRA1 transcripts level. The result from these 4 groups suggests that variants at the chromosome 10q26 locus do not likely affect the transcription of HTRA1 in the retina.
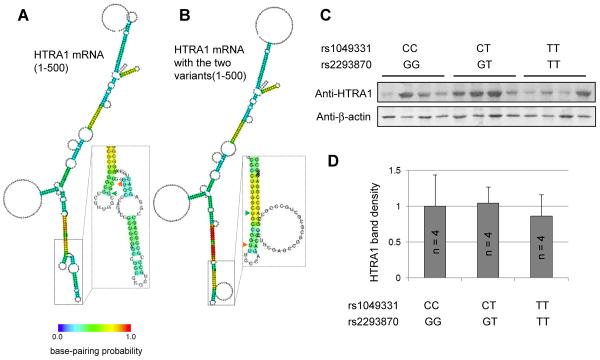
After evaluating the effect of upstream variants on HTRA1 transcription, we turned our focus on the two synonymous SNPs rs1049331 (A34A) and rs2293870 (G36G) in exon 1 of the HTRA1 gene. SNPs rs1049331 and rs2293870, located within a strong LD with rs11200638 and other variants, also associated with risk of AMD or AMD sub-phenotypes (Deangelis et al., 2008; Tam et al., 2008; Andreoli et al., 2009). Although one synonymous SNP will not result in an amino acid change, there is a possibility that the two synonymous SNPs may alter HTRA1 translation by disruptions in HTRA1 mRNA structure or tRNA preferences as previously reported in other genes (Lavner and Kotlar 2005; Chamary et al., 2006; Sauna et al., 2011). To explore this possibility, we first conducted in silico analysis. We found that SNPs rs1049331 and rs2293870 cause slight changes in the secondary structure of HTRA1 mRNA predicted by a CentroidFold algorithm (Hamada et al., 2009) (Figure 2A, B). We then applied immunoblot to semi-quantify HTRA1 protein in retina-RPE-choroids. Another set of punches was collected for protein extraction from the same 12 eye tissues used for RT-PCR analysis (Figure 1A). Genotypes at rs1049331 and rs2293870 were obtained by PCR using primers (forward: 5′-AGAGTCGCCATGCAGATCC-3′; reverse: 5′-CACAGGTTGGCGTAGGTGTT-3′) and sequencing. HTRA1 antibodies (monoclonal from R&D or polyclonal from Abcam) were used in this experiment. HTRA1 band density was quantified by AlphaImager software and normalized normalization by β-actin band densities (Figure 2C). No statistically significant difference was identified (P > 0.05) in HTRA1 protein bands among genotypes at rs1049331 and rs2293870 (Figure 2D). Risk genotype AA at rs11200638 or the aforementioned risk haplotype is not associated with a higher HTRA1 protein level in the retina-RPE-choroid either.
Figure 2.
Variants at the chromosome 10q26 locus and the level of HTRA1 protein. A. Secondary structure of HTRA1 mRNA (1–500 nt) predicted by CentroidFold. B. Secondary structure of HTRA1 mRNA (1–500 nt) carrying minor alleles at two synonymous SNPs rs1049331 (green arrow head) and rs2293870 (orange arrow head). There are slight structural changes caused by the two SNPs showing in the boxes. C. Immunoblot analysis of HTRA1 and β-actin. D. Quantification of immunoblot band densities. There is no statistical analysis among genotypes at the chromosome 10q26 locus variants.
A 1.7-fold increase of HTRA1 protein level was initially reported in wet AMD affected RPE samples with genotype AA (n = 4) compared to control RPE samples of genotype GG (n = 6) at rs11200638 by immunoblot analysis (Yang et al., 2006). However, the correlation between genotypes at rs11200638 and HTRA1 protein level is not verified in our experiments. By coordinating HTRA1 mRNA and protein level, our results further suggest that variants at chromosome 10q26, including the two synonymous SNPs, may not affect HTRA1 protein level. Further studies are needed to examine the relationship between variants at chromosome 10q26 and HTRA1 at the protein level in the retina.
A recent systemic survey for biomarkers of AMD revealed many genes that are either over- or under-expressed in AMD-affected RPE/choroid. HTRA1 was not on the list of those differentially expressed genes (Newman et al., 2012). Interestingly, overexpressing HTRA1 specifically in mouse RPE induces some phenotypes relevant to AMD in humans (Vierkotten et al., 2011; Jones et al., 2011). However, if chromosome 10q26 locus variants do not change the transcription, translation or other functions of HTRA1, it could be other genes at this locus underlying the susceptibility to AMD.
ACKNOWLEDGEMENTS
We thank all the individuals and their families who donated eyes for the study. We would also like to thank the Florida Lions Eye Bank which provided eye tissues for this research. This research was supported by National Institutes of Health grants (2R01EY012118-11 to M.A.P.-V., J.L.H., W.K.S and A.A.) and was partially supported by NIH center grant P30-EY014801 and by an unrestricted grant to the University of Miami from Research to Prevent Blindness, New York, NY.
W.K.S., M.A.P-V., A.A., and J.L.H. are co-inventors on a patent related to use of ARMS2 genotypes for diagnosis of AMD, licensed to ArcticDx. S.G.S. has served on advisory boards for Alimera Sciences, Bausch + Lomb, Eyetech, and ThromboGenics, and has received royalties from IC Labs.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
All other authors have no conflicts of interest to declare.
REFERENCES
- Andreoli MT, Morrison MA, Kim BJ, Chen L, Adams SM, Miller JW, DeAngelis MM, Kim IK. Comprehensive analysis of complement factor H and LOC387715/ARMS2/HTRA1 variants with respect to phenotype in advanced age-related macular degeneration. Am J Ophthalmol. 2009;148:869–874. doi: 10.1016/j.ajo.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan CC, Shen D, Zhou M, Ross RJ, Ding X, Zhang K, Green WR, Tuo J. Human HTRA1 in the archived eyes with age-related macular degeneration. Trans Am. Ophthalmol. Soc. 2007;105:92–97. [PMC free article] [PubMed] [Google Scholar]
- Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;2:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- Chowers I, Meir T, Lederman M, Goldenberg-Cohen N, Cohen Y, Banin E, Averbukh E, Hemo I, Pollack A, Axer-Siegel R, Weinstein O, Hoh J, Zack DJ, Galbinur T. Sequence variants in HTRA1 and LOC387715/ARMS2 and phenotype and response to photodynamic therapy in neovascular age-related macular degeneration in populations from Israel. Mol. Vis. 2008;14:2263–2271. [PMC free article] [PubMed] [Google Scholar]
- Deangelis MM, Ji F, Adams S, Morrison MA, Harring AJ, Sweeney MO, Capone A, Jr, Miller JW, Dryja TP, Ott J, Kim IK. Alleles in the HtrA serine peptidase 1 gene alter the risk of neovascular age-related macular degeneration. Ophthalmology. 2008;7:1209–1215. doi: 10.1016/j.ophtha.2007.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich U, Myers CA, Fritsche LG, Milenkovich A, Wolf A, Corbo JC, Weber BH. Risk- and non-risk-associated variants at the 10q26 AMD locus influence ARMS2 mRNA expression but exclude pathogenic effects due to protein deficiency. Hum. Mol. Genet. 2011;20:1387–1399. doi: 10.1093/hmg/ddr020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsche LG, Loenhardt T, Janssen A, Fisher SA, Rivera A, Keilhauer CN, Weber BH. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat. Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- Hamada M, Kiryu H, Sato K, Mituyama T, Asai K. Prediction of RNA secondary structure using generalized centroid estimators. Bioinformatics. 2009;25:465–473. doi: 10.1093/bioinformatics/btn601. [DOI] [PubMed] [Google Scholar]
- Kanda A, Chen W, Othman M, Branham KE, Brooks M, Khanna R, He S, Lyons R, Abecasis GR, Swaroop A. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanda A, Stambolian D, Chen W, Curcio CA, Abecasis GR, Swaroop A. Age-related macular degeneration-associated variants at chromosome 10q26 do not significantly alter ARMS2 and HTRA1 transcript levels in the human retina. Mol. Vis. 2010;16:1317–1323. [PMC free article] [PubMed] [Google Scholar]
- Jones A, Kumar S, Zhang N, Tong Z, Yang JH, Watt C, Anderson J, Amrita, Fillerup H, McCloskey M, et al. Increased expression of multifunctional serine protease, HTRA1, in retinal pigment epithelium induces polypoidal choroidal vasculopathy in mice. Proc. Natl. Acad. Sci. USA. 2011;35:14578–14583. doi: 10.1073/pnas.1102853108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;1:127–138. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- Newman AM, Gallo NB, Hancox LS, Miller NJ, Radeke CM, Maloney MA, Cooper JB, Hageman GS, Anderson DH, Johnson LV, et al. Systems-level analysis of age-related macular degeneration reveals global biomarkers and phenotype-specific functional networks. Genome Med. 2012;2:16. doi: 10.1186/gm315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;10:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Tam PO, Ng TK, Liu DT, Chan WM, Chiang SW, Chen LJ, DeWan A, Hoh J, Lam DS, Pang CP. HTRA1 variants in exudative age-related macular degeneration and interactions with smoking and CFH. Invest. Ophthalmol. Vis. Sci. 2008;6:2357–2365. doi: 10.1167/iovs.07-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuo J, Ross RJ, Reed GF, Yan Q, Wang JJ, Bojanowski CM, Chew EY, Feng X, Olsen TW, Ferris FL, 3rd, Mitchell P, Chan CC. The HTRA1 promoter polymorphism, smoking, and age-related macular degeneration in multiple case-control samples. Ophthalmology. 2008;115:1891–1898. doi: 10.1016/j.ophtha.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierkotten S, Muether PS, Fauser S. Overexpression of HTRA1 leads to ultrastructural changes in the elastic layer of Bruch's membrane via cleavage of extracellular matrix components. PLoS One. 2011;8:e22959. doi: 10.1371/journal.pone.0022959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Scott WK, Haines JL, Pericak-Vance MA. Genotype at polymorphism rs11200638 is not correlated with HTRA1 expression level. Arch. Ophthalmol. 2010a;128:1491–93. doi: 10.1001/archophthalmol.2010.256. [DOI] [PubMed] [Google Scholar]
- Wang G, Spencer KL, Scott WK, Whitehead P, Court BL, Ayala-Haedo J, Mayo P, Schwartz SG, Kovach JL, Gallins P, Polk M, Agarwal A, Postel EA, Haines JL, Pericak-Vance MA. Analysis of the 3′ UTR insertion/deletion polymorphism of ARMS2 gene in age-related macular degeneration. Hum. Genet. 2010b;127:595–602. doi: 10.1007/s00439-010-0805-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Scott WK, Agarwal A, Haines JL, Pericak-Vance MA. Coding variants in the ARMS2 gene and the risk of age-related macular degeneration. Archives of Ophthalmology. 2012 doi: 10.1001/jamaophthalmol.2013.589. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Camp NJ, Sun H, Tong Z, Gibbs D, Cameron DJ, Chen H, Zhao Y, Pearson E, Li X, Chien J, Dewan A, Harmon J, Bernstein PS, Shridhar , Zabriskie NA, Hoh J, Howes K, Zhang K. A variant of the HTRA1 gene increases susceptibility to age-related macular degeneration. Science. 2006;314:992–993. doi: 10.1126/science.1133811. [DOI] [PubMed] [Google Scholar]
- Yang Z, Tong Z, Chen Y, Zeng J, Lu F, Sun X, Zhao C, Wang K, Davey L, Chen H, London N, Muramatsu D, Salasar F, Carmona R, Kasuga D, Wang X, Bedell M, Dixie M, Zhao P, Yang R, Gibbs D, Liu X, Li Y, Li C, Li Y, Campochiaro B, Constantine R, Zack DJ, Campochiaro P, Fu Y, Li DY, Katsanis N, Zhang K. Genetic and functional dissection of HTRA1 and LOC387715 in age-related macular degeneration. PLoS. Genet. 2010;6:e1000836. doi: 10.1371/journal.pgen.1000836. [DOI] [PMC free article] [PubMed] [Google Scholar]