Abstract
BACKGROUND
There are few data on the comparative epidemiology and virology of the pandemic 2009 influenza A (H1N1) virus and cocirculating seasonal influenza A viruses in community settings.
METHODS
We recruited 348 index patients with acute respiratory illness from 14 outpatient clinics in Hong Kong in July and August 2009. We then prospectively followed household members of 99 patients who tested positive for influenza A virus on rapid diagnostic testing. We collected nasal and throat swabs from all household members at three home visits within 7 days for testing by means of quantitative reverse-transcriptase–polymerase-chain-reaction (RT-PCR) assay and viral culture. Using hemagglutination-inhibition and viral-neutralization assays, we tested baseline and convalescent serum samples from a subgroup of patients for antibody responses to the pandemic and seasonal influenza A viruses.
RESULTS
Secondary attack rates (as confirmed on RT-PCR assay) among household contacts of index patients were similar for the pandemic influenza virus (8%; 95% confidence interval [CI], 3 to 14) and seasonal influenza viruses (9%; 95% CI, 5 to 15). The patterns of viral shedding and the course of illness among index patients were also similar for the pandemic and seasonal influenza viruses. In a subgroup of patients for whom baseline and convalescent serum samples were available, 36% of household contacts who had serologic evidence of pandemic influenza virus infection did not shed detectable virus or report illness.
CONCLUSIONS
Pandemic 2009 H1N1 virus has characteristics that are broadly similar to those of seasonal influenza A viruses in terms of rates of viral shedding, clinical illness, and transmissibility in the household setting.
Households are thought to play a major role in the community spread of influenza virus during annual epidemics and occasional pandemics.1-4 As the pandemic 2009 influenza A (H1N1) virus (hereafter called pandemic virus) spread across the world, many countries implemented mitigation policies, including the recommendation that persons with confirmed or suspected infection be isolated at home.5-7 The literature contains few data on viral-shedding patterns associated with naturally acquired influenza virus infections in community settings. Although data have been published on humoral antibody responses to the pandemic virus after vaccination against seasonal influenza,8 little is known about antibody responses after naturally acquired infection or the association of such responses with viral shedding and clinical illness.
We conducted a prospective study of household transmission of influenza A in Hong Kong in July and August 2009. We assessed patterns in viral shedding, course of illness, and transmissibility associated with pandemic and seasonal influenza A virus infection.
METHODS
RECRUITMENT AND FOLLOW-UP OF PATIENTS
From 14 outpatient clinics and emergency departments in private hospitals across Hong Kong in July and August 2009, we recruited patients who presented with acute respiratory illness within 48 hours after the onset of illness and who lived with at least two other household members. We used a positive result for influenza A or B on a QuickVue Influenza A+B test (Quidel) to determine the eligibility of index patients and their household contacts for follow-up.
Diaries for recording daily symptoms were provided to all household contacts at an initial home visit, typically within 24 hours after the recruitment of the index patient. All household contacts were instructed in a simple hand-hygiene intervention9 and provided with liquid hand soap, alcohol hand rub, and a digital tympanic thermometer. The period of follow-up for secondary infections in household contacts was approximately 7 days.
Pooled specimens of nasal and throat swabs were collected from all household contacts, regardless of whether the person was ill at the initial home visit, and at two follow-up visits approximately 3 and 6 days later. A subgroup of index patients and household contacts agreed to provide a baseline serum sample at the initial home visit and a convalescent serum sample at the final home visit, after 20 to 35 days.
Written informed consent was obtained from all participants who were 18 years of age or older, and proxy written informed consent for participants under the age of 18 years was obtained from parents or legal guardians. The study protocol was approved by the institutional review board at the University of Hong Kong.
LABORATORY METHODS
Nasal and throat swabs were tested by means of a quantitative reverse-transcriptase-polymerasechain-reaction (RT-PCR) assay to detect the presence of influenza A or B virus and determine molecular viral loads, as described previously.9-12 Specimens that were found to be positive were subtyped by means of an RT-PCR assay with the use of subtype-specific primers, and quantitative viral cultures were analyzed to determine the infectious viral load in vitro, calculated as the tissue-culture infectious dose (TCID50), the quantity of virus required for a cytopathic effect in 50% of inoculated cultures.
Serum specimens were tested with a hemagglutination-inhibition assay for antibody responses to the pandemic virus A/California/4/2009 and two seasonal influenza viruses: the A/Brisbane/59/2007 (H1N1) virus and an A/Brisbane/10/2007 (H3N2)-like virus, A/Uruguay/716/2007. Serum specimens were also tested with a viral-neutralization assay for antibody responses to the pandemic virus and the A/Perth/16/2009 (H3N2)–like virus, A/HK/1985/2009. Additional details regarding our laboratory methods are provided in the Supplementary Appendix, available with the full text of this article at NEJM.org.
OUTCOME MEASURES
We defined serologically confirmed influenza virus infection as a rise by a factor of four or more in convalescent serum antibody titers to pandemic or seasonal influenza A virus, as compared with titers at baseline. We defined RT-PCR–confirmed influenza virus infection as a positive result on testing of one or more nasal and throat specimens collected during follow-up. We used a broad definition of acute respiratory illness, the presence of at least two signs or symptoms (temperature ≥37.8°C, cough, headache, sore throat, aches or pains in muscles, runny nose, and phlegm) on 1 or more days during follow-up, which is similar to the definitions used in previous studies.9,13 We also used the surveillance definition for influenza-like illness (temperature ≥37.8°C plus cough or sore throat), as recommended by the Centers for Disease Control and Prevention (CDC).14
STATISTICAL ANALYSIS
We estimated secondary attack rates, based on the proportion of household contacts in whom influenza developed (as determined by RT-PCR assay), as well as rates of acute respiratory illness and influenza-like illness. We calculated 95% confidence intervals for the crude secondary attack rates using a cluster bootstrap method with 1000 resamples.15 Households in which one or more household contacts had RT-PCR–confirmed influenza at the baseline home visit (i.e., households with one or more potential co-index patients) were excluded from the analysis of secondary attack rates.
We calculated standardized daily scores for three groups of signs and symptoms — systemic signs and symptoms (temperature ≥37.8°C, headache, and myalgia), upper respiratory symptoms (sore throat and runny nose), and lower respiratory symptoms (cough and phlegm) — by adding up the total number of signs and symptoms that were present and dividing by the highest possible score (3, 2, and 2, respectively).16,17 We plotted average symptom scores according to the time since the onset of acute respiratory illness, which was defined as the first day when the subject reported at least two of the seven signs or symptoms listed above.16,18
We defined the serial interval as the time between the onset of illness in an index patient and the onset of illness in a household contact. We estimated serial interval distributions on the basis of an underlying Weibull distribution, using methods that have been described previously.18 We estimated 95% confidence intervals for the mean serial interval, using a parametric bootstrap approach with 1000 resamples.18,19
We estimated geometric mean antibody titers and used Wilcoxon signed-rank tests to compare groups. Antibody titers below the lower limit of 1:10 were estimated to be 1:5 for calculations of geometric mean titers. Statistical analyses were performed with the use of R software, version 2.8.1 (R Development Core Team). Raw data from the study and R syntax to permit additional statistical analyses are available by contacting the corresponding author.
RESULTS
INDEX PATIENTS AND HOUSEHOLD CONTACTS
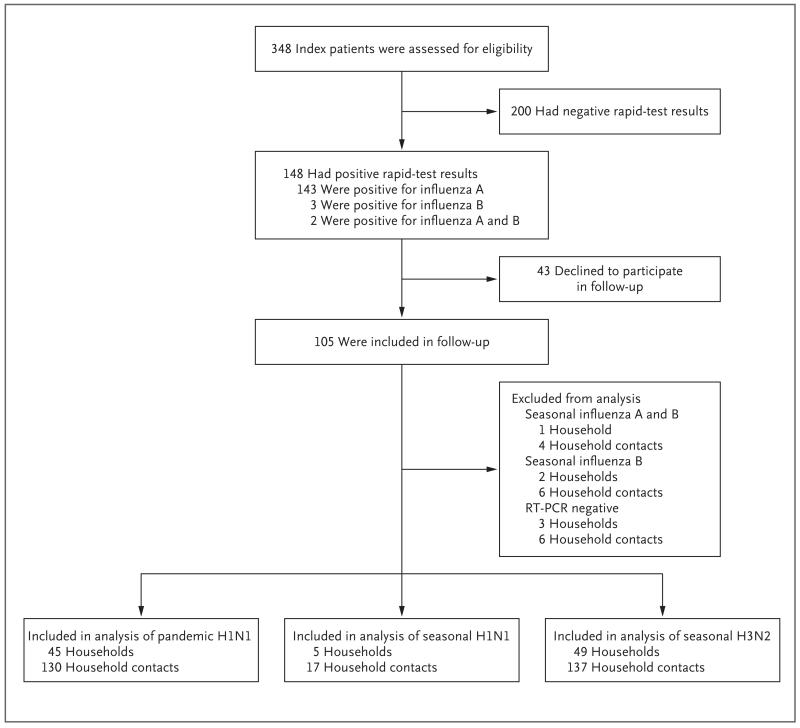
A total of 348 patients consented to participate in the study and met the inclusion criteria. Of these patients, 148 had a positive result on the rapid test. Household contacts of 43 of these patients (29%) declined home visits or could not be contacted after numerous repeated attempts. The characteristics of these 43 index patients were similar to those of the index patients whom we followed. The sensitivity of the QuickVue test was 77% (95% confidence interval [CI], 68 to 85) for seasonal influenza and 80% (95% CI, 70 to 88) for pandemic influenza, with the results of the RT-PCR assay used as the reference standard. We obtained complete follow-up data for the households of 105 index patients. We excluded the households of six index patients who did not have RT-PCR confirmation of virus infection or who had coinfection with influenza B or influenza A and B viruses. Thus, we compared the characteristics and transmissibility of pandemic and seasonal influenza A viruses in 99 households (Fig. 1).
Figure 1. Enrollment and Outcomes.
Oseltamivir was prescribed for 44% of the 99 index patients with pandemic or seasonal influenza virus infection (Table 1, and Table 1 in the Supplementary Appendix). Index patients with pandemic infection were on average younger and less likely to have presented with febrile illness than were those with seasonal influenza. The characteristics of household contacts were similar for index patients with pandemic infection and those with seasonal infection.
Table 1. Characteristics of Index Patients and Their Household Contacts with Pandemic or Seasonal Influenza.
| Characteristic | Pandemic Influenza |
Seasonal Influenza* |
P Value† |
|---|---|---|---|
| number (percent) | |||
| Index patients | |||
|
| |||
| No. of patients | 45 (100) | 54 (100) | |
|
| |||
| Age | 0.01 | ||
|
| |||
| ≤5 yr | 2 (4) | 1 (2) | |
|
| |||
| 6–15 yr | 20 (44) | 19 (35) | |
|
| |||
| 16–30 yr | 16 (36) | 10 (19) | |
|
| |||
| 31–50 yr | 6 (13) | 13 (24) | |
|
| |||
| >50 yr | 1 (2) | 11 (20) | |
|
| |||
| Male sex | 23 (51) | 29 (54) | 0.96 |
|
| |||
| Oseltamivir prescribed | 23 (51) | 21 (39) | 0.31 |
|
| |||
| Household contacts | |||
|
| |||
| No. of patients | 130 (100) | 154 (100) | |
|
| |||
| Age | 0.53 | ||
|
| |||
| ≤5 yr | 4 (3) | 4 (3) | |
|
| |||
| 6–15 yr | 18 (14) | 31 (20) | |
|
| |||
| 16–30 yr | 19 (15) | 24 (16) | |
|
| |||
| 31–50 yr | 64 (49) | 62 (40) | |
|
| |||
| >50 yr | 25 (19) | 33 (21) | |
|
| |||
| Male sex | 51 (39) | 61 (40) | 0.95 |
Seasonal influenza includes both H1N1 and H3N2 nonpandemic strains of influenza A.
P values were calculated by means of the chi-square test or Fisher’s exact test.
SECONDARY INFECTIONS
On the basis of RT-PCR results and clinical definitions of infection, secondary attack rates were similar among household contacts of index patients with pandemic influenza A virus infection and those with seasonal infection (Table 2). The attack rates were higher among household contacts of index patients who were 15 years of age or younger than among household contacts of older index patients, although the differences were not significant.
Table 2. Secondary Attack Rates among Household Contacts of Index Patients with Pandemic or Seasonal Influenza, According to the Age ofthe Index Patient and the Method of Diagnosis*.
| Age of Index Patient and Method of Diagnosis† |
Secondary Attack Rate among Household Contacts (95% CI)‡ |
|
|---|---|---|
| Pandemic Influenza | Seasonal Influenza | |
| Any age§ | ||
|
| ||
| RT-PCR assay | 0.08 (0.03–0.14) | 0.09 (0.05–0.15) |
|
| ||
| Acute respiratory illness | 0.26 (0.16–0.36) | 0.19 (0.12–0.27) |
|
| ||
| Influenza-like illness | 0.06 (0.03–0.11) | 0.04 (0.01–0.07) |
|
| ||
| <16 yr¶ | ||
|
| ||
| RT-PCR assay | 0.11 (0.02–0.23) | 0.13 (0.05–0.24) |
|
| ||
| Acute respiratory illness | 0.33 (0.20–0.47) | 0.21 (0.09–0.34) |
|
| ||
| Influenza-like illness | 0.07 (0.02–0.14) | 0.05 (0.00–0.10) |
|
| ||
| ≥16 yr∥ | ||
|
| ||
| RT-PCR assay | 0.05 (0.00–0.10) | 0.07 (0.02–0.12) |
|
| ||
| Acute respiratory illness | 0.20 (0.08–0.32) | 0.17 (0.09–0.27) |
|
| ||
| Influenza-like illness | 0.05 (0.00–0.11) | 0.03 (0.00–0.08) |
RT-PCR denotes reverse-transcriptase–polymerase chain reaction.
RT-PCR–confirmed influenza was defined on the basis of a positive RT-PCR assay for one or more pooled nasal and throat swabs. Acute respiratory illness was defined as the presence of at least two of the following symptoms: a temperature of at least 37.8°C, cough, headache, sore throat, myalgia, runny nose, and phlegm. Influenza-like illness was defined as the presence of a temperature of at least 37.8°C plus cough or sore throat.
All 95% confidence intervals were calculated with the use of a cluster bootstrap method.
Included in this category were 115 contacts of 41 index patients with pandemic influenza and 148 contacts of 53 index patients with seasonal influenza.
Included in this category were 54 contacts of 19 index patients with pandemic influenza and 61 contacts of 20 index patients with seasonal influenza.
Included in this category were 61 contacts of 22 index patients with pandemic influenza and 87 contacts of 33 index patients with seasonal influenza.
On the basis of eight patients with secondary infection and the corresponding index patients, the mean (±SD) serial interval for pandemic virus infection was estimated at 3.2±1.3 days (95% CI, 2.4 to 4.0). On the basis of seven patients secondary infection and the corresponding patients, the mean serial interval for seasonal H3N2 was estimated at 3.4±1.2 days (95% CI, 2.7 to 4.1).
VIRAL SHEDDING
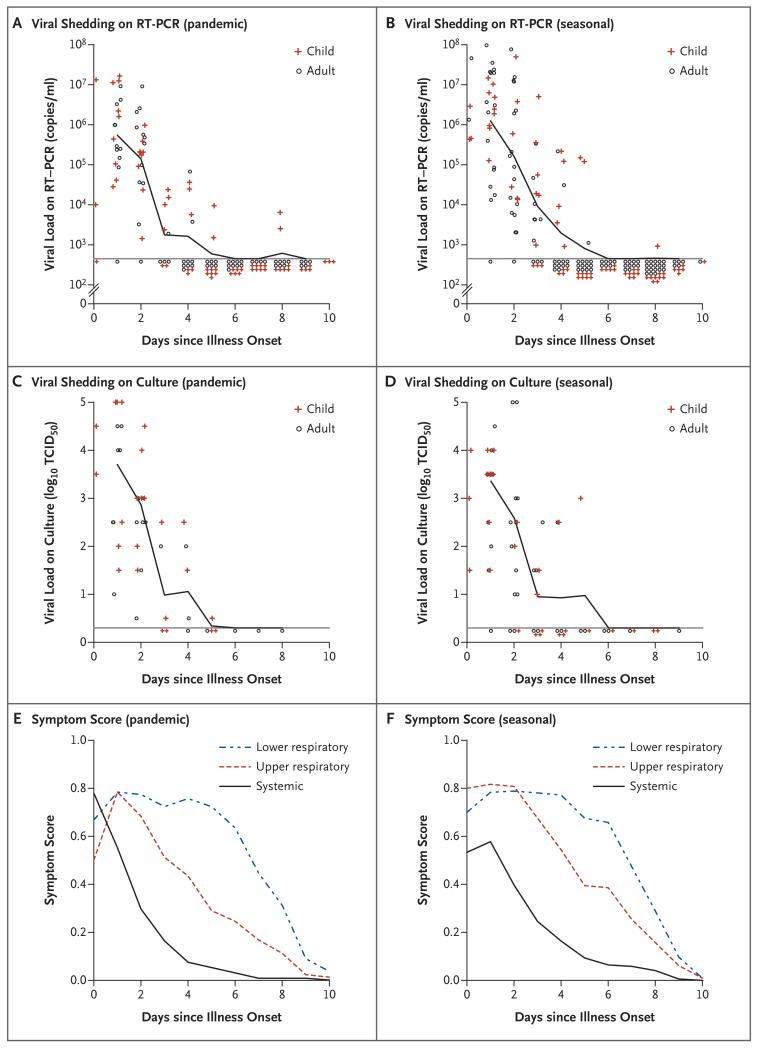
Figure 2 shows the rates of viral shedding, according to RT-PCR assay and culture, and mean standardized symptom scores throughout the course of illness among index patients with pandemic or seasonal influenza A virus infection. For both pandemic and seasonal influenza, molecular viral shedding, as seen on RT-PCR assay, generally ceased after 5 to 7 days of illness. Respiratory symptoms persisted for up to 10 days after onset of illness. The trends in viral shedding on RT-PCR assay and culture and the course of illness were similar in the subgroup of index patients who did not receive oseltamivir treatment.
Figure 2. Patterns of Viral Shedding and Course of Illness in Index Patients, According to the Onset of Acute Respiratory Illness.
Panels A and B show the geometric mean viral load on reverse-transcriptase—polymerase-chain-reaction (RTPCR) assay for index patients with pandemic influenza and those with seasonal influenza, respectively. The lower limit of detection for the RT-PCR assay was approximately 900 copies per milliliter (horizontal lines). Panels C and D show the geometric mean tissue-culture infectious dose (TCID50) (the quantity of virus required for a cytopathic effect in 50% of inoculated cultures) for index patients with pandemic influenza and those with seasonal influenza, respectively. The lower limit of detection of the quantitative culture assay was approximately 0.3 log10 TCID50 (horizontal lines). Panels E and F show the mean scores for lower respiratory, upper respiratory, and systemic symptoms in index patients with pandemic influenza and those with seasonal influenza, respectively. Symptom scores were calculated from a composite of three groups of signs and symptoms of influenza and ranged from 0 to 1, with higher scores indicating a greater severity of symptoms. In all panels, the onset of acute respiratory illness was defined as the self-reported day of illness onset before recruitment to the study.
Figure 1 in the Supplementary Appendix shows the rates of viral shedding on RT-PCR assay and culture and symptom scores throughout the course of illness in household contacts who had RT-PCR-confirmed influenza A virus infection. In this small sample, comparisons are imprecise, but on average, infections in household contacts appeared to be associated with lower levels of viral shedding and a shorter duration of clinical illness than infections in index patients.
ANTIBODY TITERS
Baseline and convalescent serum samples were available for 47 patients with RT-PCR-confirmed pandemic or seasonal influenza A infection. Most of these patients had an increase in antibody titers by a factor of four or more from baseline to convalescence (Fig. 2 in the Supplementary Appendix). The majority of household contacts for whom baseline serum samples were available had low antibody titers against the pandemic or seasonal virus that was associated with the infection in the corresponding index patient (Fig. 3 in the Supplementary Appendix). There was no significant protective effect of an increased titer of antibodies against RT-PCR–confirmed infection.
Of the 19 household contacts who had an increase by a factor of four or more in the antibody titer against pandemic (H1N1) or seasonal influenza A (H3N2) virus infection, 11 (58%) had detectable viral shedding; 10 of these 11 household contacts (91%) reported symptoms of acute respiratory illness, whereas only 5 (45%) reported symptoms of influenza-like illness (Table 3). Acute respiratory illness was reported by 27 household contacts for whom paired serum samples were available, but only 12 of these contacts (44%) had serologic evidence of infection or detectable viral shedding on RT-PCR assay (Table 2 in the Supplementary Appendix).
Table 3. Presence ofViral Shedding and Clinical Illness among 19 Household Contacts with Serologic Evidence of Pandemic or Seasonal Influenza.*.
| Variable | Increase in Antibody Titer† | |
|---|---|---|
| Pandemic Influenza (N = 11) |
Seasonal Influenza (N = 8) |
|
| no. (%) | ||
| Viral shedding detected on RT-PCR assay |
7 (64) | 4 (50) |
|
| ||
| Temperature ≥37.8°C | 3 (27) | 3 (38) |
|
| ||
| Cough | 6 (55) | 6 (75) |
|
| ||
| Acute respiratory illness‡ | 6 (55) | 6 (75) |
|
| ||
| Influenza-like illness‡ | 3 (27) | 2 (25) |
This analysis was based on the subgroup of 94 household contacts for whom serum samples were obtained during the acute and convalescent phases of illness: 54 contacts of index patients with pandemic influenza and 40 contacts of index patients with seasonal influenza. RT-PCR denotes reverse-transcrip-tase–polymerase chain reaction.
An increased antibody titer was defined as an increase by a factor of four or more. Analyses were performed with the use of viral neutralization to A/California/4/2009 (H1N1) (for index patients with RT-PCR–confirmed pandemic influenza) or A/Perth/16/09 (H3N2)–like virus A/HK/1985/2009 (for contacts of index patients with RT-PCR–confirmed seasonal influenza).
Acute respiratory illness was defined as the presence of at least two of the following symptoms: a temperature of at least 37.8°C, cough, headache, sore throat, myalgia, runny nose, and phlegm. Influenza-like illness was defined as the presence of a temperature of at least 37.8°C plus cough or sore throat.
Index patients with pandemic influenza who received oseltamivir treatment within 48 hours after the onset of illness had an increase in the geometric mean antibody titer of 4.0 from baseline to convalescence on hemagglutination-inhibition assay, whereas patients who did not receive antiviral treatment had a geometric mean titer increase of 32.0 (P = 0.03) (Fig. 4 in the Supplementary Appendix). Increases in antibody titers on viral-neutralization assay differed between treated and untreated index patients with pandemic influenza, although the difference was not significant (P = 0.08). Among index patients with seasonal influenza, there were no significant differences in titer increases or convalescent antibody titers between those who received antiviral treatment and those who did not receive such treatment.
DISCUSSION
In this study, the pandemic 2009 H1N1 virus and seasonal influenza A viruses had similar rates of household transmission. However, the observed attack rate for pandemic influenza was higher than that for seasonal influenza, particularly among children. This difference in attack rates could be associated with the lack of preexisting immunity against the pandemic influenza virus20 rather than an inherent difference in transmissibility. Most patients had low levels of immunity to pandemic and seasonal influenza A viruses, as indicated by humoral antibody titers. Pandemic and seasonal influenza A virus infections were associated with similar patterns of viral shedding and clinical illness.
Secondary attack rates were approximately double the rates that we observed in the hand-hygiene intervention group of our controlled trial in 2008.9 One possible explanation for this difference is that immunity among household contacts was lower both to pandemic H1N1 virus and to seasonal H3N2 virus. The predominant seasonal H3N2 viruses that were circulating in Hong Kong during our study period were antigenically drifted A/Perth/16/2009–like viruses. Secondary attack rates for pandemic and seasonal influenza, as measured by rates of influenza-like illness, were approximately 6% and 4%, respectively, which are slightly lower than secondary attack rates observed in the United States4,21 and Kenya22 during this pandemic and in previous studies of the transmission of seasonal influenza in households.23,24 In our study, the rates of transmission could have differed because of the hand-hygiene intervention that was implemented in all households.9 Secondary attack rates based on clinical criteria for influenza-like illness were substantially lower than secondary attack rates based on positive results of RT-PCR assay or serologic evidence of infection. Attack rates based on clinical criteria for acute respiratory illness were higher than rates based on the criteria for influenza-like illness, but the incidence of such respiratory illness was poorly correlated with infection confirmed on RT-PCR assay or serologic analysis.
There are few quantitative data on viral-shedding patterns associated with naturally acquired influenza virus infection in community settings, although quantitative data on shedding after infection are available from volunteer challenge studies17 and those involving hospitalized patients.25,26 We used a rapid test to screen index patients who sought outpatient care. Since an increased level of viral shedding is associated with increased rapid-test sensitivity,27 our data on viral shedding and course of illness may represent infections associated with a generally increased level of viral shedding. The rapid test that we used had moderate sensitivity for detecting both pandemic and seasonal influenza strains.28,29 The virologic and clinical data on secondary cases should be more representative of naturally acquired influenza virus infections in general than are data on index cases, and these data show that influenza A is associated with mild illness that is often afebrile.
Oseltamivir was prescribed for 44% of outpatients who had a positive rapid-test result in our study, which was approximately double the rate among patients with similar characteristics seen in the same clinics in 2007 and 2008.9,30 Antiviral prophylaxis for close contacts was not prescribed during our study period, although it was used during the containment phase earlier in the pandemic.31 Our data suggest that patients with pandemic H1N1 infection may have reduced convalescent antibody titers after treatment with oseltamivir, as compared with no treatment. Further studies are warranted to confirm or refute this potential association and to investigate the degree to which patients who are infected with 2009 H1N1 virus and are treated with antiviral agents are protected against reinfection in sub-sequent pandemics. A recent report from Chile described three cases of reinfection with 2009 H1N1 virus, and in each case reinfection occurred a few weeks after the first infection was identified and treated with oseltamivir.32 In a controlled trial involving 374 patients with seasonal influenza virus infection who were randomly assigned to receive either oseltamivir or placebo, the antibody titer increased from baseline to convalescence by a factor of approximately 16 on hemagglutination-inhibition assay, with no significant difference between the two study groups.33 It is possible that a reduced amount of viral antigen is sufficient to elicit a strong antibody response when the patient’s immune system has been primed with a closely related antigen, as is the case in persons who have had previous seasonal influenza but not in those with initial exposure to the 2009 pandemic virus. Thus, although the use of oseltamivir may not blunt the antibody response to seasonal influenza, it might do so with pandemic influenza.
Of 11 household contacts in our study who had detectable viral shedding on RT-PCR assay, only 1 did not report the occurrence of acute respiratory illness, although 6 of the 11 contacts did not report the occurrence of influenza-like illness. Some household contacts who had serologic evidence of infection with pandemic or seasonal influenza did not have viral shedding and did not report illness, and the presence of viral shedding was closely correlated with illness. Viral shedding before the onset of acute respiratory illness was rare in household contacts who had RT-PCR–confirmed influenza virus infection.
Our study had a number of limitations. First, we used a case-ascertainment study design in which index patients were recruited from outpatient clinics,9,34,35 and we used a rapid test to screen these patients for influenza virus infection. Such recruitment leads to bias in the selection of index patients, although secondary cases among household contacts should be representative of influenza A virus infections in the community. The index patients in our study had illness that was severe enough to warrant medical attention and had a positive result on a rapid test. If more severe illness or a higher level of viral shedding in association with a positive rapid-test result27 reflects greater infectiousness, we may have overestimated household secondary attack rates in the general community. Second, the hand-hygiene intervention that was provided to all households may have reduced the rate of transmission,9 perhaps to a similar extent for pandemic and seasonal viruses. Finally, we collected nose and throat swabs during home visits at 3-day intervals, and we may have missed some RT-PCR–positive infections if peak viral shedding in the respiratory tract occurred between these visits. We may also have missed secondary infections that occurred more than 7 days after the recruitment of index patients.
In spite of the differences in the age groups affected and occasional complications in patients with underlying disease, most of the epidemiologic data from affected countries suggest that pandemic 2009 H1N1 virus infection is not, on average, a more severe illness than seasonal influenza. Experimental data from in vitro cultures of primary human cells and ex vivo cultures of human respiratory tissue suggest that viral tropism and replication kinetics are similar for pandemic and seasonal H1N1 viruses.36 However, data from animal models suggest that the 2009 H1N1 virus causes more severe illness and may be less transmissible than seasonal influenza viruses.37,38 Our study suggests that pandemic and seasonal influenza A viruses are associated with similar viral-load dynamics, severity of clinical illness, and transmissibility.
Supplementary Material
Acknowledgments
Supported by grants from the National Institute of Allergy and Infectious Diseases (HHS-N266200700005C and N01-AI-70005) and the Hong Kong University Grants Committee (AoE/M-12/06).
We thank all the doctors, nurses, and staff members at the participating centers for facilitating recruitment; the dedicated team of health care workers who conducted the home visits; and Chan Kit Man, Calvin Cheng, Lai-Ming Ho, Ho Yuk Ling, Dennis Ip, Lam Yiu Pong, Max Lau, Tom Lui, Tong Hok Leung, Alfred Yeung, Eileen Yeung, Jenny Yuen, and Zhou Ying for research support.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Ferguson NM, Cummings DA, Cauchemez S, et al. Strategies for containing an emerging influenza pandemic in Southeast Asia. Nature. 2005;437:209–14. doi: 10.1038/nature04017. [DOI] [PubMed] [Google Scholar]
- 2.Ferguson NM, Cummings DA, Fraser C, Cajka JC, Cooley PC, Burke DS. Strategies for mitigating an influenza pandemic. Nature. 2006;442:448–52. doi: 10.1038/nature04795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Longini IM, Jr, Nizam A, Xu S, et al. Containing pandemic influenza at the source. Science. 2005;309:1083–7. doi: 10.1126/science.1115717. [DOI] [PubMed] [Google Scholar]
- 4.Yang Y, Sugimoto JD, Halloran ME, et al. The transmissibility and control of pandemic influenza A (H1N1) virus. Science. 2009;326:729–33. doi: 10.1126/science.1177373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention [Accessed May 14, 2010];Home care guidance: physician directions to patient/parent. 2009 Sep 24; at http://www.cdc.gov/h1n1flu/guidance_homecare_directions.htm.
- 6.Idem [Accessed May 14, 2010];CDC recommendations for the amount of time persons with influenzalike illness should be away from others. 2009 Oct 23; at http://www.cdc.gov/h1n1flu/guidance/exclusion.htm.
- 7.Idem [Accessed May 14, 2010];Guidance for businesses and employers to plan and respond to the 2009–2010 influenza season. 2009 Aug 19; at http://www.flu.gov/professional/business/guidance.pdf.
- 8.Hancock K, Veguilla V, Lu X, et al. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med. 2009;361:1945–52. doi: 10.1056/NEJMoa0906453. [DOI] [PubMed] [Google Scholar]
- 9.Cowling BJ, Chan KH, Fang VJ, et al. Facemasks and hand hygiene to prevent influenza transmission in households: a cluster randomized trial. Ann Intern Med. 2009;151:437–46. doi: 10.7326/0003-4819-151-7-200910060-00142. [DOI] [PubMed] [Google Scholar]
- 10.Chan KH, Peiris JSM, Lim W, Nicholls JM, Chiu SS. Comparison of nasopharyngeal flocked swabs and aspirates for rapid diagnosis of respiratory viruses in children. J Clin Virol. 2008;42:65–9. doi: 10.1016/j.jcv.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Lambert SB, Whiley DM, O’Neill NT, et al. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122(3):e615–e620. doi: 10.1542/peds.2008-0691. [DOI] [PubMed] [Google Scholar]
- 12.Peiris JS, Tang WH, Chan KH, et al. Children with respiratory disease associated with metapneumovirus in Hong Kong. Emerg Infect Dis. 2003;9:628–33. doi: 10.3201/eid0906.030009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowling BJ, Fung ROP, Cheng CKY, et al. Preliminary findings of a randomized trial of non-pharmaceutical interventions to prevent influenza transmission in households. PLoS One. 2008;3(5):e2101. doi: 10.1371/journal.pone.0002101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Babcock HM, Merz LR, Fraser VJ. Is influenza an influenza-like illness? Clinical presentation of influenza in hospitalized patients. Infect Control Hosp Epidemiol. 2006;27:266–70. doi: 10.1086/501539. [DOI] [PubMed] [Google Scholar]
- 15.Field CA, Welsh AH. Bootstrapping clustered data. J R Stat Soc [B] 2007;69:369–90. [Google Scholar]
- 16.Lau LLH, Cowling BJ, Fang VJ, et al. Viral shedding and clinical illness in naturally acquired influenza virus infections. J Infect Dis. 2010;201:1509–16. doi: 10.1086/652241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carrat F, Vergu E, Ferguson NM, et al. Time lines of infection and disease in human influenza: a review of volunteer challenge studies. Am J Epidemiol. 2008;167:775–85. doi: 10.1093/aje/kwm375. [DOI] [PubMed] [Google Scholar]
- 18.Cowling BJ, Fang VJ, Riley S, Malik Peiris JS, Leung GM. Estimation of the serial interval of influenza. Epidemiology. 2009;20:344–7. doi: 10.1097/EDE.0b013e31819d1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shao J, Tu D, editors. Bootstrap confidence sets and hypothesis tests. The jackknife and bootstrap. Springer-Verlag; New York: 1995. pp. 129–89. [Google Scholar]
- 20.Miller E, Hoschler K, Hardelid P, Stanford E, Andrews N, Zambon M. Incidence of 2009 pandemic influenza A H1N1 infection in England: a cross-sectional serological study. Lancet. 2010;375:1100–8. doi: 10.1016/S0140-6736(09)62126-7. [DOI] [PubMed] [Google Scholar]
- 21.Cauchemez S, Donnelly CA, Reed C, et al. Household transmission of 2009 pandemic influenza A (H1N1) virus in the United States. N Engl J Med. 2009;361:2619–27. doi: 10.1056/NEJMoa0905498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Introduction and transmission of 2009 pandemic influenza A (H1N1) virus — Kenya, June–July 2009. MMWR Morb Mortal Wkly Rep. 2009;58:1143–6. [PubMed] [Google Scholar]
- 23.Hayden FG, Belshe R, Villanueva C, et al. Management of influenza in households: a prospective, randomized comparison of oseltamivir treatment with or without postexposure prophylaxis. J Infect Dis. 2004;189:440–9. doi: 10.1086/381128. [DOI] [PubMed] [Google Scholar]
- 24.Welliver R, Monto AS, Carewicz O, et al. Effectiveness of oseltamivir in preventing influenza in household contacts: a randomized controlled trial. JAMA. 2001;285:748–54. doi: 10.1001/jama.285.6.748. [DOI] [PubMed] [Google Scholar]
- 25.Lee N, Chan PK, Hui DS, et al. Viral loads and duration of viral shedding in adult patients hospitalized with influenza. J Infect Dis. 2009;200:492–500. doi: 10.1086/600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li IW, Hung IF, To KK, et al. The natural viral load profile of patients with pandemic 2009 influenza A (H1N1) and the effect of oseltamivir treatment. Chest. 2010;137:759–68. doi: 10.1378/chest.09-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng CK, Cowling BJ, Chan KH, et al. Factors affecting QuickVue influenza A+B rapid test performance in the community setting. Diagn Microbiol Infect Dis. 2009;65:35–41. doi: 10.1016/j.diagmicrobio.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 28.Evaluation of rapid influenza diagnostic tests for detection of novel influenza A (H1N1) virus — United States, 2009. MMWR Morb Mortal Wkly Rep. 2009;58:826–9. [PubMed] [Google Scholar]
- 29.Vasoo S, Stevens J, Singh K. Rapid antigen tests for diagnosis of pandemic (Swine) influenza A/H1N1. Clin Infect Dis. 2009;49:1090–3. doi: 10.1086/644743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng S, Cowling BJ, Fang VJ, et al. Effects of oseltamivir treatment on duration of clinical illness and viral shedding and household transmission of influenza virus. Clin Infect Dis. 2010;50:707–14. doi: 10.1086/650458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JT, Cowling BJ, Lau EH, et al. School closure and mitigation of pandemic (H1N1) 2009, Hong Kong. Emerg Infect Dis. 2010;16:538–41. doi: 10.3201/eid1603.091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez CM, Ferres M, Labarca JA. Pandemic (H1N1) 2009 reinfection, Chile. Emerg Infect Dis. 2010;16:156–7. doi: 10.3201/eid1601.091420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treanor JJ, Hayden FG, Vrooman PS, et al. Efficacy and safety of the oral neuraminidase inhibitor oseltamivir in treating acute influenza: a randomized controlled trial. JAMA. 2000;283:1016–24. doi: 10.1001/jama.283.8.1016. [DOI] [PubMed] [Google Scholar]
- 34.Yang Y, Longini IM, Jr, Halloran ME. Design and evaluation of prophylactic interventions using infectious disease incidence data from close contact groups. Appl Stat. 2006;55:317–30. doi: 10.1111/j.1467-9876.2006.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Viboud C, Boëlle PY, Cauchemez S, et al. Risk factors of influenza transmission in households. Br J Gen Pract. 2004;54:684–9. [PMC free article] [PubMed] [Google Scholar]
- 36.Chan MCW, Chan RW, Yu WCL, et al. Tropism and innate host responses of the 2009 pandemic H1N1 influenza virus in ex vivo and in vitro cultures of human conjunctiva and respiratory tract. Am J Pathol. 2010;176:1828–40. doi: 10.2353/ajpath.2010.091087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh Y, Shinya K, Kiso M, et al. In vitro and in vivo characterization of new swineorigin H1N1 influenza viruses. Nature. 2009;460:1021–5. doi: 10.1038/nature08260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munster VJ, de Wit E, van den Brand JM, et al. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science. 2009;325:481–3. doi: 10.1126/science.1177127. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.