Abstract
Thalidomide-like drugs such as lenalidomide are clinically important treatments for multiple myeloma and show promise for other B cell malignancies. The biochemical mechanisms underlying their antitumor activity are unknown. Thalidomide was recently shown to bind to, and inhibit, the cereblon ubiquitin ligase. Cereblon loss in zebrafish causes fin defects reminiscent of the limb defects seen in children exposed to thalidomide in utero. Here we show that lenalidomide-bound cereblon acquires the ability to target for proteasomal degradation two specific B cell transcription factors, Ikaros family zinc finger proteins 1 and 3 (IKZF1 and IKZF3). Analysis of myeloma cell lines revealed that loss of IKZF1 and IKZF3 is both necessary and sufficient for lenalidomide's therapeutic effect, suggesting that the antitumor and teratogenic activities of thalidomide-like drugs are dissociable.
Fifty years ago, thalidomide was used for insomnia and morning sickness but was later banned because of its teratogenicity, manifest as profound limb defects. Thalidomide and the related drugs lenalidomide and pomalidomide (IMiDs) have regained interest, however, as immunomodulators and antineoplastics, especially for multiple myeloma and other B cell malignancies (1–3). Nonetheless, the biochemical mechanisms underlying their teratogenic and therapeutic activities, and whether they are linked, are unknown.
In this regard, thalidomide was recently shown to bind to cereblon, which is the substrate-recognition component of a cullin-dependent ubiquitin ligase, and to inhibit its autoubiquitination activity (4). Treatment of zebrafish with cereblon morpholinos or thalidomide caused fin defects (4), suggesting that IMiDs act by stabilizing cereblon substrates. However, myeloma cells rendered IMiDs-resistant have frequently down-regulated cereblon (5–8). Conversely, high cereblon concentrations in myeloma cells are associated with increased responsiveness to IMiDs (9, 10). Collectively, these observations suggest that IMiDs are not simply cereblon antagonists but, instead, alter the substrate specificity of cereblon to include proteins important in myeloma.
To look for such proteins, we made a plasmid library encoding 15,483 open reading frames (ORFs) fused to firefly luciferase (Fluc), knowing that the stabilities of such fusions are usually influenced by the ubiquitin ligase(s) for the corresponding unfused ORF (11–13). Indeed, Elledge and co-workers used a green fluorescence protein (GFP)–ORF library to monitor the stabilities of thousands of ORFs after specific perturbations (13). Partly on the basis of their work, we inserted a renilla luciferase (Rluc) reporter into each ORF-luciferase cDNA for normalization purposes and placed both reporters under internal ribosome entry site (IRES) control (Fig. 1A and fig. S1).
Fig. 1. Down-regulation of IKZF1 and IKZF3 by lenalidomide.
(A) Vector schematic. (B) Distribution of fold change in Fluc/Rluc ratios after lenalidomide (LEN) (2 μM) treatment. (C and D) Fluc/Rluc ratios (top panels) and immunoblots (bottom panels) of 293FT cells transfected to produce the indicated IKZF proteins fused to Fluc (top panels) or HA tag (bottom panels). Where indicated, cells were treated with lenalidomide (2 μM), MLN4924 (1 μM), or MG132 (10 μM) for 12 hours. Fluc/Rluc ratios were normalized to corresponding dimethyl sulfoxide (DMSO)–treated cells. Data are presented as mean ± SD (n = 4). (E) Immunoblot analysis of MM1S and L363 cells treated with LEN (2 μM) and MLN4924 (1 μM), as indicated, for 12 hours.
In pilot experiments 293FT embryonic kidney cells grown in multiwell plates were transfected with the ORF-luciferase library (one ORF per well) and treated with the proteasome inhibitor MG132, the hydroxylase inhibitor dimethyloxalylglycine (DMOG), or vehicle. Fluc/Rluc values measured 36 to 48 hours later were stable over a wide range of input plasmid concentrations (fig. S2). As expected, MG132 stabilized many proteasomal substrates and DMOG stabilized HIF1α, which is rapidly degraded when prolyl hydroxylated (fig. S3).
Next, we used this approach to identify changes in protein stability in 293FT cells treated with lenalidomide (Fig. 1A and fig. S4). A total of 2113 ORF-luciferase fusions produced luciferase signals that were undetectable or highly variable (>50% SD), leaving 13,370 for analysis. As expected, most ORFs were unaffected by lenalidomide (Fig. 1B, fig. S5, and table S1). The 107 ORFs that were >3 SDs from the mean (46 ORFs plus 61 ORFs displaying decreased or increased Fluc/Rluc ratios after lenalidomide treatment, respectively) were retested in secondary assays. One down-regulated ORF (IKZF3) and one up-regulated ORF (C11orf65) retested positively (table S2).
C11orf65 was unaffected by lenalidomide when fused to a hemagglutinin (HA) epitope tag instead of Fluc and so was not studied further (fig. S6). By contrast, lenalidomide down- regulated IKZF3 and its paralog IKZF1, which had fallen just outside the 3-SD cut-off in the primary screen, fused to either Fluc or HA (Fig. 1, B and C, and table S1). These effects were specific because lenalidomide did not affect exogenous IKZF2, IKZF4, IKZF5, or the B cell transcription factor IRF4 (Fig. 1C and fig. S7). Similar results were obtained with two common splice variants (V1 and V2) of IKZF1 and IKZF2 (Fig. 1C and fig. S7) and with pomalidomide (fig. S8). Down-regulation of exogenous IKZF1 was blocked by MG132 and by MLN4924, which inhibits cullin-dependent ubiquitin ligases (Fig. 1D) (14, 15).
Consistent with these findings, lenalidomide down-regulated endogenous IKZF1 in U937 leukemia cells (fig. S9), which do not express IKZF3, and both IKZF1 and IKZF3 in MM1S and L363 myeloma cells (Fig. 1E) unless the cells were pretreated with MG132 or MLN4924 (Fig. 1E and fig. S9). Multiple IKZF1 bands were detected by immunoblot analysis, presumably due to alternative splicing. Lenalidomide did not alter IKZF1 and IKZF3 mRNA levels, consistent with it acting posttranscriptionally (fig. S10).
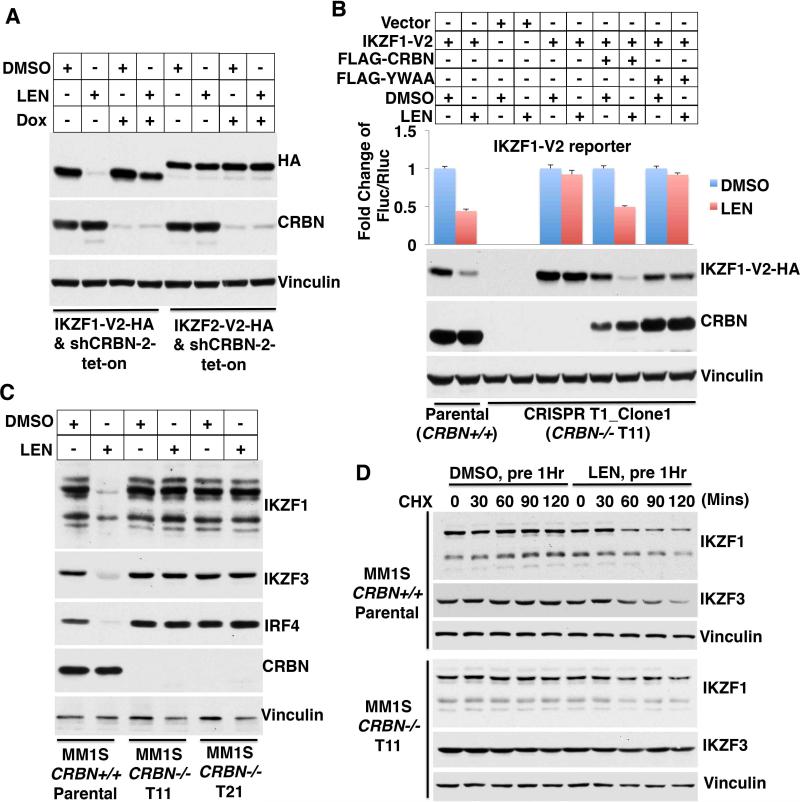
Down-regulation of cereblon in 293FT cells with a doxycycline-inducible short hairpin RNA (shRNA) prevented the destabilization of exogenous, HA-tagged, IKZF1, by lenalidomide (Fig. 2A and fig. S11). Similarly, cereblon shRNA blocked the down-regulation of endogenous IKZF1 by lenalidomide in U937 leukemia cells (U937) and myeloma cells (L363 and KMS34) (fig. S12, A to C). We also used clustered regularly interspaced short palindromic repeats (CRISPR)–based gene editing (16, 17) to generate CRBN−/− 293FT cells, which were then transfected to produce IKZF1 fused to Fluc (Fig. 2B, top panel, and fig. S13) or HA (Fig. 2B, bottom panel). Exogenous IKZF1 was not down-regulated in CRBN−/− 293FT cells (Fig. 2B and fig. S13). This defect was rescued by wild-type cereblon, but not a lenalidomide-resistant cereblon mutant (YWAA) (4) (Fig. 2B). Similar results were obtained with a second CRBN−/− 293FT subclone, but not in subclones with detectable amounts of cereblon (fig. S13). Moreover, endogenous IKZF1 and IKZF3 were not degraded by lenalidomide in two independent CRBN−/− MM1S myeloma cell lines generated with CRISPR (Fig. 2, C and D).
Fig. 2. Down-regulation of IKZF1 and IKZF3 by lenalidomide requires cereblon.
(A) Immunoblot analysis of 293FT cells stably infected with lentiviral vectors expressing the indicated IKZF-HA proteins and a doxycycline-inducible CRBN shRNA. Where indicated, LEN (2 μM) and doxcycyline (Dox) (1 μg/ml) were added for 12 and 60 hours, respectively. (B) Fluc/Rluc ratios (top panels) and immunoblots (bottom panels) of CRBN+/+ and CRBN−/− 293FT cells transfected to produce IKZF1 fused to Fluc (top panel) or HA tag (bottom panels). Where indicated cells were treated with LEN (2 μM) for 12 hours. Fluc/Rluc ratios were normalized to corresponding DMSO-treated cells. Data are presented as mean ± SD (n = 4). (C and D) Immunoblot analysis of CRBN+/+ and CRBN–/– MM1S myeloma cells. Where indicated, cells were treated with LEN (2 μM) for 24 hours (C) or 1 hour before the addition of cyclohexamide (CHX) (100 μg/ml) for the indicated periods (D).
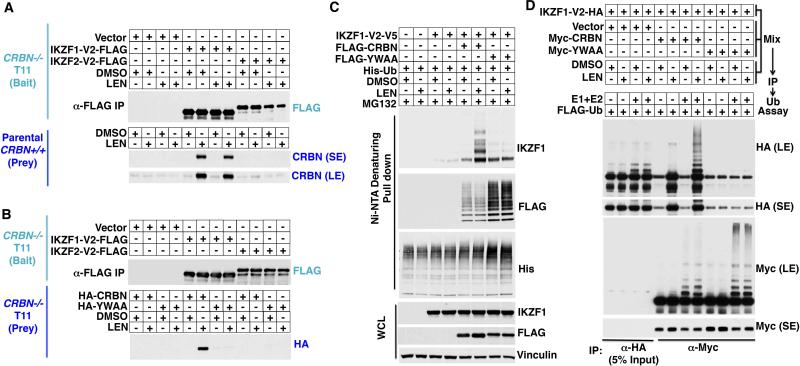
Lenalidomide enhanced the binding of IZKF1 and IKZF3, but not IKZF2 and IKZF5, to the cereblon ubiquitin ligase in cotransfection experiments using MG132-treated 293FT cells (fig. S14, A and B). Next, we immunoprecipitated FLAG-tagged IKZF1 and IKZF2 from CRBN−/− cells that were or were not treated with lenalidomide. The immobilized immunoprecipitates were then used to capture endogenous cereblon from CRBN+/+ cells (Fig. 3A) or exogenous cereblon from CRBN−/− cells transfected to produce wild-type cereblon or the YWAA variant (Fig. 3B). In both cases, the cells producing cereblon were or were not treated with lenalidomide before lysis. Wild-type cereblon, but not the YWAA variant, bound specifically to IKZF1 provided that it was exposed to lenalidomide, consistent with lenalidomide binding directly to cereblon rather than to IKZF1 (Fig. 3, A and B, and fig. S15). Lenalidomide also promoted the binding of cereblon to exogenous IKZF1 (fig. S16) and to endogenous IKZF1 and IKZF3 (fig. S17) when added directly to binding assays performed with cell extracts. Moreover, wild-type cereblon, but not the YWAA variant, promoted the ubiquitylation of IKZF1 in vivo (Fig. 3C) and in vitro (Fig. 3D) after exposure to lenalidomide.
Fig. 3. Lenalidomide promotes ubiquitylation of IKZF1 and IKZF3 by cereblon.
(A and B) FLAG-IKZF was immunoprecipitated from CRBN−/− 293FT cells stably infected to produce the indicated IKZF proteins and used to capture cereblon from CRBN+/+ 293FT cells (A) or CRBN−/− 293FT cells transfected to produce the indicated CRBN variants (B). Cells were treated with LEN (2 μM) for 12 hours before lysis, as indicated. Bound proteins were detected by immunoblot analysis. (C) Immunoblot analysis of proteins captured with nickel Sepharose from 293FT cells transfected to produce the indicated FLAG-, His-, and V5-tagged proteins. The cells were treated with MG132 (10 μM) and, where indicated, with LEN (2 μM) for 12 hours. (D) CRBN−/− 293FT cells were transfected to produce IKZF1-HA and the indicated Myc-cereblon variants and lysed. The extracts were mixed, treated with LEN (2 μM) or DMSO, and immunoprecipitated with antibodies against HA (anti-HA) or anti-Myc. The immunoprecipitates were incubated with recombinant E1, E2, and ubiquitin (Ub) and subjected to immunoblot analysis.
We analyzed a series of IKZF1/2 chimeras and determined that the region of IKZF1 corresponding to residues 108 to 197 of IKZF1(V2) mediated lenalidomide-dependent binding to cereblon (fig. S18). Within this region, there are only seven amino acid differences between IKZF1 and IKZF2 (fig. S19, A and B). Changing IKZF1 residue Q146 (or IKZF3 Q147) to the corresponding residue in IKZF2 (histidine) abrogated cereblon binding and lenalidomide-induced degradation (figs. S19C and S20). Conversely, the reciprocal change in IKZF2 rendered it partially sensitive to lenalidomide (fig. S19D).
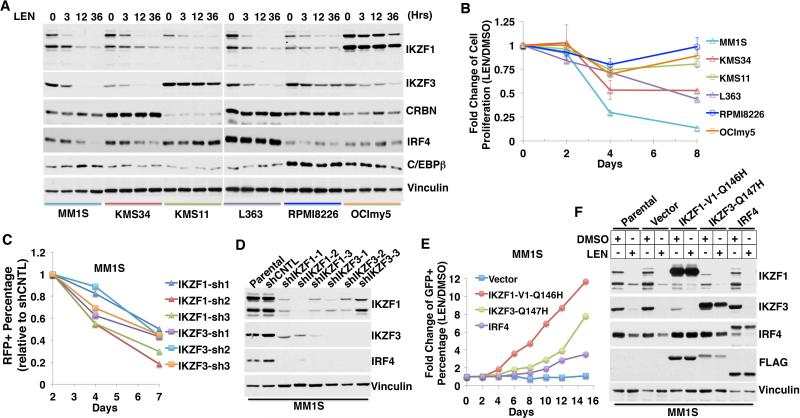
Next, we tested six myeloma cell lines for their sensitivity to lenalidomide in vitro (Fig. 4, A and B). Previous studies showed that lenalidomide, at least indirectly, down-regulates IRF4 and linked this to its antimyeloma activity (5–8). MM1S, KMS34, and L363 cells were sensitive to lenalidomide in vitro whereas KMS11, RPMI8226, and OCImy5 cells were relatively resistant (Fig. 4B). In the three sensitive lines, IKZF1 and IKZF3 were down-regulated by lenalidomide (Fig. 4A). In two of these lines (MM1S and KMS34), loss of IKZF1 and IKZF3 was followed by a decrease in IRF4, consistent with IRF4 acting downstream of IKZF1 and/or IKZF3 in these cells (Fig. 4A and fig. S21A). Indeed, we confirmed decreased IRF4 mRNA and decreased binding of IKZF1 to the IRF4 locus by chromatin immunoprecipitation (ChIP) in MM1S cells treated with lenalidomide (fig. S21). The third sensitive cell line, L363, expressed high basal amounts of IRF4 that were unaffected by lenalidomide, providing evidence that the antiproliferative effects of this drug involves at least one target other than IRF4 (Fig. 4A and fig. S21A).
Fig. 4. Antimyeloma activity of lenalidomide linked to loss of IKZF1 and IKZF3.
(A and B) Immunoblot analysis (A) and proliferation (B) of myeloma cell lines treated with LEN (2 μM) for the indicated periods. In (B), data are presented as mean ± SD (n = 4). (C) Change in % red fluorescent protein (RFP) positivity over time in MM1S cells infected with viruses encoding RFP and the indicated shRNAs. The day 2% RFP for each virus was normalized to 1, and subsequent values were expressed relative to cells infected with a virus encoding RFP and a control (CNTL) shRNA. (D) Immunoblot analysis of MM1S cells transiently infected with lentiviruses expressing the indicated shRNAs for 72 hours. (E) MM1S cells were infected with lentiviral vectors encoding GFP and the indicated FLAG-tagged proteins. Shown for each protein is the percentage of GFP positivity for cells treated with LEN (2 μM) for the indicated duration compared to DMSO. (F) Immunoblot analysis of MM1S cells infected as in (E) and treated with DMSO or LEN (2 μM) for 24 hours.
Two of the resistant lines had relatively high basal amounts of IKZF1 (OCImy5) or IKZF3 (KMS11) and corresponding low amounts of cereblon compared to the sensitive lines, and down-regulation of IKZF1 and IKZF3 by lenalidomide was attenuated in the third (RPMI8226) (Fig. 4A). IRF4 was not down-regulated by lenalidomide in the three resistant lines.
Next, we performed competition experiments with cells in which IKZF1 or IKZF3 was suppressed with shRNAs or enhanced through expression of lenalidomide-resistant versions of IKZF1 or IKZF3. Down-regulation of either IKZF1 or IKZF3 in the lenalidomide-sensitive cell lines MM1S and KMS34 markedly decreased cellular fitness compared to cells expressing a control shRNA and was associated with down-regulation of IRF4 (Fig. 4, C and D, and fig. S22). Notably, down-regulation of either IKZF protein led to loss of the other. Conversely, expression of the stabilized versions of IKZF1(Q146H) or IKZF3(Q147H) conferred lenalidomide resistance to MM1S cells (Fig. 4, E and F, and fig. S23) and KMS34 cells (fig. S24). Ectopic expression of a T cell–specific Ikaros family member, IKZF2, which is naturally lenalidomide resistant (Fig. 1C), had similar effects (figs. S23G and S25). The effects of expressing IRF4 itself were much less pronounced, again suggesting that IKZF1 and IKZF3 have additional targets that are relevant for lenalidomide's antimyeloma activity (Fig. 4E and fig. S23H). It remains to be seen whether lenalidomide-resistance conferred by IKZF family members is due primarily to transcriptional activation of their target genes or to noncanonical functions.
Our findings link lenalidomide's antimyeloma activity to down-regulation of IKZF1 and IKZF3, two transcription factors that play critical roles in B cell development and are highly expressed in B cell malignancies, including myeloma (fig. S26) (18–21). There are many other examples of cancers that become addicted to transcription factors that specify cell lineage (22, 23). Although IKZF1 is a tumor suppressor in some other B cell malignancies (24), there is precedence for the same gene acting as either a tumor suppressor or an oncogene in different contexts.
Ikaros family members can serve as transcriptional activators or repressors in different settings. For example, IKZF1 and IKZF3 repress interleukin-2 (IL-2) expression in T cells, thus explaining how IMiDs induce IL-2 production in vivo (19, 25, 26).
The proteasomal inhibitor bortezomib has antimyeloma activity, alone and in combination with lenalidomide, although the pertinent proteasomal substrates are debated (27, 28). This creates a paradox because proteasomal blockade prevents the destruction of IKZF1 and IKZF3 by lenalidomide. Proteasomal blockade by bortezomib is incomplete with therapeutic dosing, however, which might allow sufficient clearance of IKZF1 and IKZF3 while retaining bortezomib's other salutary effects. It is also possible that these two proteins, once polyubiquitylated, are inactive or dominant-negatives.
Earlier work suggested that thalidomide's teratogenic effects reflected cereblon inactivation, whereas our findings indicate that the therapeutic effects of the IMiDs reflect a cereblon gain of function. Notably, cereblon might have additional substrates that were not in our fusion library, could not be recognized as luciferase fusions, or require accessory proteins or signals absent in 293FT cells. Regardless, our findings create a path to uncouple the therapeutic and teratogenic activities of the IMiDs.
It is not yet clear whether lenalidomide's effect on cereblon is hypermorphic or neomorphic. Precedence for the latter is provided by rapamycin, which converts FKBP12 into a TORC1 kinase inhibitor and cyclosporine, which converts cyclophylin into a calcineurin antagonist (29). Perhaps oncoproteins currently deemed undruggable, such as c-Myc or β-catenin, could be destroyed by drugs that, like lenalidomide, repurpose ubiquitin ligases.
Supplementary Material
Acknowledgements
We thank M. Vidal and A. MacWilliams at Dana-Farber Cancer Institute for the ORF cDNA collection and for help with recombination cloning. This work was supported by a grant (W.G.K.) and F32 fellowship (G.L.) from NIH. C.J.O. is supported by a Leukemia and Lymphoma Society Fellowship award, and J.E.B. and C.S.M. are supported by the Multiple Myeloma Research Foundation. W.G.K. is a Howard Hughes Medical Institute Investigator. C.S.M. has received consulting fees from Celgene, the manufacturer of lenalidomide and pomalidomide. The primary and secondary luciferase screen data are presented in the supplementary materials.
References and Notes
- 1.Martiniani R, Di Loreto V, Di Sano C, Lombardo A, Liberati AM. Biological activity of lenalidomide and its underlying therapeutic effects in multiple myeloma. Adv Hematol. 2012;2012:842945. doi: 10.1155/2012/842945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Terpos E, Kanellias N, Christoulas D, Kastritis E, Dimopoulos MA. Pomalidomide: a novel drug to treat relapsed and refractory multiple myeloma. OncoTargets and therapy. 2013;6:531. doi: 10.2147/OTT.S34498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhu YX, Kortuem KM, Stewart AK. Molecular mechanism of action of immune-modulatory drugs thalidomide, lenalidomide and pomalidomide in multiple myeloma. Leukemia & lymphoma. 2013 Apr;54:683. doi: 10.3109/10428194.2012.728597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito T, et al. Identification of a primary target of thalidomide teratogenicity. Science. 2010 Mar 12;327:1345. doi: 10.1126/science.1177319. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Girona A, et al. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012 Nov;26:2326. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang LH, et al. Lenalidomide efficacy in activated B-cell-like subtype diffuse large B-cell lymphoma is dependent upon IRF4 and cereblon expression. Br J Haematol. 2013 Feb;160:487. doi: 10.1111/bjh.12172. [DOI] [PubMed] [Google Scholar]
- 7.Yang Y, et al. Exploiting synthetic lethality for the therapy of ABC diffuse large B cell lymphoma. Cancer Cell. 2012 Jun 12;21:723. doi: 10.1016/j.ccr.2012.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhu YX, et al. Cereblon expression is required for the antimyeloma activity of lenalidomide and pomalidomide. Blood. 2011 Nov 3;118:4771. doi: 10.1182/blood-2011-05-356063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broyl A, et al. High cereblon expression is associated with better survival in patients with newly diagnosed multiple myeloma treated with thalidomide maintenance. Blood. 2013 Jan 24;121:624. doi: 10.1182/blood-2012-06-438101. [DOI] [PubMed] [Google Scholar]
- 10.Heintel D, et al. High expression of cereblon (CRBN) is associated with improved clinical response in patients with multiple myeloma treated with lenalidomide and dexamethasone. Br J Haematol. 2013 Jun;161:695. doi: 10.1111/bjh.12338. [DOI] [PubMed] [Google Scholar]
- 11.Zhang GJ, et al. Bioluminescent imaging of Cdk2 inhibition in vivo. Nat Med. 2004 Jun;10:643. doi: 10.1038/nm1047. [DOI] [PubMed] [Google Scholar]
- 12.Safran M, et al. Mouse model for noninvasive imaging of HIF prolyl hydroxylase activity: assessment of an oral agent that stimulates erythropoietin production. Proc Natl Acad Sci U S A. 2006 Jan 3;103:105. doi: 10.1073/pnas.0509459103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008 Nov 7;322:918. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 14.Soucy TA, et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009 Apr 9;458:732. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 15.Ohh M, et al. An intact NEDD8 pathway is required for Cullin-dependent ubiquitylation in mammalian cells. EMBO reports. 2002 Feb;3:177. doi: 10.1093/embo-reports/kvf028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013 Feb 15;339:819. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013 Feb 15;339:823. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Merkenschlager M. Ikaros in immune receptor signaling, lymphocyte differentiation, and function. FEBS Lett. 2010 Dec 15;584:4910. doi: 10.1016/j.febslet.2010.09.042. [DOI] [PubMed] [Google Scholar]
- 19.Thompson EC, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007 Mar;26:335. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 20.Ferreiros-Vidal I, et al. Genome-wide identification of Ikaros targets elucidates its contribution to mouse B-cell lineage specification and pre-B-cell differentiation. Blood. 2013 Mar 7;121:1769. doi: 10.1182/blood-2012-08-450114. [DOI] [PubMed] [Google Scholar]
- 21.Monticelli S, Sallusto F. Negative regulators take center stage. Nature immunology. 2012 Aug;13:719. doi: 10.1038/ni.2377. [DOI] [PubMed] [Google Scholar]
- 22.Garraway LA, Sellers WR. Lineage dependency and lineage-survival oncogenes in human cancer. Nat Rev Cancer. 2006 Aug;6:593. doi: 10.1038/nrc1947. [DOI] [PubMed] [Google Scholar]
- 23.Shah SP, et al. Mutation of FOXL2 in granulosa-cell tumors of the ovary. N Engl J Med. 2009 Jun 25;360:2719. doi: 10.1056/NEJMoa0902542. [DOI] [PubMed] [Google Scholar]
- 24.Kastner P, et al. Function of Ikaros as a tumor suppressor in B cell acute lymphoblastic leukemia. American journal of blood research. 2013;3:1. [PMC free article] [PubMed] [Google Scholar]
- 25.Quintana FJ, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nature immunology. 2012 Aug;13:770. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Avitahl N, et al. Ikaros sets thresholds for T cell activation and regulates chromosome propagation. Immunity. 1999 Mar;10:333. doi: 10.1016/s1074-7613(00)80033-3. [DOI] [PubMed] [Google Scholar]
- 27.Laubach J, Richardson P, Anderson K. Multiple myeloma. Annu Rev Med. 2011;62:249. doi: 10.1146/annurev-med-070209-175325. [DOI] [PubMed] [Google Scholar]
- 28.Richardson PG, et al. Multicenter, phase I, dose-escalation trial of lenalidomide plus bortezomib for relapsed and relapsed/refractory multiple myeloma. J Clin Oncol. 2009 Dec 1;27:5713. doi: 10.1200/JCO.2009.22.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crabtree GR, Schreiber SL. Three-part inventions: intracellular signaling and induced proximity. Trends Biochem Sci. 1996 Nov;21:418. doi: 10.1016/s0968-0004(96)20027-1. [DOI] [PubMed] [Google Scholar]
- 30.Campanero MR, Flemington EK. Regulation of E2F through ubiquitin-proteasome-dependent degradation: stabilization by the pRB tumor suppressor protein. Proc Natl Acad Sci U S A. 1997 Mar 18;94:2221. doi: 10.1073/pnas.94.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiederschain D, et al. Single-vector inducible lentiviral RNAi system for oncology target validation. Cell Cycle. 2009 Feb 1;8:498. doi: 10.4161/cc.8.3.7701. [DOI] [PubMed] [Google Scholar]
- 32.Ott CJ, et al. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012 Oct 4;120:2843. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002 Apr;1:237. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- 34.Loven J, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell. 2013 Apr 11;153:320. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.