Abstract
NMR fingerprinting of the components of finely divided plant cell walls swelled in DMSO has been recently described. Cell wall gels, produced directly in the NMR tube with perdeutero-dimethylsulfoxide, allowed the acquisition of well resolved/dispersed 2D 13C–1H correlated solution-state NMR spectra of the entire array of wall polymers, without the need for component fractionation. That is, without actual solubilization, and without apparent structural modification beyond that inflicted by the ball milling and ultrasonication steps, satisfactorily interpretable spectra can be acquired that reveal compositional and structural details regarding the polysaccharide and lignin components in the wall. Here, the profiling method has been improved by using a mixture of perdeuterated DMSO and pyridine (4:1, v/v). Adding pyridine provided not only easier sample handling because of the better mobility compared to the DMSO-d6-only system but also considerably elevated intensities and improved resolution of the NMR spectra due to the enhanced swelling of the cell walls. This modification therefore provides a more rapid method for comparative structural evaluation of plant cell walls than is currently available. We examined loblolly pine (Pinus taeda, a gymnosperm), aspen (Populus tremuloides, an angiosperm), kenaf (Hibiscus cannabinus, an herbaceous plant), and corn (Zea mays L., a grass, i.e., from the Poaceae family). In principle, lignin composition (notably, the syringyl : guaiacyl : p-hydroxyphenyl ratio) can be quantified without the need for lignin isolation. Correlations for p-coumarate units in the corn sample are readily seen, and a variety of the ferulate correlations are also well resolved; ferulates are important components responsible for cell wall cross-linking in grasses. Polysaccharide anomeric correlations were tentatively assigned for each plant sample based on standard samples and various literature data. With the new potential for chemometric analysis using the 2D NMR fingerprint, this gel-state method may provide the basis for an attractive approach to providing a secondary screen for selecting biomass lines and for optimizing biomass processing and conversion efficiencies.
Introduction
Developing robust, relatively rapid, yet detailed methods for plant cell wall structural characterization has become important for evaluating the escalating number of plant cell wall samples produced in biotechnology research aimed at converting biomass to biofuels. Various degradative methods, as well as non-destructive analytical spectroscopic methods (UV, IR, NIR and solid-state NMR), can be applied to plant cell wall analysis, but solution-state NMR, using modern instruments coupled with modern solution-state NMR pulse experiments, is unparalleled for providing detailed chemical structural information.1 Recent advances in dissolution of cell wall materials, admittedly currently still limited to finely divided (“ball-milled”)walls, have allowed unfractionated walls to be examined without the need for component isolation; polymer isolation approaches at best fractionate the polymer of interest, not always representatively, and at worst alter that polymer. The incredible complexity of the wall naturally results in complex spectra, but the considerable dispersion provided by 2D NMR methods, such as 13C–1H correlation spectroscopy, allows for at least some key resonances from many of the components to be sufficiently resolved to allow substantive interpretation. Obviously, much of the ability to interpret resonances in such spectra draws on the decades of painstakingly assigned NMR spectra from a plethora of model compounds, synthetic model polymers, and isolated components. To take advantage of the power of solution-state NMR, we recently developed a convenient method in which the plant cell wall need not be actually dissolved, but simply swollen in dimethylsulfoxide-based gels, to obtain well-resolved spectra.2
Plant cell walls are comprised principally of cellulose, hemicelluloses, and lignins. Secondary walls of higher plants have cellulose as the major component. Cellulose has a polymer chain of linear β-(1→4)-linked d-glucosyl (β-1,4-glucan) units, with a degree of polymerization (DP) of ~7000–15 000.3 Highly crystalline cellulose fibers are distributed across all of earth’s terrestrial plants, and can even be found in the animal kingdom—the tunicin from tunicates for example.4 Hemicelluloses (a complex class of polyoses) also exist in most plants, comprising typically 20–30% of dried wood weight5 and up to 50% of the biomass of annual and perennial plants.6 Hemicelluloses can be generally categorized into four different classes: xylans, mannans, mixed-link β-glucans, and xyloglucans.6 They are often branched polysaccharides with lower DPs containing, in addition to glucosyl units, xylose, mannose, galactose, rhamnose, and arabinose.7 The other major component of the cell wall is the lignins. Unlike the polysaccharides that are synthesized by linking (activated) monomeric sugars via enzymatic processes, lignins are synthesized from their monomers, principally the three monolignols (the hydroxycinnamyl alcohols; p-coumaryl, coniferyl, and sinapyl alcohols) by radical coupling chemistry. Many other phenolic components are also implicated in lignification.8–11
In the past, although some compositional characterization could be accomplished on unfractionated material using certain chemical degradation methods, isolation/fractionation of the plant cell wall polymers was a necessary step before characterization by NMR. Isolation of the each component is non-trivial, however, and representative isolations without fractionation (e.g., of syringyl-rich from guaiacyl-rich fractions in lignins) or structural alteration (such as deacetylation of hemicelluloses that occurs with mild base extraction) may be impossible. Lignins can be isolated as so-called milled wood lignins (MWL) from finely divided (ball-milled) materials by their dissolution into solvent mixtures with the appropriate solvent properties, such as dioxane :water (96 : 4, v/v) mixture12 or acetone : water (9 : 1, v/v). MWLs are not entirely free of polysaccharide contaminants. The yields, relative to the total lignin in the plant sample, range from ~10% to 65%, depending on the nature of the plant material; for example, typical softwoods yield ~15% MWL whereas hardwoods such as poplar or dicots such as kenaf may yield much higher levels. Cellulolytic enzyme lignins (CEL) can be obtained from treatment of the ball-milled cell walls with crude polysaccharidase enzyme mixtures;13–15 CELs also retain considerable amounts of the polysaccharide.
Isolation methods for polysaccharides are varied because of their wide range of chemical and physical characteristics. Numerous solvents, such as DMSO, alkali (KOH or NaOH), and sodium borate in alkali, can be used to fractionate and isolate hemicelluloses.5 Xylans exist as several different structural types in many terrestrial plants and algae, and can be generally isolated by two-step procedures with a NaOH/H2O2 delignification step.16,17 Galactoglucomannans and glucomannans are the major hemicellulosic components in the secondary cell walls of softwoods, but occur as minor components in hardwoods.6 Water-soluble O-acetylated galactoglucomannan can be isolated from spruce thermomechanical pulp (TMP) by hot water extraction.18,19 Xyloglucan is a major building component of the primary cell walls of higher plants, and can be extracted with aqueous alkali (1Mto 4MKOH or NaOH).20,21 The mixed-linkage-β-d-glucans can be extracted from barley and oat.22,23 It is often costly to remove lignin, especially in industrial fractionation of plant cell walls to cellulose, e.g., via chemical pulping to produce pulp and paper, or via ethanolysis24 to produce cellulose for saccharification to glucose for fermentation to ethanol. In other processes that ferment polysaccharide-derived sugars to biofuels, lignin remains a limiting factor due to its association with the polysaccharides in the wall.
A ball-milling step is an unavoidable procedure in many isolation methods for cell wall components. Significant bond cleavage results from such vigorous milling. Vibratory ball milling of cotton cellulose results in a decrease in particle size to about 10 µm and conversion of crystalline to amorphous cellulose, but in virtually insignificant oxidation of the cellulose.25 Lignin polymers fragment via cleavage of β-ether bonds,26,27 and trivial development of carbonyl structures during the ball milling.14 Ball-milled plant cell walls can be totally dissolved, not always fully intact, in various solvents such as 2 N sodium hydroxide, 50% aqueous sodium thiocyanate, 60% aqueous lithium bromide, and formic acid.13
Developing new cellulose dissolution methods has been important for the characterization of cellulosic material, and also for industrial applications. Ionic liquids (ILs), such as 1-n-butyl-3-methylimidazolium chloride (BMIMCl; [C4mim]+Cl−), 1-n-butyl-3-methylimidazolium acetate (BMIMAc), 1-ethyl-3-methylimidazolium chloride (EMIMCl; [C2mim]+Cl−), 1-ethyl-3-methylimidazolium acetate (EMIMAc), 1-n-butyldimethyl-imidazolium chloride (BDMIMCl; [C4dmim]+Cl−), 1-allyl-2,3-dimethylimidazolium bromide ([Admim]+Br−) were tested for their abilities to dissolve cellulose,28,29 and also to use as reaction media for cellulose derivatization30,31 and wood modification.32 Ionic liquids have also been applied to bioenergy research. Partial dissolution of wood has been successfully realized with BMIMCl.33 Hydrolysis kinetics in EMIMCl were studied with Miscanthus,34 and EMIMCl was also tested for corn stover recently.35 Wood liquefaction has been studied with 3,3′-ethane-1,2-diylbis(1-methyl-1H-imidazol-3-ium) dichloride, and 3,3′-ethane-1,2-diylbis(1-methyl-1H-imidazol-3-ium) dichloroaluminate.36 However, whereas ionic liquids have the ability to dissolve plant cell walls, they have not yet been found useful for lignin structural studies because the lignin components are degraded during the dissolution, which is often conducted at elevated temperatures.37
Even though complete dissolution of the entire cell wall without disrupting the genuine chemical structure is not an easy task, whole-cell-wall dissolution systems based on hydrogen-bond-disrupting solvents perform well to dissolve finely divided (ball-milled) plant cell wall material.38–40 Dimethylsulfoxide (DMSO) is a key component of the mixed solvents, which include DMSO/tetrabutylammonium fluoride (TBAF), and DMSO/N-methylimidazole (NMI). Acetylated cell wall materials, readily prepared following dissolution in DMSO/NMI, are soluble in CDCl3. High-resolution 2D39 and 3D41 NMR spectra can be acquired on the entire cell wall fraction. However, even simple acetylation of the wall causes the loss of some original information. For example, the presence of natural acetyl groups on wall components is masked when the sample is peracetylated. Also, although the dissolution and acetylation steps are simple, acetylation workup requires the (also simple but) time consuming steps of precipitation into water, isolation and washing of the precipitate (by centrifugation or ultrafiltration), drying, and re-dissolution into the NMR solvent.
To overcome such limitations, two simplified sample preparation methods to produce non-derivatized cell walls in a form amenable to solution-state NMR have been developed. The first method is logically to use deuterated DMSO and NMI directly for dissolution.39 The original barrier was that NMI-d6 was not commercially available, but its synthesis allowed development of the method.42 The resulting spectra readily revealed the presence and regiochemistry of acetates acylating cell wall hemicelluloses, notably on mannosyl and xylosyl residues, and exhibited perhaps better dispersion and resolution of the polysaccharide anomeric signals. Two significant drawbacks are the expense of NMI-d6, which is now commercially available, and the obliteration of information regarding the p-hydroxyphenyl units in lignins due to overlap of their important correlations with residual solvent peaks from incompletely deuterated NMI. It also appears that d-exchange with water may be occurring over time, resulting in larger residual solvent peaks in the NMR spectra even if highly deuterated NMI is used (unpublished data).
The second method is to simply swell the cell wall in a suitable solvent to produce a gel, from which surprisingly good spectra can still be obtained.2 Gel-samples for NMR can be prepared without the usual sample preparation requirements, such as dissolving samples in complex solvents, filtering, or drying, and with apparently no significant loss of structural information compared to the original cell wall dissolution method,39 with the possible exception that cellulose may be under-represented due to the ‘invisibility’ of crystalline cellulose in solution-state NMR. DMSO-d6 was chosen for the first version of the gel-state NMR method, because it is not only one of the more popular NMR solvents43–45 but is also an excellent swelling reagent for cellulose and other wall components. This method has already been successfully applied to the analysis of whole cell wall samples.46,47
Improvements in the current gel-sample 2D NMR method will help to deliver a more streamlined method that can be considered as a secondary screening tool to provide relatively rapid structural information on the complex polysaccharides and lignins for plant biomass research. An NMR solvent system that improves the swelling of the plant cell walls, and makes the gel-samples easier to handle is sought. The solvent system should not have residual peaks that interfere with the correlation contours from any of the components; commercially available, preferably cheap, deuterated NMR solvents are an advantage. Optimizing 2D NMR methods that allow the crucial 2D-HSQC/HMQC NMR whole-cell-wall spectra to be acquired in a short period of time (e.g., under 1 h) would be a huge advantage for the increasingly demanding bioenergy research.
Here, we introduce an improved gel-sample 2D NMR method, utilizing pyridine-d5 in DMSO-d6 for ball-milled plant cell walls. The mixture of DMSO-d6 and pyridine-d5 provides better quality 2D NMR spectra over using DMSO-d6 alone and, more importantly, allows considerably easier sample handling. This mixed solvent system is currently being evaluated and, in some cases has already been chosen, for cell wall structural research projects in the Great Lake Bioenergy Research Center (GLBRC) and at the US Dairy Forage Research Center (USDFRC) in Madison, Wisconsin, USA.
Results and discussion
Sample preparation
Ball milled plant materials were prepared as previously described,2 using grinding times summarized in Table 1. The ground particle size was predominantly <5 µm, as determined previously;42 material of approximately this degree of milling is required for optimal gelling/dissolution. The ball-milled cell wall material was directly swelled with DMSO-d6/pyridine-d5 in the NMR tube, as described in the Experimental section. The samples prepared with this solvent mixture were easier to handle, because of their better mobility and lower viscosity, than the DMSO-d6-only samples used previously (Fig. 1a). The actual solubility of components also improved. Ball milled pine cell walls, for example, can be ~28–39% dissolved in the DMSO/pyridine solvent mixture vs. ~9–19% in DMSO only. However, NMR shimming still remains insensitive due to the insoluble components in the tube; shimming is scarcely necessary for such samples.
Table 1.
Grinding times used for various samples and sample amounts. Cell wall ball-milling was performed using a Retsch PM100 planetary ball mill spinning at 600 rpm with zirconium dioxide (ZrO2) vessels (50 ml) containing ZrO2 ball bearings (10 mm × 10). The ball-milling times are fairly well optimized; extended milling time may cause additional oxidation of cell wall components
| Sample amount | Pine Interval: 20 m Interval break: 10 m |
Aspen Interval: 10 m Interval break: 5 m |
Corn & kenaf Interval: 5 m Interval break: 5 m |
|---|---|---|---|
| 100 mg | 1 h 20 m | 40 m | 25 m |
| 200 mg | 2 h 20 m | 1 h 10 m | 45 m |
| 300 mg | 3 h 20 m | 1 h 40 m | 1 h 5 m |
| 400 mg | 4 h 20 m | 2 h 10 m | 1 h 25 m |
| 500 mg | 5 h 20 m | 2 h 40 m | 1 h 45 m |
| 1 g | 10 h 20 m | 5 h 10 m | 3 h 25 m |
| 2 g | 20 h 20 m | 10 h 10 m | 6 h 45 m |
Fig. 1.
a) Gel-samples of ball-milled cell walls; i) gel-sample (~70 mg) in a 5 mm NMR tube in 1 ml of DMSO-d6; ii) a gel-sample in DMSO in a vial (upside down to demonstrate its viscosity); iii) a DMSO/pyridine (4 : 1) gel-sample in a vial demonstrating its improved mobility; iv) gel-sample (~70 mg) in a 5 mm NMR tube in 1 ml of DMSO-d6/pyridine-d5 (4 : 1). b) Short acquisition time (17 min) experiment on aspen cell walls in DMSO-d6/pyridine-d5 (4 : 1) using a 750 MHz cryoprobe-equipped NMR. The spectrum is adequate for many purposes—chemometrics, and S/G ratio estimation, for example.
2D NMR Experiments
By using the mixed DMSO-d6/pyridine-d5 solvent system, the gel-sample provided more dispersed and better resolved cell wall spectra than the original method. However, even though the sample became much easier to handle and provides better results, it still remained largely in the gel-state; the cell wall does not fully dissolve. An advantage, however, is that the rapid relaxation due to the high viscosity and presumably insoluble components allows for short acquisitions. We were able to optimize the repetition times, settling on single-scan acquisition times as short as 100 ms in F2 (1H), and interscan relaxation delays (D1) of 500 ms, as explained in the Experimental section. A total experiment time of just a few minutes already provides strikingly useful spectra. Experiments for 1–6 h with this set up provided excellent quality spectra on the 500 MHz cryoprobe instrument; we typically used 5–6 h acquisitions here. Despite resolution reduction with the short relaxation time (producing broader peaks), sufficient resolution and dispersion from these gel-state samples provides an advantage over true solution-state NMR on actual cell wall solutions; the time required to acquire spectra can be significantly reduced. Our 17 min experiment (Fig. 1b) using a 750 MHz cryoprobe-equipped NMR instrument provided a spectrum which is adequate for most purposes, for chemometrics and S/Gratio estimation, for example. This result clearly shows that we can acquire a satisfactory 2D NMR spectrum of the whole cell wall in under an hour, implying that acquiring spectra is not the barrier to obtaining data from over 20 samples per day.
We are already acquainted with the origin of the spectral signals—they derive not only from the completely soluble component, but from the insoluble swollen gel component as well. Acquisition from a DMSO or DMSO-pyridine gel prepared from the insoluble residue remaining after extensive extraction of the cell wall with DMSO, as described in the previous publication,2 provides essentially the same quality spectrum as from the whole cell wall sample.
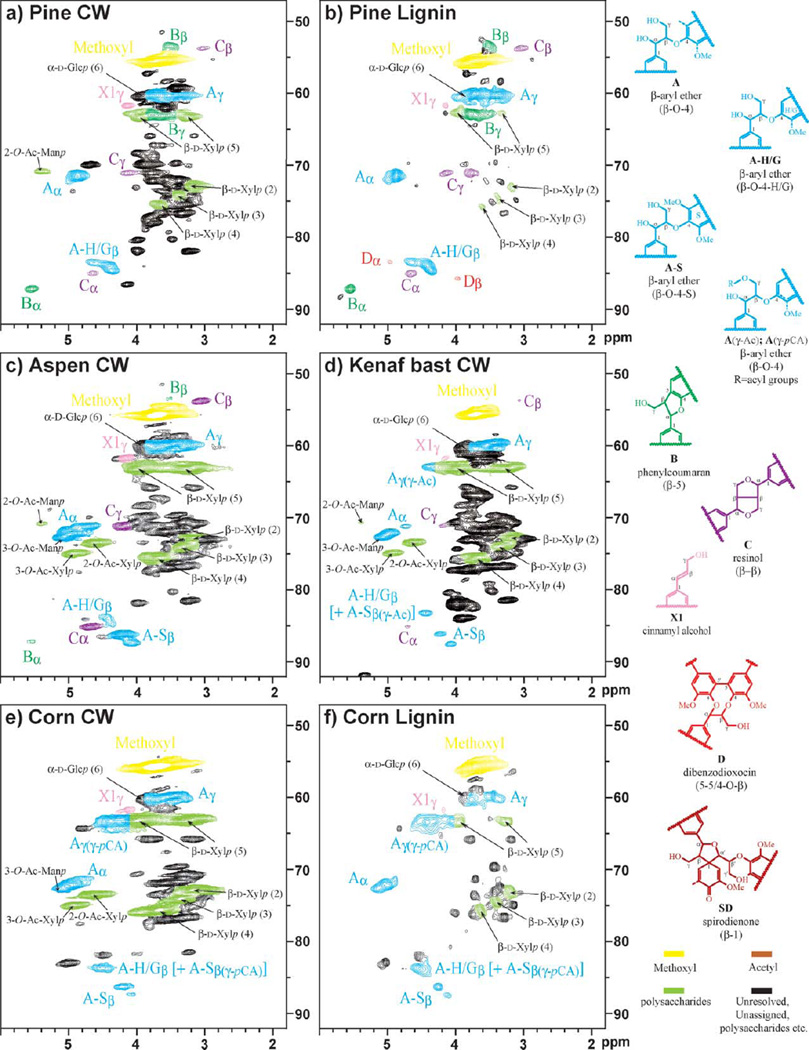
Four different plant species were used here to illustrate the usefulness of the method for the variety of plant materials of interest: loblolly pine (Pinus taeda), aspen (Populus tremuloides), kenaf (Hibiscus cannabinus), and corn (Zea mays L.). Despite the large amounts of polysaccharides (cellulose and hemicelluloses) in the sample, the lignin resonances are extremely well resolved in the 2DNMRspectra. The full HSQC (1-bond 13C–1H correlation) spectra (provided in the Supplementary Material, Fig. S1) show essentially the entire spectra (but not, for example, the aldehyde region above 8 ppm in the proton dimension) of the whole cell wall samples. Fig. 2 shows the lignin aliphatic region, along with the polysaccharide resonances in that region (the acetyl regions are included in the Supplementary Material, Fig. S2). Aromatic regions, shown in Fig. 3, represent the entire lignin component. For comparison in the pine case, we show the spectrum of an isolated (fully soluble) pine lignin in the same solvent. The polysaccharide anomeric regions are shown in more detail in Fig. 4.
Fig. 2.
Aliphatic (sidechain) regions of 2D 13C–1H correlation (HSQC) spectra from cell wall gels and soluble lignins from various samples in DMSO-d6/pyridine-d5 (4 : 1). a) Pine, b) pine isolated lignin, c) aspen, d) kenaf bast fiber, e) corn stems, f) corn isolated lignin. Correlations from lignin components, and some of the characteristic polysaccharide correlations such as those from O-acetylated xylans and mannans, are well isolated from the other densely packed polysaccharides peaks; color coding is according to the structures shown.
Fig. 3.
Aromatic regions of 2D 13C–1H correlation (HSQC) spectra from cell wall gels and soluble lignins from various samples in DMSO-d6/pyridine-d5 (4 : 1). a) Pine, b) pine isolated lignin, c) aspen, d) kenaf bast fiber, e) corn stems, f) corn isolated lignin. Correlations from the aromatic rings are well dispersed and can be categorized by the type of aromatic units (syringyl S, guaiacyl G, p-hydroxyphenyl H, p-hydroxybenzoate PB, p-coumarate pCA, and ferulate FA; color coding is according to the structures shown. Corn samples used high-d (“100%”) pyridine.
Fig. 4.
Polysaccharide anomeric regions of 2D 13C–1H correlation (HSQC) spectra from cell wall gels from various samples in DMSO-d6/pyridine-d5 (4 : 1). a) Pine, b) aspen, c) kenaf bast fiber, d) corn stems. The tentative assignments are based on actual NMR data from limited sources of polysaccharides in DMSO-d6/pyridine-d5 (4 : 1) and other references in which samples were mostly in D2O. α-d-Glcp, α-d-glucopyranoside; β-d-Glcp, β-d-glucopyranoside; α-d-Manp, α-d-mannopyranoside; β-d-Manp, β-d-mannopyranoside; β-d-Xylp, β-d-xylopyranoside; α1-d-Galp, α-d-galactopyranoside; β-d-Galp, β-d-galactopyranoside; α-l-Araf , α-l-arabinofuranoside; β-d-Araf, β-d-arabinofuranoside; α-l-Rhap, α-l-rhamnopyranoside; 2-O-Ac-β-d-Manp, acetylated β-d-Manp; 3-O-Ac-β-d-Manp, acetylated β-d-Manp; 2-O-Ac-β-d-Xylp, acetylated β-d-Xylp; 3-O-Ac-β-d-Xylp, acetylated β-d-Xylp; 4-O-MeGlcA, 4-O-methyl-α-d-glucuronic acid; R, reducing end, NR; non-reducing end.
Lignin sidechain regions
The structural types and distribution of interunit bonding patterns of the lignin fraction can be found in the aliphatic sidechain region (Fig. 2). This area also contains plentiful polysaccharides, but at least one of the correlations for each of the structures in lignin is well isolated. The important correlations, such as those for methoxyl groups, β-ether (β–O–4) units A, phenylcoumaran (β–5) units B, resinol (β–β) units C, and dibenzodioxocin (5–5/4–O–β) units D can be readily assigned by comparison with model data48 and from prior lignin and cell wall spectra. The relative volumes of the contour peaks, and the implied relative concentrations of the each component, are dependent on the plant species. The NMR chemical shift data for the phenolic components are given in Table 2.
Table 2.
Assigned NMR data for lignin and other phenolic plant cell wall components in DMSO-d6/pyridine-d5 (4 : 1). A-H/G, β–O–4-H/G; A-S, β–O–4-S; B, β–5; C, β–β; D, dibenzodioxocine (5–5/4–O–β); SD, spirodienone (β–1); X1, cinnamyl alcohol end group; G, guaiacyl; G ′, guaiacyl (oxidized α-ketone); S, syringyl; S′, syringyl (oxidized α-ketone); H, p-hydroxyphenyl; PB, p-hydroxybenzoates; FA, ferulate; pCA; p-coumarates
| α(7) | β(8) | γ(9) | 2 | 3 | 5 | 6 | |
|---|---|---|---|---|---|---|---|
| A-H/G | 71.3/4.87 | 84.4/4.39 (threo) | 60.1/3.73 | ||||
| 83.9/4.45 (erythro) | 60.1/3.40 | ||||||
| A-S | 71.3/4.87 | 87.2/4.09 (threo) | 60.1/3.73 | ||||
| 86.2/4.22 (erythro) | 60.1/3.40 | ||||||
| A-γ-Ac | 63.3/4.42 | ||||||
| 63.3/3.94 | |||||||
| B | 87.1/5.55 | 54.3/3.57 | 62.8/3.80 | ||||
| C | 85.0/4.67 | 53.7/3.06 | 71.0/4.14 | ||||
| 71.0/3.77 | |||||||
| D | 83.4/4.97 | 85.7/3.97 | |||||
| SD | 81.2/5.13 | 60.2/2.82 (β) | |||||
| 79.7/4.20 (β′) | |||||||
| X1 | 128.5/6.49 | 128.7/6.38 | 61.7/4.15 | ||||
| G | 110.8/7.02 | 115.2/6.90 | 115.2/6.90 | ||||
| 119.0/6.87 | 119.0/6.87 | ||||||
| G′ | 111.3/7.52 | 122.9/7.57 | |||||
| S | 103.7/6.73 | 103.7/6.73 | |||||
| S′ | 106.7/7.24 | 106.7/7.24 | |||||
| H | 127.9/7.27 | 127.9/7.27 | |||||
| PB | 131.4/7.71 | 131.4/7.71 | |||||
| FA | 144.9/7.53 | 115.6/6.39 | 111.1/7.36 | 123.4/7.14 | |||
| pCA | 144.9/7.53 | 113.9/6.29 | 130.2/7.49 | 115.8/6.84 | 115.8/6.84 | 130.2/7.49 |
The isolated pine lignin, a guaiacyl lignin typical of gymnosperms, was easily dissolved in DMSO-d6/pyridine-d5 (Fig. 2b). β-Ether (β–O–4) units A are the major interunit structure of lignins, followed by phenylcoumaran (β–5) units B. Minor amounts of pinoresinol (β–β) units C, and dibenzodioxocin (5–5/4–O–β) units D are also well resolved. Cinnamyl alcohol end-groups X1 are recognizable by γ-C/H correlation, although we have noted previously that this correlation may not be purely attributable to such structures. However, as only traces of polysaccharides remain in this lignin sample, we can reasonably assign the cinnamyl alcohol endgroup X1 to the entire contour at 61.7/4.15 ppm.
Pine cell walls, Fig. 2a, even though they are run as a gel-sample, provide a well resolved spectrum that has better quality than the prior DMSO-d6-only spectrum;2 the resolution and dispersion of various correlations for the lignin components in this spectrum coincide with those from the isolated pine lignin HSQC spectrum, although the cell wall spectrum is more congested with polysaccharide peaks. Minor components, such as the dibenzodioxocin (5–5/4–O–β) D and spirodienone (β–1) SD structures, may only be seen at lower contour levels (not shown). This information itself may be revealing, suggesting that the dibenzodioxocins are concentrated in the low molecular weight lignins that extract most easily during the lignin isolation. An acetate peak at δC/δH 20.8/1.98 ppm (Fig. S1), which is slightly shifted from in the DMSO-d6-only spectrum (δC/δH 20.9/2.01 ppm), likely belongs to polysaccharides rather than to lignin, as only a trace of the acetate contour can be found in the extracted pine lignin. Pine (and general softwood) hemicelluloses are known to be partially acetylated, particularly on xylan and mannan components.19,49,50
Aspen, an angiosperm, has a typical guaiacyl-syringyl type lignin that is biosynthesized from both coniferyl and sinapyl alcohols. The HSQC spectrum (Fig. 2c) of the aspen cell wall resolves extra correlations for the various lignin linkage types over the peaks already seen in pine. There are clear differences in interunit proportions between the different species, due to the syringyl units, in which both the 3-and 5-positions are substituted with methoxyl groups. Phenylcoumaran (β–5) units B can only be made from coupling of a monolignol with a guaiacyl (G) or p-hydroxyphenyl (H) unit. As a result, there are relatively fewer phenylcoumaran units B compared to the major β-ether (β–O–4) units A in aspen lignins. Similarly, dibenzodioxocin D units, resulting from coupling of a monolignol with 5–5-coupled (biphenyl) units, are minor components because, again, biphenyl units cannot be formed from syringyl units. In contrast, resinol (β–β) C correlations are easily detected because sinapyl alcohol favors forming the syringaresinol dehydrodimer, which begins most of the polymer chains. Enhanced β-ether (β–O–4) A levels in the aspen is another result of the existence of syringyl units; basically, monolignol addition to a syringyl unit has essentially only a single pathway available, β–O–4-coupling; a minor β–1-coupling pathway will be noted below. Two well dispersed β-position correlations are seen for the β-ether units A, depending on the sidechain substitution. In the case of a β–O–4-guaiacyl (or -p-hydroxyphenyl) unit (β–O–4-H/G) A-H/G, the 13Cβ/1Hβ correlation is centered at 84.2/4.40 ppm (threo: 84.4/4.39 ppm and erythro: 83.9/4.45 ppm). In the case of a β–O–4-syringyl unit (β–O–4-S) A-S, the 13Cβ/1Hβ correlation is centered at 86.7/4.15 ppm (threo: 87.2/4.09 ppm and erythro: 86.2/4.22 ppm). The A-S correlations are much more intense than those from A-H/G in this aspen cell wall sample, implying that more of β-ether units in aspen are assembled via monolignols cross-coupling with syringyl units than with guaiacyl units. Several diagnostic spirodienone (β–1) SD peaks were also detected at near the noise level (not shown); it is not possible to see all of the 13C–1H correlations but characteristic peaks are clear: SD-α at 81.2/5.13 ppm, SD-β′ at 79.7/4.20 ppm, and SD-β at 60.2/2.82 ppm. Such units were only rather recently discovered in lignins; their 13C–1H correlation assignments here are based on previous assignments.43,51,52
Kenaf bast fiber lignin is a guaiacyl-syringyl lignin that is particularly syringyl-rich,53 as noted in the cell wall spectrum of Fig. 2d. The lignin is seen as being rich in ²-ether units A, with no visible phenylcoumaran B or dibenzodioxocin D units. Only a small amount of resinol C can be detected even though the plant is syringyl-rich. The reason is that the γ-positions of sinapyl alcohol monomers in kenaf are extensively acetylated, so that the major β–β-coupling products involve at least one sinapyl acetate, precluding the formation of the bis-tetrahydrofuran (the resinol C) and instead resulting in a number of mono-tetrahydrofuran type structures.53 Also, β-ether units in kenaf bast fiber lignins are heavily γ-acetylated according to previous studies.54–60 However, recent assignments of the 13Cβ/1Hβ correlations have likely overestimated the γ-acylated structures. The entire correlation at around 83.3/4.44 ppm has been attributed to the β-position of γ-acetylated β–O–4-structures A(γ-Ac),42,60 but normal β–O–4-H/G units A-H/G also have their 13Cβ/1Hβ correlations in this region. Also, the correlations at 63.1/3.82–4.40 ppm were assigned as belonging to the γ-acylated β–O–4-structure A(γ-Ac), even though the area contains theC6/H6 resonances of various hexoses that remain in the samples.
As an example of a grass, the spectrum from corn stalk cell walls, shown in Fig. 2e, provides another example of a guaiacyl-syringyl lignin. The lignin is rich in β-ether units A with, surprisingly, no phenylcoumaran B, dibenzodioxocin D, or resinol C units visible in this spectrum. An NMR spectrum from isolated corn lignin is given in Fig. 2f for comparison. There is a pair of γ-position peaks from the γ-p-coumaroylated β-ether units A(γ-pCA) clearly seen at 63.3/4.42 and 63.3/3.94 ppm, but the correlation at 63.3/3.94 ppm overlaps with one of the 6-position peaks of xylan.
Acetylated hemicelluloses in the aliphatic regions
Plant cell walls have abundant acetyl groups. Acetylation is important to assay as the hydroxyl acylation can inhibit saccharification of polysaccharides.61 Released acetate is also somewhat inhibitory to fermentation of the sugars to ethanol.62,63
The acetate methyl can be seen at ~20.7/1.97 ppm in Fig. 1 (and Supplementary Material Fig. S1 and S2). Most of the acetyl groups belong to hemicellulosic components, except in plants such as kenaf60 which have highly acetylated lignins. Naturally acetylated mannans and xylans can be easily detected in the aliphatic regions of the spectra (Fig. 2, Table 3). The O-acetylated galactoglucomannans in softwood have 20–25% mannans,49 and the galactoglucomannans contain acetyl groups mostly on (1→4)-β-mannosyl units at position C2 and C3.64 The chemical shifts of the C2/H2 peak of 2-O-Ac-Manp (average value: 72.8/5.45 ppm) and C3/H3 peak of 3-O-Ac-Manp (average value: 74.6/5.08 ppm) have been reported in D2O.65,66 The pine cell wall spectrum here, Fig. 2a, shows a 2-O-Ac-Manp C2/H2 peak at 70.7/5.41 ppm in DMSO-d6/pyridine-d5 (4 : 1), but the 3-O-Ac-Manp C3/H3 peak (that may appear at ~72.2/5.00 ppm) is not clearly resolved from the Cα/Hα peak from the β–O–4-ether lignin component Aα. The 2-O-Ac-Manp C2/H2 peak is completely absent in the isolated pine lignin sample (Fig. 2b) as the hemicellulosic components have been removed. O-Acetylated glucomannan has also been reported in aspen and birch wood.66 Indeed, the 2-O-Ac-Manp peak is also present in the aspen cell wall spectrum (Fig. 2c) but was relatively less prevalent than in the pine cell wall (Fig. 2a). Kenaf (Fig. 2d) also shows a small correlation for the same component, but the peak is not detected in the corn cell wall (Fig. 2e). Aspen, kenaf, and corn however possess large amounts of O-acetylated xylan, unlike pine. Generally, softwood xylans, in which arabinofuranose units are attached by β-(1→3)-glycosidic bonds to the xylan,49 lack acetyl substitution. However, acetylated 4-O-methylgluconoxylan is a major hemicellulosic component in hardwoods, and acetyl groups frequently acylate the C2 and C3 positions. Teleman et al. reported NMR data (in D2O) for O-acetyl-(4-O-methyl-glucurono)-xylan in hardwoods, including aspen.50,67 Aspen, kenaf, and corn cell wall samples show a strong 2-O-Ac-β-d-Xylp C2/H2 correlation at 73.5/4.64 ppm and a 3-O-Ac-β-d-Xylp C3/H3 correlation at 75.0/4.94 ppm (Fig. 2c–e). The anomeric (C1/H1) correlations from the O-acetylated glucomannan and the O-acetylated xylan will be discussed later in the polysaccharide anomeric correlations section.
Table 3.
NMR data for polysaccharide components in the plant cell walls in D2O, DMSO-d6 and DMSO-d6/pyridine-d5 (DP, 4 : 1)
| Saccharide (unit) | C1/H1 | C2/H2 | C3/H3 | Solvents | Ref. |
|---|---|---|---|---|---|
| Cellulose; | 102.1–102.9/4.40–4.53 | D2O | 81,82 | ||
| (1→4)-β-d-Glcp | |||||
| Cellulose; | 103.1/4.39 | DP | Cellulose | ||
| (1→4)-β-d-Glcp | |||||
| (1→3)- & | 103.8/4.49 | D2O | 19,66,92,93 | ||
| (1→4)-β-d-Glcp | |||||
| (1→3)-Glcp | 103.2/4.53 | DMSO-d6 | 92 | ||
| (1→6)-β-d-Glcp | 105.1/4.53 | D2O | 93 | ||
| (1→6)-β-d-Glcp | 103.2/4.22 | DMSO-d6 | 92 | ||
| (1→4)-β-d-Glcp (R) | 96.7/4.66 | D2O | 91 | ||
| (1→4)-β-d-Glcp (R) | 97.1/4.44 | DP | Cellobiose, cellotriose | ||
| (1→4)-α-d-Glcp | 101.8/4.32 | DP | Starch | ||
| (1→4)-α-d-Glcp (R) | 92.7/5.22 | D2O | 91 | ||
| (1→4)-α-d-Glcp (R) | 92.2/5.10 | DP | Cellobiose, cellotriose | ||
| Xylan; | 101.8/4.32 | DP | Xylan from oat spelts | ||
| (1→4)-β-d-Xylp | |||||
| (1→4)-β-d-Xylp (R) | 97.6/4.58 | D2O | 67 | ||
| (1→4)-β-d-Xylp (R) | 97.5/4.38 | DP | |||
| (1→4)-α-d-Xylp (R) | 93.0 (?)/5.18 | D2O | 67 | ||
| (1→4)-α-d-Xylp (R) | 92.4/5.10 | DP | |||
| 2-O-Ac-β-d-Xylp | 101.1/4.64 | 74.8/4.69 | D2O | 50,67 | |
| 2-O-Ac-β-d-Xylp | 99.7/4.58 | 73.5/4.64 | DP | ||
| 3-O-Ac-β-d-Xylp | 102.7/4.58 | 76.5/4.98 | D2O | 50,67 | |
| 3-O-Ac-β-d-Xylp | 101.7/4.51 | 75.0/4.94 | DP | ||
| (1→4)-β-d-Manp | 101.8/4.75 | D2O | 84 | ||
| 100.3/- | 85 | ||||
| 102.2/4.73 | 66 | ||||
| (1→4)-β-d-Manp | 100.7/4.63 | DP | |||
| α-d-Manp (I) | 93.4–103.1/5.04–5.15 | D2O | 87 | ||
| 96.4–102.5/4.50–5.43 | 88 | ||||
| α-d-Manp (I) | 94.0/5.05 | DP | |||
| Mannans? | 101.0/4.66 | DP | |||
| (1→4)-α-d-Manp (R) | 95.2/5.19 | D2O | 66 | ||
| (1→4)-α-d-Manp (R) | 94.0/5.05 | DP | |||
| 2-O-Ac-Manp | 72.7/5.45 | D2O | 65 | ||
| 2-O-Ac-Manp | 100.4/4.86–4.94 | 72.9–73.1/5.38–5.48 | D2O | 66 | |
| 2-O-Ac-Manp | 98.9/4.86 | 70.7/5.41 | DP | ||
| 3-O-Ac-Manp | 74.5/5.10 | D2O | 65 | ||
| 3-O-Ac-Manp | 100.9/4.81–4.84 | 74.7/5.02–5.08 | D2O | 66 | |
| 3-O-Ac-Manp | 99.9/4.78 | 72.2/5.00 (?) | DP | ||
| 4-O-methyl-α-d-glucuronic acid (MeGlcA) | 98.8/5.26 | D2O | 67 | ||
| 98.4/5.19 | 83 | ||||
| 4-O-methyl-α-d-glucuronic acid (MeGlcA) | 97.5/5.31 | DP | |||
| (1→4)-β-d-Galp | 105.2/4.64 | D2O | 89 | ||
| 106.5/4.65 | 90 | ||||
| (1→4)-β-d-Galp | 105.5/4.38 | DP | |||
| (1→4)-β-d-Galp (R) | 97.3/4.61 | D2O | 89 | ||
| (1→4)-β-d-Galp (R) | 97.3/4.34 | DP | |||
| (1→4)-α-d-Galp (R) | 93.2/5.27 | D2O | 89 | ||
| (1→4)-α-d-Galp (R) | 92.6/5.10 | DP | |||
| (1→6)-α-d-Galp (NR) | 100.2/5.02 | D2O | 84 | ||
| 99.1/— | 85 | ||||
| 100.8/5.02 | 86 | ||||
| (1→6)-α-d-Galp (NR) | 99.1/4.82 | DP | |||
| α-l-Araf | 105.4–110.4/4.99–5.26 | D2O | 90,94–97 | ||
| α-l-Araf | 106.0–109.0/4.70–5.70 | DP | |||
| β-l-Araf | 101.4–104.3/5.10 | D2O | 90,94 | ||
| β-l-Araf | 100.5–103.5/5.35–5.00 | DP | |||
| (1→2)-α-l-Rhap | 99.16–99.18/5.29–5.31 | D2O | 99 | ||
| 108.8/5.20 | 100 | ||||
| (1→2)-α-l-Rhap | 99.3/5.24 | DP | |||
| α-l-Fucp | 100.7/5.25 | D2O | 20,101 | ||
| α-l-Fucp | 100.5/5.21 | DP |
Aromatic region (and H:G: S ratios)
Aromatic regions of the 2D 13C–1H correlation (HSQC) spectra highlight the differences in the p-hydroxyphenyl: guaiacyl : syringyl (H :G: S) distributions in the lignins. Fig. 3 shows the significant variations in the compositions of the lignin polymers and other aromatic constituents in the wall. Simple guaiacyl : syringyl (G : S) and even p-hydroxyphenyl : guaiacyl : syringyl (H :G: S) integral ratios for the cell wall spectra can be obtained readily by integrating the contours (see Experimental). Integrals from the well-dispersed 2- or 2,6-positions of each kind of aromatic types can be used (with guaiacyl integrals being logically doubled since they involve only a single (2-) correlation rather than the two in the symmetrical H and S units).
Pine lignin is a guaiacyl (G) lignin (derived from coniferyl alcohol) with traces of p-hydroxyphenyl (H) units (derived from p-coumaryl alcohol), as seen in the spectra from the isolated lignin (which fully dissolved in the solvent, Fig. 3b) or the cell wall (which formed a gel, Fig. 3a). The C/H correlations from the G aromatic rings are well resolved in DMSO-d6/pyridine-d5. The correlation for the G2-position is at 110.9/7.03 ppm, except for the 2-position of oxidized α-ketone structures G′2 (~111.1/7.50 ppm). The G5 C/H-correlation is at ~115.1/6.90 ppm, and most of the 6-position correlations are at 119.0/6.88 ppm except for the 6-position of the phenylcoumaran (β–5) units B at about 115.1/6.49 ppm, which is basically overlapped with the most of G5 correlations, so it is not possible to differentiate clearly between theG5 and G6 correlation peaks. Correlation peaks from the oxidized α-ketone structure G′6 (~122.8/7.58 ppm), which is resolved from a pyridine peak in this spectrum, also separated from the major G6 correlations. A minor H2,6-aromatic correlation from p-hydroxyphenyl (H) units is barely visible at 128.0/7.27 ppm, but the H3,5-position correlations overlap with those from guaiacyl 5-positions. The measured p-hydroxyphenyl : guaiacyl (H: G) integral value in the isolated lignin spectrum (Fig. 3b) was 0.017 (i.e., 1.7% H-units), whereas in the cell wall it was 0.021 (i.e., 2.1%). The difference between these samples suggests that lignin polymer chains in which H-units are involved are of higher molecular weight and/or are more cross-linked, and therefore do not extract as readily into the isolable lignin fraction.
Aspen lignin’s balanced syringyl-guaiacyl composition is clearly evidenced in the aromatic region in this gel-sample spectrum (Fig. 3c). All of the guaiacyl components which are in pine lignin can be found, but only a negligible amount of the oxidized α-ketone structures G′ are seen in the aspen. There is, however, a significant correlation for the oxidized (α-ketone) structures S′ at 106.8/7.24 ppm, along with the normal S2,6-correlations at 103.8/6.76 ppm. Syringyl units are known to oxidize more readily; it is not yet clear to what degree this oxidation occurs in the lignification process in the plant vs. in the sample preparation steps (ball milling and sonication in the NMR solvents). A measured S :Gintegral of 2.53 (which includes both the normal and oxidized S components) is somewhat higher than the 1.8 determined in other aspen samples from acetylated cell walls39 and the 2.2 by thioacidolysis.68 Another readily evidenced feature of aspen lignin is the p-hydroxybenzoates (PB); the PB2/6 correlations are at 131.4/7.71 ppm.69 PB is considered to exclusively acylate the γ-position of lignin side chains, analogously with p-coumarates (pCA) in grasses.56 As with natural lignin acetylation, evidence is compelling that lignin acylation by PB occurs via lignification using p-hydroxybenzoylated lignin monomers.56 Integral response factors have not been measured, but the contour volumes for PB amount is about 2.8% of those for the S2/6 and G2 lignin contours, suggesting about this level of substitution.
Kenaf bast cell walls have extremely high syringyl levels.53 The syringyl correlations predominate over the weak guaiacyl correlations (Fig. 3d). Kenaf’s high S :G ratio (the integral ratio is 5.46 here) has been noted previously.55 Spectra from the freshly harvested and milled Tainung2 kenaf also appeared to have considerable amounts of H units. It remains to be confirmed whether these apparent H-units truly arise from p-coumaryl alcohol in lignification, or are due to other p-hydroxyphenyl units such as tyrosine or tyramine. Kenaf’s H:G integral was 0.50, and the total H:G: S ratio was 7.2 : 14.4 : 78.4. H units have been detected by pyrolysis-gas chromatography/mass spectroscopy (Py-GC/MS) previously in kenaf at a lower level.58 The S:G value from the Py-GC/MS was 5.4 and the H:G: S ratio was 1.3 : 15.4 : 83.3. Kenaf was also examined with DFRC (Derivatization Followed by Reductive Cleavage) methods, which detected only trace levels of H-units.70 The H-unit correlations in this kenaf cell wall have a unique extra peak at 129.1/7.23 ppm compared to the normal H-units from other plants, although the correlations have a similarity to those obtained from HCT-deficient pine tracheids,71 which have increased H-levels in the lignin.
Corn stalks have even more correlations in the aromatic region compared to the other samples because of the hydroxycinnamates, p-coumarate (pCA) and ferulate (FA) present in grass cell walls (Fig. 3e). Syringyl correlations dominate those from guaiacyl units, with an S :G integral of 1.41. There are also oxidized α-ketone structures of syringyl units, S′. However, the most interesting features are the clearly revealed intense pCA and FA peaks.
Ferulates, largely acylating arabinoxylans, are readily seen in the corn spectrum here and in other grasses (Marita, Kim, 2009, unpublished). Ferulates are also involved in lignification, cross-coupling with lignin monomers and possibly oligomers resulting in lignin-polysaccharide cross-linking that is an important feature of grass cell walls that also limits polysaccharide digestibility.72–74 Although the plethora of cross-linked ferulates are not, yet, identified in this spectrum, being able to profile the levels of free ferulates on polysaccharides (the ones in the correlations labeled here) will presumably provide a valuable measure of cross-linking and therefore recalcitrance to saccharification. The peak at 123.3/7.13 ppm belongs to 6-position of FA, and the FA8 correlation at 115.6/6.41 ppm is also partially resolved. The FA7 correlation coincides with that of pCA7 at 144.8/7.51 ppm. The FA2 correlation appears at 111.1/7.36 ppm which is the same as for the G2 α-ketone, G′2; however the FA2 correlation can be considered as a pure peak because there does not appear to be any oxidized guaiacyl units in this corn sample—G′6 is not detectable. As noted above in other S/G lignins (kenaf and aspen), oxidation of G-units is minor. The FA5 correlation overlaps with G5 correlations.
p-Coumarates also acylate arabinoxylans,75,76 to a lesser degree than ferulates, but are mainly found acylating lignin sidechains in grasses. Again, they arise via lignification using p-coumaroylated monolignols.77 The pCA2/6-correlations are at 130.2/7.48 ppm, and the 3,5-correlations at 115.8/6.83 ppm overlap with those from guaiacyl units. The pCA7 and pCA8 correlations (144.8/7.51 and 113.9/6.29 ppm) are also noted.
The integral ratio between cinnamates and normal lignin units was 55 : 45. This ratio suggests that pCA might be over-represented in the NMR spectrum since the corn lignin is only about 19% p-coumarate by weight;78 response factors are required for the cinnamates estimation. We hope to improve the reliability of the integrals with determination of relative response factors in the future, along with an exploration of other NMR integration analysis methods, such as those in Sparky79 or NMR Pipe.80
An isolated corn lignin was also examined in the DMSO-d6/pyridine-d5 solvent system (Fig. 3f). The NMR spectrum shows a similar profile as the whole cell wall, but with much smaller FA correlations. As noted above, ferulates acylate arabinoxylans, but their presence in the lignin fraction suggests the association between lignins and polysaccharides mediated by ferulates. The S/G ratio of the corn lignin is 2.18, somewhat higher than the value from the whole cell wall (1.41). The cinnamates ratio presumably overrepresent pCA (as noted above) as 51 : 49 in the isolated lignin. At this time, we simply report that the proportional values define the characteristic structural differences between each species.
More importantly, ferulate signals are well resolved from p-coumarate signals. They are unable to be differentiated from other guaiacyl units and p-coumarate units in acetylated samples. The ready determination of ferulates and p-coumarates makes gel-sample NMR of underivatized whole cell wall materials particularly valuable for grasses.
Polysaccharide anomeric correlations
Most of the correlations in the 90–110/3.5–6.0 ppm region, Fig. 4, belong to polysaccharide anomerics. We have attempted to assign the peaks relating to polysaccharides as much as possible (Table 3), but more substantial assignment of the many dispersed contours needs to be made in future studies using models and a variety of well-characterized polysaccharide polymers. The provisional peak assignments in this paper are based on hemicellulose data reported in numerous publications and the NMR data for various oligomers and polysaccharides that were collected directly in our lab from various sources (Table 3), such as locust bean gum (galactomannan polysaccharide, from seeds of Ceratonia Siliqua L.), xylan from oat spelts, d-(+)-cellobiose, d-(+)-cellotriose, cellulose powder (microcrystalline, ~20 micron), mannan from Saccharomyces cerevisiae, and (+)-arabinogalactan from larch wood. The NMR chemical shifts of the hemicelluloses in DMSO-d6/pyridine-d5 (4 : 1) solvents system are slightly upfield (about 1–2 ppm for 13C, and 0.1–0.2 ppm for 1H) from the chemical shifts that were obtained in D2O, the solvent normally used for hemicelluloses NMR studies.
Cellulose [(1→4)-β-d-Glcp], the most important plant cell wall component, appears in spectra from all of the plants at 103.1/4.39 ppm. The same correlation peak is also noted from cellulose oligomers.81,82 However, xylan [(1→4)-β-d-Xylp] at 101.8/4.32 ppm share the same chemical shift with starch [(1→4)-α-d-Glcp]. Another ubiquitous component in all of these plant samples is 4-O-methyl-α-d-glucuronic acid (MeGlcA). This well-isolated correlation at 97.5/5.31 ppm matches with the NMR data of the isolated hemicelluloses from aspen and Argania spinosa fruit.67,83
The pine cell wall (Fig. 4a) reveals the anomeric from β-d-mannosyl [(1→4)-β-d-Manp] residues at 100.7/4.63 ppm. These values match with the NMR data from locust bean gum, and other literature.84,85 The anomeric positions (C1/H1) of 2-O-Ac-Manp at 98.9/4.86 ppm and 3-O-Ac-Manp at 99.9/4.78 ppm were also detected.65,66 The 2-O-Ac-Manp correlation is not well isolated from the α-d-galactosyl [(1→6)-α-d-Galp] nonreducing end branch units at 99.1/4.82 ppm.84–86 The anomeric peak of the reducing end of α-d-mannosyl [(1→4)-α-d-Manp] residues appears at 94.0/5.05 ppm.66 The internal units of α-d-Manp (which was obtained from the Saccharomyces cerevisiae sample) at 94.0/5.05 ppm can be detected near the noise level.87,88 Pine also has β-d-galactosyl [(1→4)-β-d-Galp] internal units at 105.5/4.38 ppm.89,90 The reducing end of β-d-galactosyl [(1→4)-β-d-Galp] residue is at 97.3/4.34 ppm89 which is close to the reducing end of β-d-glucosyl [(1→4)-β-d-Glcp] units at 97.1/4.44 ppm.91 However the correlation from the reducing end of (1→4)-β-d-Galp shares the region with the reducing end of (1→4)-β-d-Xylp at 97.5/4.38 ppm.67 The correlations from α-d-galactosyl [(1→4)-α-d-Galp] reducing end units at 92.6/5.10 ppm89 and β-d-glucosyl [(1→4)-α-d-Glcp] reducing end units at 92.2/5.10 ppm overlap.91 This peak also has similar chemical shifts as the reducing end of (1→4)- α-d-Xylp units.67 The β-d-glucosyl [(1→3)-& (1→4)-β-d-Glcp] units of glucomannan may appear in the same correlation region as cellulose at 103.0/4.46 ppm.19,66,92,93 There are several peaks in the area of 106–109/4.7–5.7 ppm, and those peaks may all be considered to be α-l-arabinofuranosyl (α-l-Araf) residues.90,94–97 Each correlation appears to represent different connectivity, such as (1→3)-, (1→5)-, and (1→6)-, but we are not currently able to assign them conclusively. The small correlations in the area of 100.5–103.5/5.35–5.00 ppm are most likely β-l-arabinofuranosyl (β-l-Araf) units.90,94 A suspected trace of (1→2)-α-l-rhamnopyranosyl [(1→2)-α-l-Rhap] residues at 99.3/5.24 ppm appears in pine, and the correlation is bigger in aspen; rhamnose units are known in aspen.98
The aspen cell wall spectrum (Fig. 4b) shows huge correlations for cellulose at 103.1/4.39 ppm and xylan at about 101.9/4.38 ppm as in other plant cell wall spectra. As noted above, acetylated 4-O-methylgluconoxylan is a major hemicellulosic component in hardwoods, such as aspen, birch and beech, and the acetyl groups are on the C2 and C3 positions.50,67 The corresponding anomeric (C1/H1) peak of 2-O-Ac-β-d-Xylp at 99.7/4.58 ppm, and the one for 3-O-Ac-β-d-Xylp at 101.7/4.51 ppm can be found near the cellulose peak. The reducing end of (1→4)-β-d-Xylp is at 97.5/4.38 ppm;67 however the trace amount of the reducing end of (1→4)-β-d-Galp may share this region.89 The correlations from the (1→4)-α-d-Xylp reducing end at about 92.4/5.10 ppm, and the α-d-galactosyl [(1→4)-α-d-Galp] reducing end also have similar chemical shifts.67 The β-d-mannosyl [(1→4)-β-d-Manp] residues appear at 100.5/4.63 ppm, the anomeric peak of 2-O-Ac-Manp is at 99.0/4.79 ppm, and a trace of the 3-O-Ac-Manp is at 99.9/4.79 ppm. The internal unit of α-d-Manp is at 94.0/5.05 ppm as a minor component. An unassigned peak at 101.0/4.66 ppm may be related to mannan structures.88 Only one of the α-l-arabinosyl (α-l-Araf) residues appears at 108.1/4.90 ppm. The cellulose correlation appears at about 103.0/4.46 ppm, and an extra correlation which could not be detected in the pine cell wall spectrum appears at 103.4/4.25 ppm. (1→6)-β-Glucan [(1→6)-β-d-Glcp] units may also appear in this region.92,93 4-O-Methyl-α-d-glucuronic acid (MeGlcA), with its anomeric at 97.5/5.31 ppm, is also a major component in aspen as in the other plants. There is an extra component in aspen cell wall at 99.3/5.24 ppm, and the peak is considered to be the anomeric position of (1→2)-α-l-rhamnopyranosyl [(1→2)-α-l-Rhap] residue of rhamnogalacturonan, as in pectins isolated from aspen.98 The chemical shift matches those from an NMR study of rhamnogalacturonans.99,100
Kenaf’s major polysaccharides (Fig. 4c) are similar to those of aspen. Kenaf has major peaks for cellulose (including β-d-glucan), xylan, 4-O-methyl-α-d-glucuronic acid (MeGlcA), O-acetyl-xylosyl residues (2-O-Ac-β-d-Xylp and 3-O-Ac-β-d-Xylp), and α-l-arabinosyl (α-l-Araf ) residues with extra correlations. O-Acetyl-mannosyl residues (2-O-Ac-Manp) and β-d-galactosyl [(1→4)-α-d-Galp] residues appear to have lower correlation peaks than other plant materials, but still clearly appear. Only a trace of β-d-mannosyl [(1→4)-β-d-Manp] residues appear at 100.5/4.60 ppm. There is also an unassigned peak at 101.0/4.70 ppm that, as we have seen in aspen, that may be related to mannan structures. The reducing end of β-d-glucosyl [(1→4)-β-d-Glcp] units at 97.1/4.44 ppm appears close to the reducing end of (1→4)-β-d-Xylp. A correlation at 92.3/5.10 ppm is likely the α-d-glucosyl [(1→4)-α-d-Glcp] reducing end. The kenaf cell wall spectrum shows a correlation at 100.5/5.21 ppm, which fits with the anomeric position of α-l-fucopyranosyl residues in fractions that have been isolated from apple pomace,20 and kenaf.101
Corn (Fig. 4d) has much simpler polysaccharide components compared to the other plants in this paper. As usual, the major peaks for cellulose and xylan can be easily seen, and the correlations should also contain the anomerics of 3-O-acetylmannosyl residues (3-O-Ac-β-d-Xylp) because we have seen the C3/H3 peak in the corn spectrum above. Also a huge peak of 2-O-Ac-β-d-Xylp anomeric appears at 99.7/4.58 ppm. 4-OMethyl-α-d-glucuronic acid (MeGlcA) is also prevalent. Cellulose and β-d-glucans are also at 103.0/4.43 ppm to 103.3/4.25 ppm. A small peak at 101.0/4.70 ppm may be related to mannan structures, but only a trace of β-d-mannosyl [(1→4)-β-d-Manp] residues appears at 100.6/4.54 ppm. The reducing ends of (1→4)-β-d-Xylp and (1→4)-α-d-Xylp are also apparent.
Despite the limited assignments made here for polysaccharides, it is clear from Fig. 4 that the polysaccharide anomeric correlations are well dispersed and, in principle, interpretable. The anomeric correlation profiles are significantly different between the various plant types suggesting that the data will be valuable for characterization of polysaccharide polymers as well as for lignins. One caveat: most of the cellulose component is considered to remain crystalline and not fully swollen in the gel. It is likely that crystalline cellulose is ‘invisible’ to solution-state NMR. Cellulose may therefore be under-represented in these spectra compared to the more mobile components, the hemicelluloses, non-crystalline cellulose, and lignin.
Conclusions
The ball-milled cell wall gel-samples in DMSO-d6/pyridine-d5 (4 : 1) were used to obtain detailed 13C–1H correlation spectra via solution-state 2D HSQC NMR experiments. The quality of the spectra is almost equivalent to the spectra from the original cell wall dissolution method (with acetylation),39 and the DMSO/NMI dissolution method.42
One of the significant advantages of this new NMR solvent system is that the gel-samples swell more effectively than in DMSO-d6 alone,2 resulting in better resolution of the 2D NMR spectra. Another advantage is that the solvent mixture provides better mobility to the gel-samples, so the sample handling during the preparation has been much improved. One of the general advantages of this method, as described in the original method, is the simple and rapid sample preparation. Emerging biomass research demands treatment of large numbers of plant samples. If necessary, this DMSO-d6/pyridine-d5 system also allows further solubilization methods to be used sequentially by simply adding NMI and acetic anhydride followed by normal workup,39 to produce the acetylated cell wall materials that dissolve in CDCl3 or DMSO-d6 for further NMR work. The next advantage is that the spectra from underivatized walls in DMSO-d6/pyridine-d5 or DMSO-d6 have better dispersion of some correlations (than acetylated samples), and can allow more substantive assignments. In the case of ferulates, these important cross-linking agents in grass cell walls74 are readily apparent. And although correlations of incompletely deuterated pyridine appear in the aromatic area when the DMSO-d6/pyridine-d5 system is used, there are no major cell wall components that are compromised. Using 100% pyridine-d5 is recommended for corn (or any grass) samples to avoid the possible interference by the residual solvent peaks.
A major limitation of these NMR methods is in the process of ball-milling, rather than in the NMR methods themselves. Fine milling breaks the weakest bonds—glycosidic linkages in polysaccharides and α-aryl ethers in lignins. As a result, the degree of polymerization is markedly decreased and some oxidation occurs during milling, even though the major polymer structures remain intact. Ultrasonication to produce the gel could also induce minor structural changes.102 Also the ball-milling process is time consuming and labor intensive. Another major limitation is that the prepared gel-samples are suitable only for short-range NMR experiments, i.e., HMQC or HSQC. Long-range experiments (e.g., HMBC)are impossible because magnetization is lost by the rapid relaxation before the required long-range 13C–1H coupling delay (typically 60–100 ms) is complete. However, the HMQC/HSQC fingerprint is incredibly valuable to rapidly detail the composition/structure of the complex cell wall, and gel samples allow a shorter total experiment time for each sample.
Expanding biofuel and biomass research and transgenic plant production areas need to systematically analyze the plant cell wall composition and structure in an efficient way. NMR profiling provides what appears to be the best tool for the detailed structural study for the complex cell wall polymers, and this gel-sample NMR method can rapidly provide total profiles of the cell wall components. It also can be considered as an ideal secondary screening method, because the method allows speedier processing and analysis of samples than could be previously accomplished in the past. In addition, there is promising potential for chemometric analysis of biological samples using the 2D NMR fingerprint.103
Experimental section
General
All Reagents and polysaccharides were purchased from Aldrich and Sigma (Milwaukee, WI, USA) unless otherwise noted. Solvents used were AR grade and supplied by Fisher Scientific (Atlanta, GA, USA). The ultrasonic bath was a Branson (Danbury, CT, USA; 3510EMT, tank capacity 5.7 L, with mechanical timer), and it was used for homogenization for the gel-state NMR samples. Preparative thin layer chromatography (TLC) plates were from Macherey-Nagel (Bethlehem, PA, USA).
Plant materials
Plant samples were prepared as previously published.2 Dried plant cell walls were cryogenically pre-ground for 2 min at 30 Hz using a Retsch (Newtown, PA, USA) MM301 mixer mill with corrosion-resistant stainless steel screw-top grinding jars (50 ml) containing stainless a single steel ball bearing (30 mm). The pre-ground cell walls were extracted with distilled water (3 ×) and 80% ethanol (3 ×) using ultrasonication. Isolated cell walls were then finely milled using a Retsch PM100 planetary ball mill spinning at 600 rpm with zirconium dioxide (ZrO2) vessels (50 ml) containing ZrO2 ball bearings (10 mm × 10). The grinding times are dependent on the plant cell wall type and the amount, as described in Table 1.
Loblolly pine. Pre-ground cell wall material (1 g) was ball-milled as described above for 10 h 20 min in 20 min intervals with 10 min interval breaks to avoid excessive heating during the ball-milling.
Aspen. Pre-ground cell wall material (800 mg) was ball-milled for 4 h 10 min in 10 min intervals with 5 min interval breaks.
Kenaf bast fiber. Pre-ground cell wall material (200 mg) from freshly harvested 1 year old Tainung2 kenaf55 was ball-milled for 45 min in 5 min intervals with 5 min interval breaks.
Corn Stalks. Pre-ground cell wall material (507 mg) was ball-milled for 1 h 45 min in 5 min intervals with 5 min interval breaks. The source material was that used in a prior study.78
Lignin extraction
Isolated lignins from pine and corn were isolated from ball milled cell walls by extraction with 96 : 4 dioxane :water as previously described.78,104
Lignin model compounds
Model compounds were prepared and their NMR spectra were acquired in DMSO to enable the assignments made in a previous paper.2 Coniferyl alcohol and sinapyl alcohol were prepared from commercially available coniferaldehyde and sinapaldehyde using borohydride exchange resin.105 p-Coumaryl alcohol was synthesized from p-coumaric acid.106 Coniferyl alcohol dimers were synthesized from in vitro radical coupling reactions using MnO2 in dioxane :H2O (1 : 1, v/v).107 Sinapyl alcohol dimers were prepared using FeCl3·6H2O in dioxane :H2O (5 : 2, v/v).108 p-Coumaryl alcohol dimers were synthesized with horseradish peroxidase with hydrogen peroxide in acetone :water (1 : 10, v/v) or with FeCl3·6H2O in acetone: H2O (5 : 1, v/v). Each metallic oxidative radical reaction was stirred for 1 to 4 h, and the metal salts were filtered off through a silica gel bed in fine sintered glass filters. The peroxidase reactions were conducted for about 15 h. Reaction solutions were poured into EtOAc, and washed with satd. aqueous NH4Cl. EtOAc extracts were dried over anhydrous MgSO4, and concentrated under reduced pressure. Model dimers were separated on preparative TLC plates with CHCl3–MeOH (10 : 1, v/v). The fully authenticated NMR data for model compounds will be deposited in the “NMR Database of lignin and cell wall model compounds” available via the internet.48
Polysaccharide samples and preparation
Locust Bean Gum (polymers of β-D-mannopyranose + α-D-galactose) and xylan from oat spelts. Locust bean gum (enriched in galactomannan polysaccharides, from the seeds of Ceratonia siliqua L.) and xylan from oat spelts were ground with a Retsch PM100 ball mill as described above. The grinding time required was twice as long as for pine cell walls to obtain adequately fine material that produced acceptable 2D NMR signals.
D-(+)-Cellobiose, D-(+)-cellotriose, mannan from Saccharomyces cerevisiae, and (+)-arabinogalactan from larch wood (Fluka; Milwaukee, WI, USA). These compounds or polymers were directly used to obtain adequate 2D NMR spectra.
Cellulose powder (microcrystalline, ~20 micron). Cellulose was ground with a Retsch PM100 ball mill as described above, and dissolved in DMSO. The collected solution was dried, and examined by NMR.
NMR sample preparation
Gel-samples were prepared directly in the NMR tube via the simple method originally described, as summarized here. In general, ~50–100 mg of ball-milled cell walls were directly transferred into 5mm NMR tubes. About 1 ml of pre-mixed DMSO-d6/pyridine-d5 (4 : 1) was directly added into the NMR tubes for each sample. The sample was distributed as well as possible off the bottom and up the sides of the horizontally positioned NMR tube. The NMR solvent mixture was carefully introduced (via a syringe), spreading it from the bottom of the NMR tube, along the sides, and towards the top of the sample. The NMR tubes were then placed in an ultrasonic bath and sonicated for 1–5 h (depending on the sample), until the gel became apparently homogeneous; the final sample height in the tube was ~4–5 cm. Pyridine-d5 with a purity of 99.5 atom% D was used for the most cell wall samples, but pyridine-d5 of enhanced purity (“100”; min. 99.94 atom% D) was used for the corn stalk sample to minimize interference between the residual solvent peaks and the correlations from ferulate and p-coumarate moieties.
NMR experiments
NMR spectra from the gel-samples were acquired on a 750 MHz (DMX-750) Bruker Biospin (Rheinstetten, Germany) instrument equipped with an inverse (proton coils closest to the sample) gradient 5 mm TXI 1H/13C/15N cryoprobe. The central DMSO solvent peak was used as internal reference (δC 39.5, δH 2.49 ppm). The 13C–1H correlation experiment was an adiabatic HSQC experiment (Bruker standard pulse sequence ‘hsqcetgpsisp.2’; phase-sensitive gradient-edited-2D HSQC using adiabatic pulses for inversion and refocusing)109 typically had the following parameters for the plant cell wall samples: spectra were acquired from 11 to −1 ppm in F2 (1H) using 1078 data points for an acquisition time (AQ) of 60 ms, an interscan delay (D1) of 750 ms, 196 to −23 ppm in F1 (13C) using 480 increments (F1 acquisition time 5.78 ms) of 16 scans, with a total acquisition time of 6 h. The same version of the adiabatic HSQC experiment (hsqcetgpsisp.2)was used on a Bruker 500 MHz (DMX-500) NMR with a cryogenically cooled 5 mm gradient cryoprobe with inverse geometry, and the parameter set was more effectively optimized for gel-samples: spectra were acquired from 10 to 0 ppm in F2 (1H) using 1000 data points for an acquisition time (AQ) of 100 ms, an interscan delay (D1) of 500 ms, 200 to 0 ppm in F1 (13C) using 320 increments (F1 acquisition time 6.36 ms) of 100 scans, with a total acquisition time of 5 h 34 m. The number of scans can, of course, be adjusted as usual depending on the signal-to-noise required from a sample. We also used a Bruker Avance 360 MHz instrument equipped with an inverse (proton coils closest to the sample) gradient 5 mm 1H/broadband gradient probe for structural elucidation and assignment authentication for the model compounds. The standard Bruker implementations of the traditional suite of 1D and 2D (gradient-selected, 1H-detected, e.g., COSY, HMQC/HSQC, HSQC-TOCSY, HMBC) NMR experiments were used. Normal HMQC (inv4gptp) experiments at 360 MHz were used for model compounds and had the following parameters: spectra were acquired from 10 to 0 ppm in F2 (1H) using 1400 data points for an acquisition time of ≤200 ms, 200 to 0 ppm in F1 (13C) using 128 (or 256) increments (F1 acquisition time 35.3 ms) of 32 scans, with a total acquisition time of 1 h 23 min. Processing used typical matched Gaussian apodization in F2 and a squared cosine-bell in F1. Interactive integrations of contours in 2D HSQC/HMQC plots were carried out using Bruker’s TopSpin 2.5 software, as was all data processing.
Supplementary Material
Acknowledgements
We are grateful to Paul Schatz, Takuya Akiyama, Ron Hatfield, Jane Marita, Dan Yelle, Fachuang Lu, and Dino Ress for various discussions. This research was supported by the Office of Science (BER), U.S. Dept. of Energy, Interagency agreement No. DEAI02- 06ER64299, and was also funded in part by the DOE Great Lakes Bioenergy Research Center (www.greatlakesbioenergy.org), which is supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through Cooperative Agreement DE-FC02-07ER64494 between The Board of Regents of the University of Wisconsin System and the U.S. Department of Energy. This study made use of 750 MHz instruments at the National Magnetic Resonance Facility at Madison, which is supported by National Institutes of Health grants P41RR02301 (Biomedical Research Technology Program, National Center for Research Resources) and P41GM66326 (National Institute of General Medical Sciences). Equipment in the facility was purchased with funds from the University of Wisconsin, the National Institutes of Health (P41GM66326, P41RR02301, RR02781, RR08438), the National Science Foundation (DMB-8415048, BIR-9214394), and the U.S. Department of Agriculture.
Footnotes
References
- 1.Ralph J, Landucci LL. In: Lignins. Heitner C, Dimmel DR, editors. New York, NY: Marcel Dekker; 2010. in press. [Google Scholar]
- 2.Kim H, Ralph J, Akiyama T. Bio Energy Research. 2008;1:56–66. [Google Scholar]
- 3.Fengel D, Wegener G. Wood: Chemistry, Ultrastructure, Reactions. Berlin-New York: Walter De Gruyter; 1989. pp. 66–105. [Google Scholar]
- 4.Wardrop AB. Protoplasma. 1970;70:73–86. [Google Scholar]
- 5.Sjöström E. Wood Chemistry: Fundamentals and applications. Second edition edn. New York: Academic press, Inc.; 1993. [Google Scholar]
- 6.Ebringerová A, Hromádková Z, Heinze T. Adv. Polym. Sci. 2005;186:1–67. [Google Scholar]
- 7.Fengel D, Wegener G. Wood: Chemistry, Ultrastructure, Reactions. Berlin-New York: Walter De Gruyter; 1989. pp. 106–131. [Google Scholar]
- 8.Sederoff RR, MacKay JJ, Ralph J, Hatfield RD. Curr. Opin. Plant Biol. 1999;2:145–152. doi: 10.1016/S1369-5266(99)80029-6. [DOI] [PubMed] [Google Scholar]
- 9.Boerjan W, Ralph J, Baucher M. Annu. Rev. Plant Biol. 2003;54:519–549. doi: 10.1146/annurev.arplant.54.031902.134938. [DOI] [PubMed] [Google Scholar]
- 10.Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH, Boerjan W. Phytochem. Rev. 2004;3:29–60. [Google Scholar]
- 11.Ralph J, Brunow G, Harris PJ, Dixon RA, Schatz PF, Boerjan W. In: Recent Advances in Polyphenol Research. Daayf F, El Hadrami A, Adam L, Balance GM, editors. Oxford, UK: Wiley-Blackwell Publishing; 2008. pp. 36–66. [Google Scholar]
- 12.Björkman A. Nature. 1954;174:1057–1058. [Google Scholar]
- 13.Pew JC. Tappi. 1957;40:553–558. [Google Scholar]
- 14.Chang H-M, Cowling EB, Brown W, Adler E, Miksche G. Holzforschung. 1975;29:153–159. [Google Scholar]
- 15.Hu Z, Yeh T-F, Chang H.-m, Matsumoto Y, Kadla JF. Holzforschung. 2006;60:389–397. [Google Scholar]
- 16.Lawther JM, Sun L, Banks WB. J. Appl. Polym. Sci. 1996;60:1827–1837. [Google Scholar]
- 17.Ebringerová A, Heinze T. Macromol. Rapid Commun. 2000;21:542–556. [Google Scholar]
- 18.Willför S, Sjöholma R, Laine C, Roslund M, Hemminga J, Holmbom B. Carbohydr. Polym. 2003;52:175–187. [Google Scholar]
- 19.Hannuksela T, Hervé du Penhoat C. Carbohydr. Res. 2004;339:301–312. doi: 10.1016/j.carres.2003.10.025. [DOI] [PubMed] [Google Scholar]
- 20.Watt DK, Brasch DJ, Larsen DS, Melton LD. Carbohydr. Polym. 1999;39:165–180. [Google Scholar]
- 21.Busato AP, Vargas-Rechia CG, Reicher F. Phytochemistry. 2001;58:525–531. doi: 10.1016/s0031-9422(01)00217-5. [DOI] [PubMed] [Google Scholar]
- 22.Temelli F. J. Food Sci. 1997;62:1194–1201. [Google Scholar]
- 23.Symons LJ, Brennan CS. J. Food Sci. 2004;69:257–261. [Google Scholar]
- 24.Pan XJ, Arato C, Gilkes N, Gregg D, Mabee W, Pye K, Xiao ZZ, Zhang X, Saddler J. Biotechnol. Bioeng. 2005;90:473–481. doi: 10.1002/bit.20453. [DOI] [PubMed] [Google Scholar]
- 25.Forziati FH, Stone WK, Rowen JW, Appel WD. J. Res. Natl. Inst. Stand. Technol. 1950;45:109–113. [Google Scholar]
- 26.Ikeda T, Holtman K, Kadla JF, Chang HM, Jameel H. J. Agric. Food Chem. 2002;50:129–135. doi: 10.1021/jf010870f. [DOI] [PubMed] [Google Scholar]
- 27.Fujimoto A, Matsumoto Y, Chang HM, Meshitsuka G. J. Wood Sci. 2005;51:89–91. [Google Scholar]
- 28.Swatloski RP, Spear SK, Holbrey JD, Rogers RD. J. Am. Chem. Soc. 2002;124:4974–4975. doi: 10.1021/ja025790m. [DOI] [PubMed] [Google Scholar]
- 29.Kosan B, Michels C, Meister F. Cellulose. 2008;15:59–66. [Google Scholar]
- 30.Barthel S, Heinze T. Green Chem. 2006;8:301–306. [Google Scholar]
- 31.Liebert T, Heinze T. BioResources. 2008;3:576–601. [Google Scholar]
- 32.Xie H, King A, Kilpelainen I, Granstrom M, Argyropoulos DS. Biomacromolecules. 2007;8:3740–3748. doi: 10.1021/bm700679s. [DOI] [PubMed] [Google Scholar]
- 33.Fort DA, Remsing RC, Swatloski RP, Moyna P, Moyna G, Rogers RD. Green Chem. 2007;9:63–69. [Google Scholar]
- 34.Vanoye L, Fanselow M, Holbrey JD, Atkinsb MP, Seddona KR. Green Chem. 2009;11:390–396. [Google Scholar]
- 35.Binder JB, Raines RT. J. Am. Chem. Soc. 2009;131:1979–1985. doi: 10.1021/ja808537j. [DOI] [PubMed] [Google Scholar]
- 36.Honglu X, Tiejun S. Holzforschung. 2006;60:509–512. [Google Scholar]
- 37.Kubo S, Hashida K, Yamada T, Hishiyama S, Magara K, Kishino M, Ohno H, Hosoya S. J. Wood Chem. Technol. 2008;28:84–96. [Google Scholar]
- 38.Heinze T, Dicke R, Koschella A, Kull AH, Klohr EA, Koch W. Macromol. Chem. Phys. 2000;201:627–631. [Google Scholar]
- 39.Lu F, Ralph J. Plant J. 2003;35:535–544. doi: 10.1046/j.1365-313x.2003.01817.x. [DOI] [PubMed] [Google Scholar]
- 40.Ass BAP, Frollini E, Heinze T. Macromol. Biosci. 2004;4:1008–1013. doi: 10.1002/mabi.200400088. [DOI] [PubMed] [Google Scholar]
- 41.Ralph J, Lu F. Org. Biomol. Chem. 2004;2:2714–2715. doi: 10.1039/B412633E. [DOI] [PubMed] [Google Scholar]
- 42.Yelle DJ, Ralph J, Frihart CR. Magn. Reson. Chem. 2008;46:508–517. doi: 10.1002/mrc.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capanema EA, Balakshin MY, Kadla JF. J. Agric. Food Chem. 2005;53:9639–9649. doi: 10.1021/jf0515330. [DOI] [PubMed] [Google Scholar]
- 44.Bardet M, Lundquist K, Parkas J, Robert D, von Unge S. Magn. Reson. Chem. 2006;44:976–979. doi: 10.1002/mrc.1877. [DOI] [PubMed] [Google Scholar]
- 45.Ibarra D, Chavez MI, Rencoret J, del Rio JC, Gutierrez A, Romero J, Camarero S, Martinez MJ, Jimenez-Barbero J, Martinez AT. J. Agric. Food Chem. 2007;55:3477–3490. doi: 10.1021/jf063728t. [DOI] [PubMed] [Google Scholar]
- 46.Grabber JH, Mertens DR, Kim H, Funk C, Lu F, Ralph J. J. Sci. Food Agric. 2009;89:122–129. [Google Scholar]
- 47.Grabber JH, Hatfield RD, Lu F, Ralph J. Biomacromolecules. 2008;9:2510–2516. doi: 10.1021/bm800528f. [DOI] [PubMed] [Google Scholar]
- 48.Ralph SA, Landucci LL, Ralph J. NMR Database of Lignin and Cell Wall Model Compounds. 2004 Available over Internet at http://www.dfrc.ars.usda.gov/software.html.
- 49.Fengel D, Wegener G. Wood: Chemistry, Ultrastructure, Reactions. Berlin-New York: Walter De Gruyter; 1989. [Google Scholar]
- 50.Teleman A, Tenkanen M, Jacobs A, Dahlman O. Carbohydr. Res. 2002;337:373–377. doi: 10.1016/s0008-6215(01)00327-5. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Gellerstedt G. Chem. Commun. 2001:2744–2745. [Google Scholar]
- 52.Zhang L, Gellerstedt G, Ralph J, Lu F. J. Wood Chem. Technol. 2006;26:65–79. [Google Scholar]
- 53.Lu F, Ralph J. Org. Biomol. Chem. 2008;6:3681–3694. doi: 10.1039/b809464k. [DOI] [PubMed] [Google Scholar]
- 54.Sarkanen KV, Chang H-M, Allan GG. Tappi. 1967;50:587–590. [Google Scholar]
- 55.Ralph J. J. Nat. Prod. 1996;59:341–342. [Google Scholar]
- 56.Ralph J, Lu F. J. Agric. Food Chem. 1998;46:4616–4619. doi: 10.1021/jf970676m. [DOI] [PubMed] [Google Scholar]
- 57.Lu F, Ralph J. Chem. Commun. 2002:90–91. doi: 10.1039/b109876d. [DOI] [PubMed] [Google Scholar]
- 58.del Río JC, Gutiérrez A, Martínez AT. Rapid Commun. Mass Spectrom. 2004;18:1181–1185. doi: 10.1002/rcm.1457. [DOI] [PubMed] [Google Scholar]
- 59.del Río JC, Marques G, Rencoret J, Martínez AT, Gutiérrez A. J. Agric. Food Chem. 2007;55:5461–5468. doi: 10.1021/jf0705264. [DOI] [PubMed] [Google Scholar]
- 60.del Río JC, Rencoret J, Marques G, Gutiérrez A, Ibarra D, Santos JI, Jiménes-Barbero J, L Z, Martínez AT. J. Agric. Food Chem. 2008;56:9525–9534. doi: 10.1021/jf800806h. [DOI] [PubMed] [Google Scholar]
- 61.Pauly M, Scheller HV. Planta. 2000;210:659–667. doi: 10.1007/s004250050057. [DOI] [PubMed] [Google Scholar]
- 62.Ferrari MD, Neirotti E, Albornoz C, Saucedo E. Biotechnol. Bioeng. 1992;40:753–759. doi: 10.1002/bit.260400702. [DOI] [PubMed] [Google Scholar]
- 63.Taherzadeh MJ, Eklund R, Gustafsson L, Niklasson C, Lidén G. Ind. Eng. Chem. Res. 1997;36:4659–4665. [Google Scholar]
- 64.Willför S, Sundberg K, Tenkanen M, Holmbom B. Carbohydr. Polym. 2008;72:197–210. [Google Scholar]
- 65.Capek P, Alföldi J, Lišková D. Carbohydr. Res. 2002;337:1033–1037. doi: 10.1016/s0008-6215(02)00090-3. [DOI] [PubMed] [Google Scholar]
- 66.Teleman A, Nordström M, Tenkanen M, Jacobs A, Dahlman O. Carbohydr. Res. 2003;338:525–534. doi: 10.1016/s0008-6215(02)00491-3. [DOI] [PubMed] [Google Scholar]
- 67.Teleman A, Lundqvist J, Tjerneld F, Stålbrand H, Dahlman O. Carbohydr. Res. 2000;329:807–815. doi: 10.1016/s0008-6215(00)00249-4. [DOI] [PubMed] [Google Scholar]
- 68.Li L, Zhou Y, Cheng X, Sun J, Marita JM, Ralph J, Chiang VL. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4939–4944. doi: 10.1073/pnas.0831166100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lu F, Ralph J, Morreel K, Messens E, Boerjan W. Org. Biomol. Chem. 2004;2:2888–2890. doi: 10.1039/B411428K. [DOI] [PubMed] [Google Scholar]
- 70.Lu F, Ralph J. J. Agric. Food Chem. 1998;46:547–552. doi: 10.1021/jf970676m. [DOI] [PubMed] [Google Scholar]
- 71.Wagner A, Ralph J, Akiyama T, Flint H, Phillips L, Torr KM, Nanayakkara B, Te Kiri L. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11856–11861. doi: 10.1073/pnas.0701428104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jung HG, Allen MS. J. Anim. Sci. 1995;73:2774–2790. doi: 10.2527/1995.7392774x. [DOI] [PubMed] [Google Scholar]
- 73.Ralph J, Helm RF. In: Forage Cell Wall Structure and Digestibility. Jung HG, Buxton DR, Hatfield RD, Ralph J, editors. Madison, WI: ASA-CSSA-SSSA; 1993. pp. 201–246. [Google Scholar]
- 74.Ralph J, Bunzel M, Marita JM, Hatfield RD, Lu F, Kim H, Schatz PF, Grabber JH, Steinhart H. Phytochem. Rev. 2004;3:79–96. [Google Scholar]
- 75.Lam TBT, Iiyama K, Stone BA. Phytochemistry. 1994;36:773–775. [Google Scholar]
- 76.Lam TBT, Kadoya K, Iiyama K. Phytochemistry. 2001;57:987–992. doi: 10.1016/s0031-9422(01)00052-8. [DOI] [PubMed] [Google Scholar]
- 77.Hatfield RD, Marita JM, Frost K, Grabber J, Ralph J, Lu F, Kim H. Planta. 2009;229:1253–1267. doi: 10.1007/s00425-009-0900-z. [DOI] [PubMed] [Google Scholar]
- 78.Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG. J. Am. Chem. Soc. 1994;116:9448–9456. [Google Scholar]
- 79.Goddard TD, Kneller DG. Sparky - NMR Assignment and integration software. San Francisco, San Francisco: University of California; 2008. Available over Internet at http://www.cgl.ucsf.edu/home/sparky/ [Google Scholar]
- 80.Delaglio F. NMR Pipe spectral processing and analysis system. 2008 Available over Internet at http://spin.niddk.nih.gov/NMRPipe/
- 81.Flugge LA, Blank JT, Petillo PA. J. Am. Chem. Soc. 1999;121:7228–7238. [Google Scholar]
- 82.Moulthrop JS, Swatloski RP, Moyna G, Rogers RD. Chem. Commun. 2005:1557–1559. doi: 10.1039/b417745b. [DOI] [PubMed] [Google Scholar]
- 83.Habibi Y, Vignon MR. Carbohydr. Res. 2005:1431–1436. doi: 10.1016/j.carres.2005.01.039. [DOI] [PubMed] [Google Scholar]
- 84.Ramesh HP, Yamaki K, Ono H, Tsushida T. Carbohydr. Polym. 2001;45:69–77. [Google Scholar]
- 85.Garros-Rosa I, Reicher F, Petkowicz CLO, Sierakowski MR, Moreira RA. Polímeros: Ciência e Tecnologia. 2006;16:99–103. [Google Scholar]
- 86.Dentini M, Caucci D, Barbetta A, Crescenzi V, Capitani D, Mannina L, Viel S. Biomacromolecules. 2006;7:54–63. doi: 10.1021/bm050341a. [DOI] [PubMed] [Google Scholar]
- 87.Kobayashi H, Shibata N, Watanabe M, Komido M, Hashimoto N, Hisamichi K, Suzuki S. Carbohydr. Res. 1992;231:317–323. doi: 10.1016/0008-6215(92)84028-q. [DOI] [PubMed] [Google Scholar]
- 88.Vinogradov E, Petersen B, Bock K. Carbohydr. Res. 1998;307:177–183. doi: 10.1016/s0008-6215(98)00042-1. [DOI] [PubMed] [Google Scholar]
- 89.Fransen CTM, Van Laere KMJ, van Wijk AAC, Brüll LP, Dignum M, Thomas-Oates JE, Haverkamp J, Schols HA, Voragen AG, Kamerling JP, Vliegenthart JFG. Carbohydr. Res. 1998;314:101–114. doi: 10.1016/s0008-6215(98)00285-7. [DOI] [PubMed] [Google Scholar]
- 90.Cardoso SM, Ferreira JA, Mafra I, Silva AMS, Coimbra MA. J. Agric. Food Chem. 2007;55:7124–7130. doi: 10.1021/jf070769w. [DOI] [PubMed] [Google Scholar]
- 91.Capek P, Kubačková M, Alföldi J, Bilisics L, Lišková D, Kákoniová D. Carbohydr. Res. 2000;329:635–645. doi: 10.1016/s0008-6215(00)00210-x. [DOI] [PubMed] [Google Scholar]
- 92.Reis RA, Tischer CA, Gorin PAJ, Iacomini M. FEMS Microbiology Letters. 2002;210:1–5. doi: 10.1111/j.1574-6968.2002.tb11152.x. [DOI] [PubMed] [Google Scholar]
- 93.Tomati U, Belardinelli M, Galli E, Iori V, Capitani D, Mannina L, Viel S, Segre A. Carbohydr. Res. 2004;339:1129–1134. doi: 10.1016/j.carres.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 94.Willför S, Sjöholm R, Laine C, Holmbom B. Wood Sci. Technol. 2002;36:101–110. [Google Scholar]
- 95.Habibi Y, Mahrouz M, Maraisa M-F, Vignon MR. Carbohydr. Res. 2004;339:1201–1205. doi: 10.1016/j.carres.2004.02.004. [DOI] [PubMed] [Google Scholar]
- 96.Lee REB, Li W, Chatterjee D, Lee RE. Glycobiology. 2005;15:139–151. doi: 10.1093/glycob/cwh150. [DOI] [PubMed] [Google Scholar]
- 97.Dourado F, Cardoso SM, Silva AMS, Gama FM, Coimbra MA. Carbohydr. Polym. 2006;66:27–33. [Google Scholar]
- 98.Ermel FF, Follet-Gueye M-L, Cibert C, Vian B, Morvan C, Catesson A-M, Goldberg R. Planta. 2000;210:732–740. doi: 10.1007/s004250050674. [DOI] [PubMed] [Google Scholar]
- 99.Cui W, Eskin MNA, Biliaderis CG, Marat K. Carbohydr. Res. 1996;292:173–183. doi: 10.1016/s0008-6215(96)91045-9. [DOI] [PubMed] [Google Scholar]
- 100.Ding K, Fang J-N, Dong T, Tsim KW-K, Wu H. J. Nat. Prod. 2003;66:7–10. doi: 10.1021/np020118o. [DOI] [PubMed] [Google Scholar]
- 101.Neto CP, Seca A, Fradinho D, Coimbra MA, Domingues F, Evtuguin D, Silvestre A, Cavaleiro JAS. Ind. Crops Prod. 1996;5:189–196. [Google Scholar]
- 102.Yoshioka A, Seino T, Tabata M, Takai M. Holzforschung. 2000;54:357–364. [Google Scholar]
- 103.Hedenström M, Wiklund S, Öman T, Lu F, Gerbner L, Schatz PF, Sundberg B, Ralph J. Mol. Plant. 2009;2:933–942. doi: 10.1093/mp/ssp047. [DOI] [PubMed] [Google Scholar]
- 104.Ralph J, MacKay JJ, Hatfield RD, O’Malley DM, Whetten RW, Sederoff RR. Science. 1997;277:235–239. doi: 10.1126/science.277.5323.235. [DOI] [PubMed] [Google Scholar]
- 105.Kim H, Ralph J. J. Agric. Food Chem. 2005;53:3693–3695. doi: 10.1021/jf047787n. [DOI] [PubMed] [Google Scholar]
- 106.Quideau S, Ralph J. J. Agric. Food Chem. 1992;40:1108–1110. [Google Scholar]
- 107.Syrjänen K, Brunow G. J. Chem. Soc., Perkin Trans. 1. 2000:183–187. [Google Scholar]
- 108.Tanahashi M, Takeuchi H, Higuchi T. Wood Res. 1976;61:44–53. [Google Scholar]
- 109.Kupče E, Freeman R. J. Magn. Reson. 2007;187:258–265. doi: 10.1016/j.jmr.2007.05.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.