Abstract
Interferon α-2a (IFNA2) is a member of the Type I interferon cytokine family, known for its antiviral and anti-proliferative functions. The role of this family in the innate immune response makes it an attractive candidate for the treatment of many viral and chronic immune-compromised diseases. Recombinant IFNA2 is clinically used to modulate hairy cell leukemia as well as hepatitis c. Historically, IFNA2 has been purified from human leukocytes as well as bacterial expression systems. In most cases, bacterial expression of IFNA2 resulted in inclusion body formation, or required numerous purification steps that decreased the protein yield. Here, we describe an expression and purification scheme for IFNA2 using a pET-SUMO bacterial expression system and a single purification step. Using SUMO protein as the fusion tag achieved high soluble protein expression. The SUMO tag was cleaved with the Ulp1 protease leaving no additional amino acids on the fusion terminus following the cleavage. Mass spectrometry, circular dichroism, 2D heteronuclear NMR, and analytical ultracentrifugation confirmed the amino acid sequence identity, secondary and tertiary protein structures, and the solution behavior of the purified IFNA2. The purified protein also has antiviral and anti-proliferative activities comparable to the WHO International Standard, NIBSC 95/650, and the IFNA2 standard available from PBL Assay Science. Combining the expression and purification protocols developed here to produce IFNA2 on a laboratory scale with the commercial fermenter technology commonly used in pharmaceutical industry may further enhance IFNA2 yields, which will promote the development of interferon-based protein drugs to treat various disorders.
Keywords: interferon alpha-2a, purification, bacterial expression, soluble protein, SUMO
Introduction
The interferon family of cytokines was discovered in the late 1950s, when soluble antiviral factors conferred protection to membranes during live influenza virus challenge [1]. The interferon family is classified by its antiviral, anti-proliferative, and immunoregulatory functions. These proteins are critical members of the innate immune response whose induction is initiated by the binding of various viral components to cell surface recognition receptors. Downstream transcription factors are activated via kinase pathways that lead to interferon production. Upon release from the cell, interferon binds to ubiquitously expressed cell surface receptors which trigger a signaling cascade involving activation of antiviral and anti-proliferative factors as well as up-regulating their own expression. Thus, interferon induction leads to a positive feedback loop characterized by high antiviral activity [2–5].
Human interferons comprise two categories: Type I and Type II. These classifications refer to the cellular receptor that is used for interferon function as well as amino acid sequence [6]. Type I interferons consist of α, β, and ω subtypes, and bind to the IFNα/β receptor (IFNAR). Type I interferons are ubiquitously induced by viral infection in many cell types. Type II interferon has a single subtype, γ, which interacts with the IFNγ receptor. T cells and natural killer cells produce interferon γ following activation by early cytokines [2, 3].
Interferon α consists of 13 functional subtypes of highly conserved sequence and structural homology (75–99%) found in humans [6]. Currently, two native and two PEGylated interferon α products exist in the pharmaceutical market [7]. Interferon α-2 products, such as interferon α-2a (IFNA2) are used to treat multiple diseases such as hairy cell leukemia and hepatitis c [4, 8–10]. These drugs are available in a variety of dosage forms including multi-dose and single use syringes.
As the biopharmaceutics industry continues to grow, the study of protein-based pharmaceuticals and their formulation stability becomes increasingly important. It is often preferable to obtain proteins from an in-house expression and purification scheme due to cost or other factors. IFNA2 was initially derived directly from human leukocytes and purified for pharmaceutical use [11]. However, because IFNA2 is not glycosylated in vivo [12], it is a perfect candidate for expression using simple systems such as bacteria. In most attempts of using Escherichia coli (E. coli), IFNA2 was regularly expressed in inclusion bodies [13–15]. Our aim in this work was to develop and optimize an expression and purification scheme of IFNA2 in E. coli that would not only result in high yields of soluble protein, but also would require simplified purification steps in order to obtain high yields of highly pure protein.
Materials and Methods
Plasmid construction and screening
E. coli codon-optimized cDNA for human IFNA2 (Protein Data Bank ID: 1ITF) was synthesized by Operon (Huntsville, Alabama) and delivered in pCR2.1 TOPO subcloning vector. It was obtained as a lyophilized powder and was reconstituted in 10 mM Tris, pH 7.0 to a concentration of 120 ng/ml. The cDNA was transformed into BL21(DE3) cells (1 µl cDNA to 1 ml competent cells), plated on kanamycin/ampicillin antibiotic agar plates, and incubated at 37°C overnight. Colonies were transferred to LB media and grown overnight at 37°C in the presence of kanamycin and ampicillin. DNA was extracted using a miniprep kit (Qiagen), and was quantified on a NanoDrop spectrophotometer (Thermo Scientific, Wilmington, Delaware). The TOPO vector contained recognition sequences for BamH1 (forward) and Xho1 (reverse) restriction sites. The IFNA2 DNA sequence was amplified with specific forward (CATATGGGATCCTGCGATTTACCGCAAACC) and reverse (GTGGTGCTCGAGTTACTCTTTCGAACGCAG) primers, followed by gel extraction and digestion with BamH1 and Xho1 restriction enzymes. The amplified DNA was inserted into our vector of choice. We used a modified pET-28a vector, pET-SUMO (a generous gift from Christopher Lima (Sloan-Kettering Institute)). Ligation of the IFNA2 gene into pET-SUMO was achieved with identical forward and reverse primers listed above. Following ligation, pET-SUMO-IFNA2 was transformed into competent DH5α E. coli cells and plated on kanamycin antibiotic agar plates. Resultant colonies were screened for successful ligation using a miniprep kit (Qiagen) and subsequent DNA sequencing. A positive clone was transformed into competent BL21(DE3) E. coli cells for protein expression.
Expression of IFNA2
In order to express IFNA2 as a soluble protein in E. coli, the following scheme was used. A primary 5 ml culture of LB media was inoculated with 5 µL kanamycin and incubated overnight at 37°C with shaking. Primary culture was transferred to secondary (100ml), inoculated with kanamycin and incubated at 37°C with shaking for 2.5 hrs. The secondary culture was transferred to full scale (1 L), treated with kanamycin, and incubated at 37°C with shaking for 3 hrs. At OD600 = 1.0, temperature was reduced to 16°C and the protein expression was induced with 0.2 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) for 16 hrs with shaking. Cells were harvested using centrifugation at 4200 g for 10 min. Cell pellets were stored at −80°C.
Purification of soluble SUMO-IFNA2
The cell pellet obtained in the previous step was thawed at room temperature and resuspended in 50 mM Tris, 100 mM NaCl, pH 7.5 with two dissolved protease inhibitor cocktail tablets (complete Mini EDTA-free, Roche). Cell lysis was achieved using probe sonication (16 cycles of one min on/off, half-power). Lysate material was removed via centrifugation at 17,000 g for 45 min at 4°C. Supernatant was loaded onto a 5 ml Nickel Sepharose 6 Fast Flow column (GE Healthcare Life Sciences, Pittsburgh, Pennsylvania) (equilibration buffer of 50 mM Tris, 100 mM NaCl, 40 mM imidazole, pH 7.5) at 1 ml/min using a centrifugal pump. The column was washed with 50 mM Tris, 100 mM NaCl, 50 mM imidazole, pH 7.5. Protein was eluted using a linear gradient of imidazole (50 – 500 mM) in 50 mM Tris, 100 mM NaCl, pH 7.5. Fractions containing protein were identified via SDS-PAGE and dialyzed into 50 mM Tris, 200 mM NaCl, pH 8. Often, dilution of positive fractions was required to prevent the highly concentrated SUMO-IFNA2 from precipitating during dialysis. SUMO-IFNA2 (33.3 kDa) concentration was determined using the extinction coefficient of ε280 = 19.54 mM−1 cm−1 calculated from ExPASy (www.expasy.org).
Removal of His6-SUMO tag
To remove the His6-SUMO tag, SUMO-IFNA2 was digested with the yeast protease Ulp1 (purified in-house) (1 µg Ulp1 to 0.25 mg SUMO-IFNA2), and the cleavage reaction was incubated at room temperature for 16 hrs. Separation of the SUMO tag and IFNA2 was achieved on a 5 ml Nickel Sepharose 6 Fast Flow column using a step gradient of imidazole (70 – 500 mM) in 50 mM Tris, 100 mM NaCl, pH 7.5. Pure IFNA2 fractions, collected in the flow-through and in wash steps, were identified using SDS-PAGE and dialyzed into formulation buffer (10 mM ammonium acetate, 120 mM NaCl, pH 5.0). IFNA2 (19.2 kDa) concentration was determined using an extinction coefficient of ε280 = 18.7 mM−1 cm−1. Aliquots of 100 µl of 100 µM IFNA2 were transferred to individual eppendorf tubes, flash frozen using liquid nitrogen, and stored at −80°C.
Deletion of N-terminal serine
The pET-SUMO plasmid leaves a serine residue at the N-terminus of IFNA2 following cleavage with Ulp1. This amino acid has been incorporated in the original expression vector to enhance the cleavage of proteins in which the first N-terminal residue is a proline, lysine, valine, or leucine [16]. The first N-terminal residue in the pharmaceutical IFNA2 is a cysteine; therefore the serine residue was unnecessary. In order to eliminate this amino acid, the entire pET-SUMO-IFNA2 plasmid was subjected to polymerase chain reaction using specific forward (CAG AGA ACA GAT TGG TGG ATG CGA TTT ACC GC) and reverse (GCG GTA AAT CGC ATC CAC CAA TCT GTT CTC TG) primers. These primers were designed to delete the TCC codon corresponding to serine. Following amplification, DNA was purified via gel extraction and transformed into BL21(DE3) cells. Colonies were transferred to LB media and grown overnight at 37°C in the presence of kanamycin. DNA was extracted from E. coli bacterial cultures and submitted for DNA sequencing. A successful sequencing result, in which the TCC codon corresponding to the extra serine residue was deleted, was transformed into BL21(DE3) cells, plated on a kanamycin antibiotic agar plate, and incubated overnight at 37°C. One resultant colony was grown in LB media overnight at 37°C and was used to create a glycerol stock for further expression use. All mentions of IFNA2 in this report refer to the purified protein with an amino acid sequence exactly identical to that of the pharmaceutical product (PDB ID 1ITF).
Thawing procedure
Prior to any experimentation, individual aliquots of IFNA2 were processed in the following manner. Flash frozen aliquots were thawed in room temperature water for a maximum of 5 min followed by a centrifugation at 21,000 g for 10 min. The supernatant was transferred to a fresh eppendorf tube and the concentration was determined using an extinction coefficient of ε280 = 18.7 mM−1 cm−1.
Analytical Ultracentrifugation (AUC)
In order to establish the assembly state of purified IFNA2, sedimentation velocity experiments were conducted using a Beckman XL-A analytical centrifuge equipped with absorbance optics and an An-60 Ti rotor (Beckman Coulter, Brea, California). IFNA2 concentrations ranging from 10 µM to 50 µM in formulation buffer were tested. Samples were spun at 50,000 rpm at 4°C and the absorbance was monitored at 280 nm. In each experiment, a two-channel Epon centerpiece was used. IFNA2 assembly, reflected by the sedimentation coefficient, was assessed using SedFit (www.analyticalultracentrifugation.com). The sedimentation coefficient distribution, c(s), was corrected to 20°C and water (s20,w) using established methods [17]. Partial specific volume of IFNA2, buffer viscosity and density were calculated using Sednterp (www.unh.edu).
Circular Dichroism (CD)
To determine IFNA2 secondary structure, CD was used. Far-UV spectra were obtained at room temperature on a Chirascan Plus spectrometer (Applied Photophysics, Surrey, United Kingdom). IFNA2 stock was diluted into water to a concentration of 10 µM and scanned between 190 nm and 250 nm. The mean residue ellipticity (MRE) was calculated using the equation,
| (Eq. 1) |
Mass Spectrometry (MS)
In order to confirm the molecular weight and sequence of the purified IFNA2, intact protein as well as its tryptic digest were subjected to MS. The tryptic digest was carried out as previously described [18]. Intact protein samples were injected on an Agilent liquid chromatography system (Santa Clara, California) in conjunction with a Waters Q-TOF2 mass spectrophotometer (Millford, Massachusetts). Tryptic digest samples were prepared using iodoacetamide and analyzed for amino-terminal residue identity using an Applied Biosystems MALDI (Carlsbad, California). The experimental results were compared with the molecular weights of the theoretical digest fragments calculated from Protein Prospector (www.ucsf.edu).
Nuclear Magnetic Resonance (NMR)
Two-dimensional 15N-1H heteronuclear single quantum coherence (HSQC) NMR experiments were used to confirm the structure of IFNA2. IFNA2 was singly labeled with 15N using M9 minimal media and expressed using cold induction for 16 hrs. For these experiments, 100 µM IFNA2 (50 mM acetic acid, 0.02% sodium azide, pH 3.5) was used. The NMR spectra were obtained on a Varian 900 MHz instrument equipped with a cryoprobe (Rocky Mountain NMR Facility). Data were processed using nmrPipe software [19]. IFNA2 residue assignments available in the literature [20] were used to confirm the IFNA2 structure.
Antiviral and anti-proliferative activity assays
For the antiviral assay, IFNA2 samples were tested in duplicate in a viral challenge using Encephalomyocarditis (EMC) virus on human A549 cells. Plates were stained with crystal violet, a visual cytopathic effect inhibition (CPE) was performed, the dye was then solubilized, and the absorbance at 570 nm was measured. The half maximal effective concentration (EC50) was determined by fitting the data to the following four parameter logistic equation,
| (Eq. 2) |
where Bottom and Top represent the minimum and maximum values of the sigmoidal variation, and [IFNA2] is the protein concentration. Commercially available interferon α-2a standard (E. coli expressed) from PBL Assay Science as well as the 2nd WHO International Standard for interferon α-2a (NIBSC 95/650) were used for comparison.
For anti-proliferation assays, the IFNA2 sample was tested in triplicate using human ovarian cancer cells, Ovcar-3. Anti-proliferation was quantified using Promega Cell Titer 96 Aqueous Nonradioactive Cell Proliferation assay reagent (Cat# G-5430; Madison, Wisconsin) and absorbance at 490 nm. The data were analyzed using a four parameter logistic equation as in Eq. 2. PBL interferon α-2a standard (E. coli expressed) and the 2nd WHO International Standard (NIBSC 95/650) were run in parallel for comparison.
Results
Prior to optimizing the protocol, we made numerous attempts to obtain soluble IFNA2 from E. coli. We initially expressed IFNA2 as a soluble protein using pET-28a vector and Codon-Plus E. coli cells. This vector introduced a His6 tag, allowing for purification on a nickel column. However, cleaving the His6tag with thrombin protease following purification left two additional amino acids at the N-terminus of the IFNA2 arising from the cleavage site. In order to ensure that the purified protein exactly matches the pharmaceutical protein on the market, the pET-SUMO plasmid was chosen as the expression vector.
Design of the IFNA2 expression construct
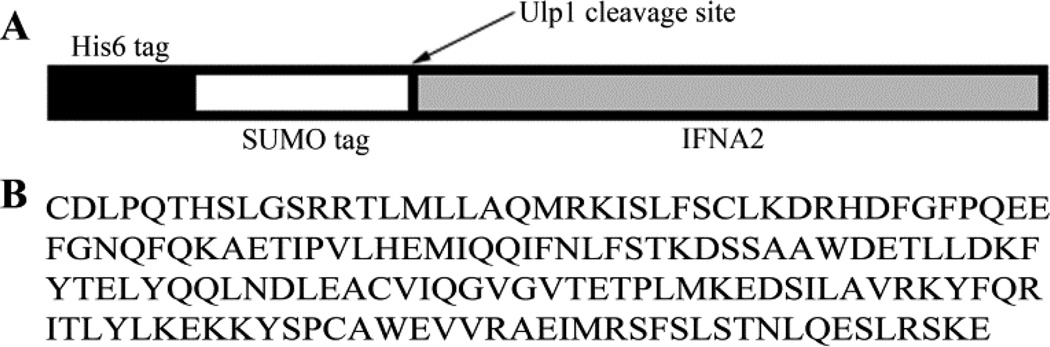
The vector for protein expression used in this study was a modified pET-28a vector, pET-SUMO. This vector contains a polyhistidine tag (His6) followed by a SUMO (small ubiquitin related modifier) (Figure 1A), a yeast-derived protein whose tertiary structure is specifically recognized by the yeast cysteine protease Ulp1 [21]. Expressed IFNA2 was amino-terminally linked to the SUMO moiety, as shown in Figure 1A. The codon-optimized cDNA of IFNA2 was used to result in the expression of a protein with the sequence shown in Figure 1B. The expression system described would result in a protein consisting of His6-SUMO-IFNA2.
Figure 1.
Post-translation fusion complex and the corresponding protein sequence. A) Pictorial representation of the translated His6-SUMO-IFNA2 fusion complex with the Ulp1 cleavage site indicated by an arrow. B) The primary protein sequence of IFNA2 following cleavage of the fusion tag from IFNA2, which matches with that of the pharmaceutical product as well as the protein for which the NMR structure has been previously determined (PDB ID 1ITF).
Expression and purification of recombinant human IFNA2
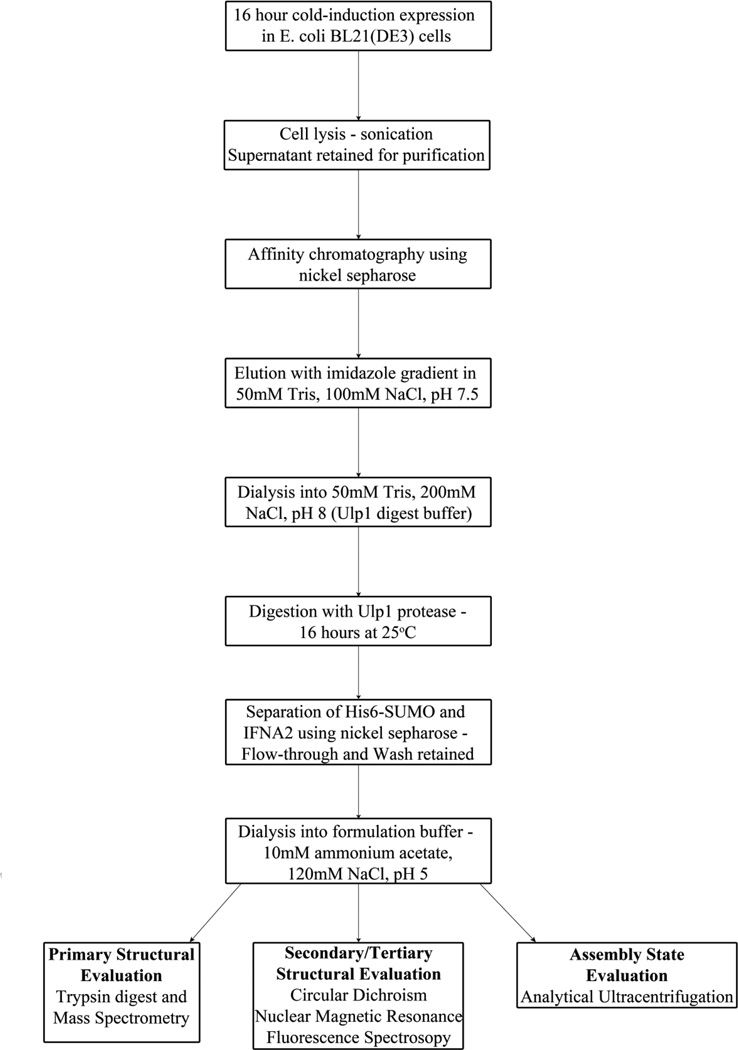
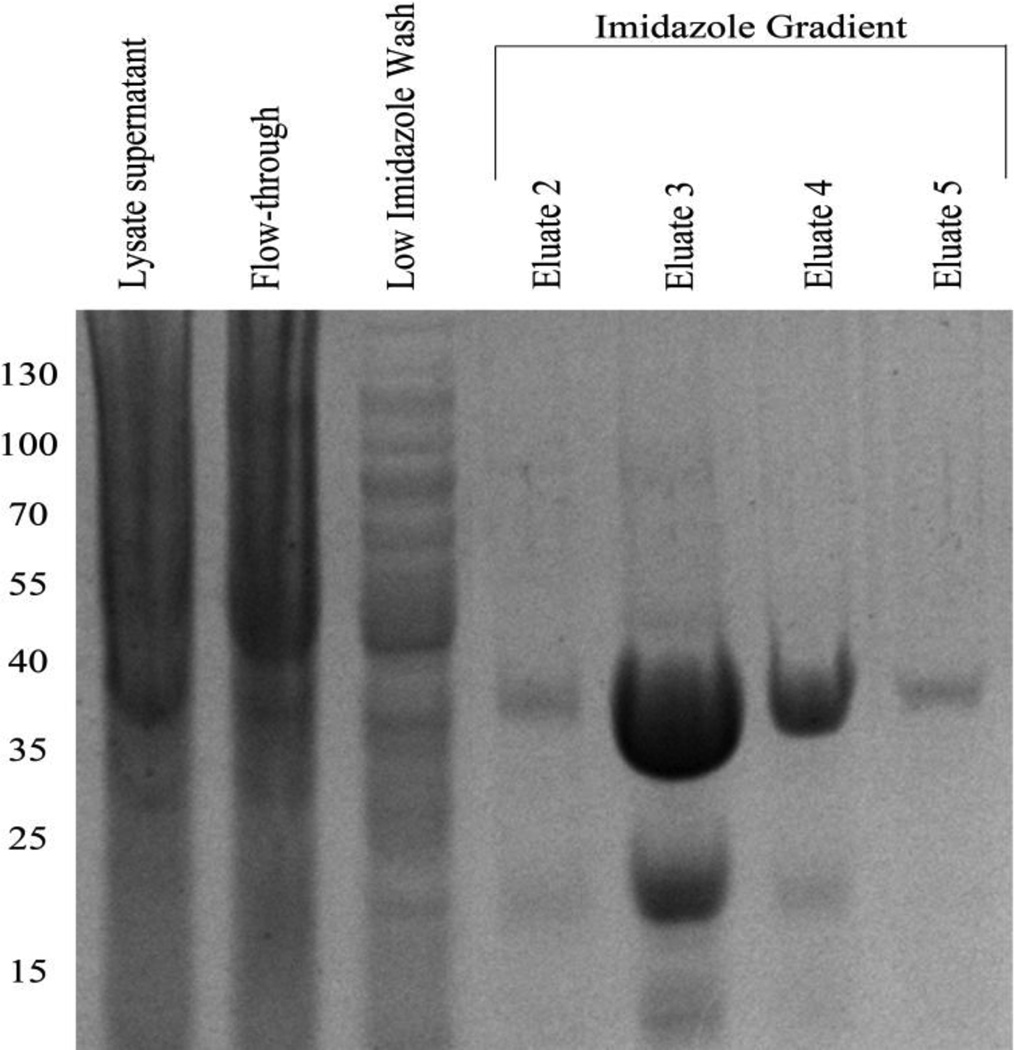
The expression and purification scheme described in this study is shown as a flowchart in Figure 2. The inclusion of the His6 tag allowed for simplified purification of the lysate supernatant using a nickel column. The His6-SUMO-IFNA2 complex resulted in a predominant species with a molecular weight of approximately 33.3 kDa (Eluate 3 lane in Figure 3).
Figure 2.
Schematic of expression and purification scheme optimized to obtain high yields of highly pure, soluble IFNA2 from E. coli.
Figure 3.
Purification of the His6-SUMO-IFNA2 fusion complex. The lysate fraction was loaded onto a nickel column and the flow-through was collected. A low concentration imidazole wash allowed for further removal of any bacterial contaminants. Eluates 2–5 show the fusion complex eluting from the nickel resin, with overexpression of the complex evident in Eluate 3 and Eluate 4 at ~33kDa. Molecular weight markers are shown on the far left.
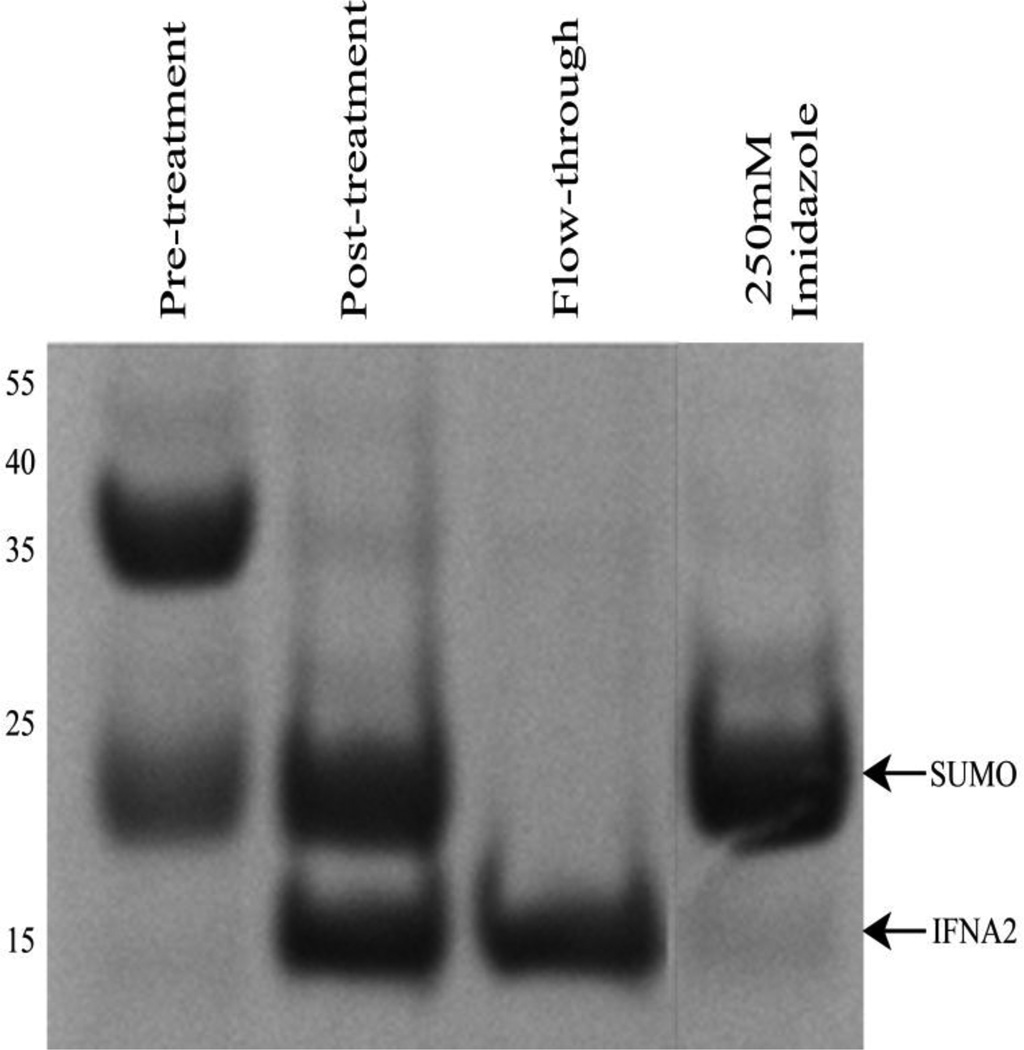
Cleavage of the recombinant protein complex was achieved using the yeast protease Ulp1. A successful cleavage reaction resulted in two distinct species observed using SDS-PAGE (post-treatment lane in Figure 4). The upper band, running at approximately 20 kDa, corresponded to the His6-SUMO tag, which has been known to migrate slowly during electrophoresis resulting in a higher molecular weight estimate than its calculated value (~14 kDa) [22]. Based on the location of SDS-PAGE migration, it is likely that the impurities observed prior to Ulp1 cleavage step were His6-SUMO-IFNA2 expression fragments. The lower band seen post-cleavage corresponds to IFNA2 at a molecular weight of 19.2 kDa.
Figure 4.
Cleavage of the His6-SUMO tag with Ulp1 protease. Pre-treatment refers to elution fractions from Figure 3 prior to addition of Ulp1 protease. Following the cleavage reaction, the post-treatment lane shows two distinct bands representing the His6-SUMO tag and untagged IFNA2. Pure IFNA2 was collected in the flow-through. A high concentration imidazole elution fraction removes the His6-SUMO tag from the nickel column.
Taking advantage of the His6 affinity for nickel resin, the His6-SUMO tag was separated from pure protein using a nickel column. Pure, untagged IFNA2 was collected in the flow-through and wash steps of the separation, retaining no residual His6-SUMO (flow-through lane in Figure 4). The cleaved His6-SUMO tag was retained on the column until elution with higher concentrations of imidazole (250 mM Imidazole lane in Figure 4).
The purification protocol introduced here yielded 16 mg of purified IFNA2 per 1 liter of expression media. This yield was achieved reproducibly with the application of the steps shown in Figure 2.
Minor loss following freeze-thaw of IFNA2 aliquots
Numerous protein systems have been shown to be sensitive to freeze-thaw damage via aggregation during freezing or thawing [23, 24]. Because our purified protein was flash frozen and subsequently stored in the frozen state, it was possible that damage could occur either during the freezing process or as a result of thawing. To test the sensitivity of our purified protein to freeze-thaw stress, individual aliquots were thawed in room temperature water and centrifuged in order to remove any potential aggregates. No visible aggregates were observed. The concentration of the resulting supernatant consistently was within 10% of the approximate 100 µM aliquot concentration.
Confirmation of IFNA2 identity using biophysical and structural methods
In order to confirm the identity of the protein obtained using our purification protocol, numerous biophysical and structural techniques were employed.
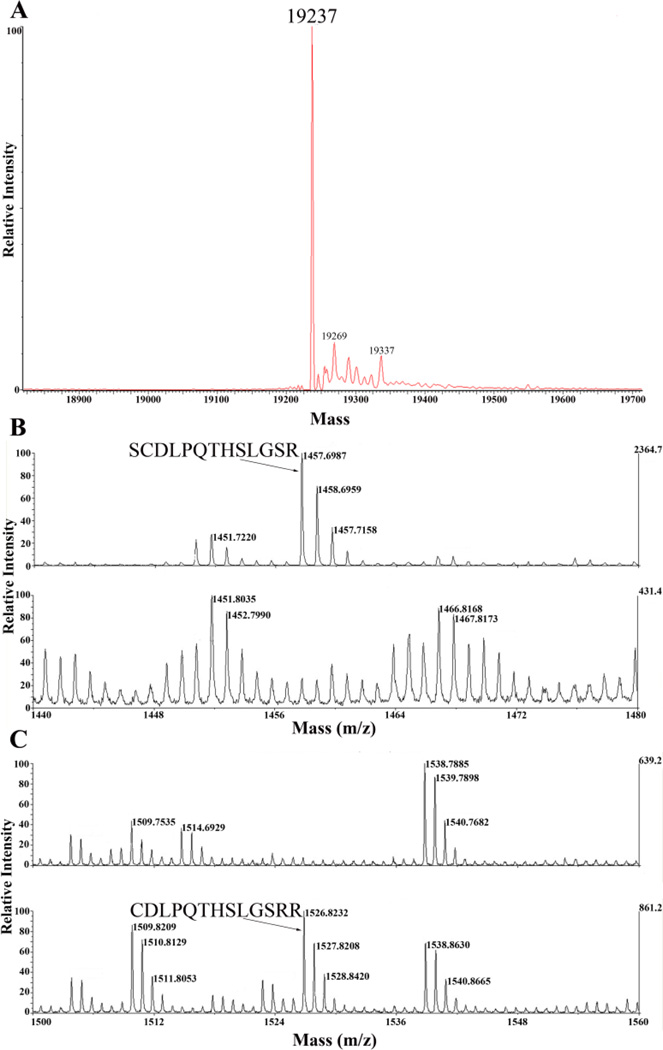
To determine the molecular weight of our purified protein with high precision, liquid chromatography - mass spectrometry (LC-MS) was utilized. The main protein peak was observed at 19.2 kDa, consistent with the calculated molecular weight from ExPASy (Figure 5A).
Figure 5.
MS analysis of IFNA2. A) Intact full-length protein injection shows a predominant peak at 19237 Da. B) Tryptically digested IFNA2 initially showed a residual serine on the N-terminus of the protein, indicated by the 1457 m/z peak. The inset text shows the N-terminal fragment corresponding to this m/z value. The upper and lower panels show pre- and post-deletion of the N-terminal serine, respectively. C) Tryptically digested IFNA2 shows the presence of the 1526 m/z peak, corresponding to the N-terminal cysteine. The inset text shows the N-terminal fragment at this m/z value. The upper and lower panels show pre- and post-deletion of the N-terminal serine, respectively.
A benefit of using the pET-SUMO vector is the lack of additional amino acids on the protein of interest as a result of the cleavage with Ulp1. To ensure that our purified protein retained no residual amino acids on its amino-terminus, tryptically digested gel fragments were subjected to MS analysis. A theoretical tryptic digest was conducted using Protein Prospector in which the protein primary sequence was used to calculate all possible digest fragments with corresponding molecular weights. Initial analysis indicated that a residual serine preceded the expected amino-terminal fragment, evidenced by a peak at 1457 m/z (Figure 5B). To express IFNA2 with the same sequence as that of the pharmaceutical protein, the serine codon was removed in a reconstructed vector. Subsequent tryptic digest analysis demonstrated an amino-terminal digest fragment corresponding to the theoretical fragment without serine at 1526 m/z, confirming the complete removal of the His6-SUMO tag leaving no additional amino acids on the purified protein (Figure 5C). These m/z values reflect carbamidomethyl modifications on the cysteine residue, due to the sample treatment with iodoacetamide during the tryptic digest protocol [18].
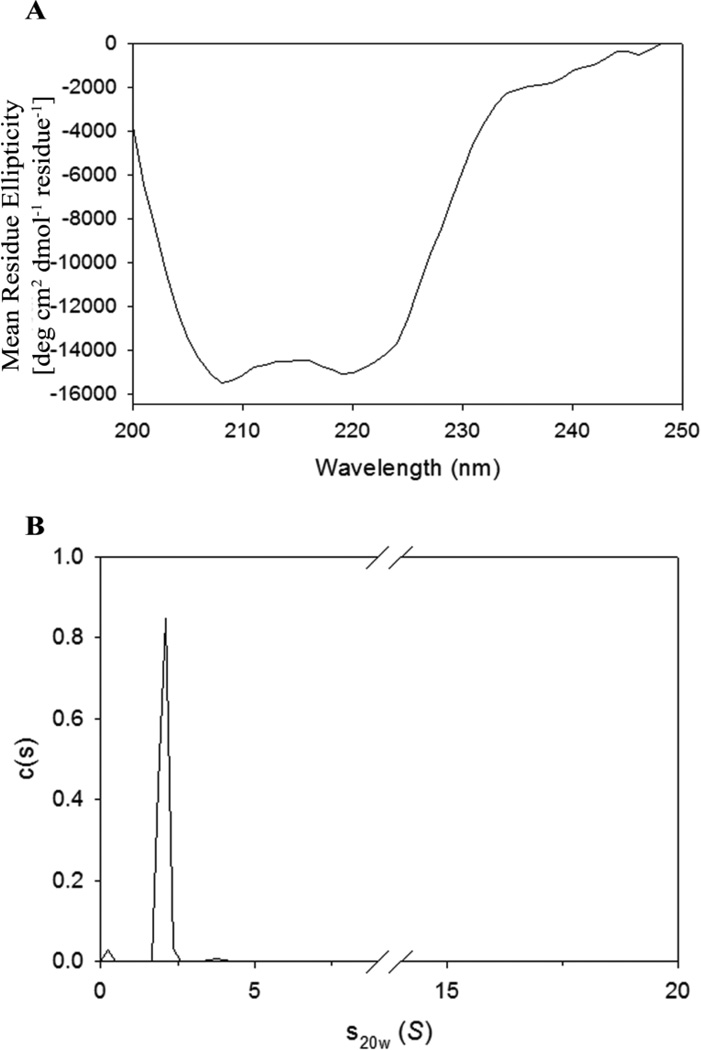
Proteins within the interferon family are characterized by predominantly α-helical structure [5]. Thus, CD was used to determine the secondary structure of our purified protein. Stock protein was diluted in water to a concentration of 10 µM, and its far-UV CD spectrum is shown in Figure 6A. The spectrum showed characteristic negative peaks at 208 and 222 nm, consistent with a well-folded α-helical structure. The MRE (Eq. 1) obtained in this study agrees well with the previously documented CD spectra for IFNA2 [25], with minor deviations attributed to differences in buffer conditions and pH.
Figure 6.
Biophysical characterization of IFNA2. A) Far-UV CD spectrum of IFNA2 shows that this protein is well-folded and α-helical. B) Sedimentation velocity AUC demonstrates that IFNA2 is a monomer in solution at pharmaceutically relevant concentrations.
Additionally, sedimentation velocity AUC experiments were used to estimate the protein molecular weight and to determine the assembly state of IFNA2.These results confirmed that the purified protein was a monomer in solution (Figure 6B). The corresponding molecular weight was 19.5 kDa, which is in good agreement with the theoretical estimate (19.2 kDa) from the amino acid sequence and the MS results (19.2 kDa).
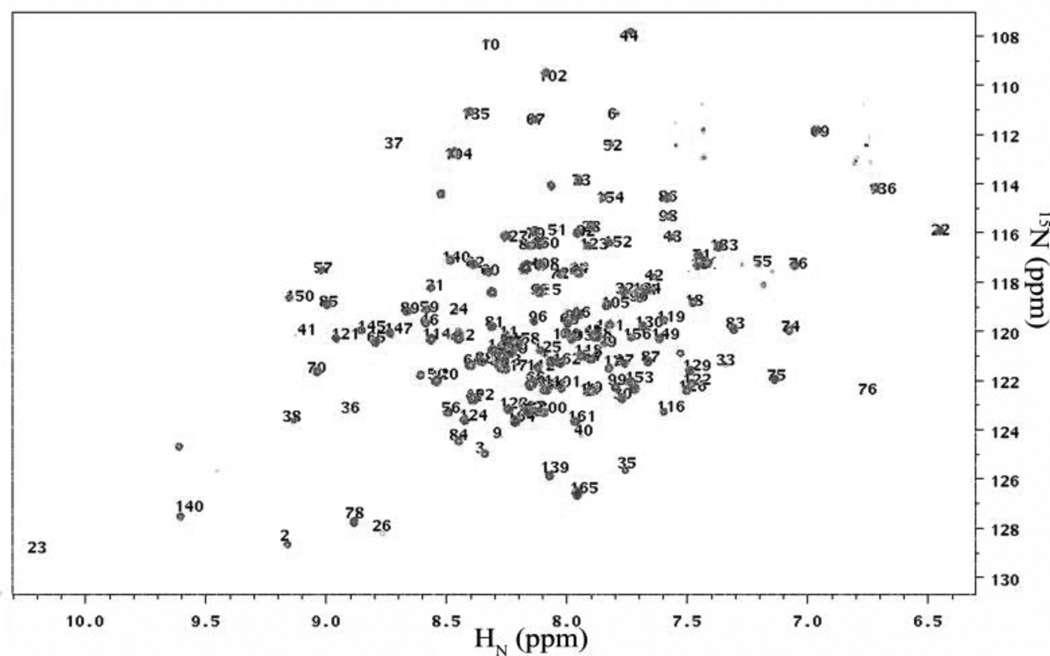
The solution structure of IFNA2 has been previously determined using 2D-NMR techniques [20]. We performed 2D-NMR HSQC experiments at pH 3.5 to confirm the structure of our purified protein. These pH conditions were chosen to reflect the solution conditions previously used to determine the IFNA2 structure. The overlay of the HSQC spectrum of our protein with the published crosspeak assignments is shown in Figure 7. The assigned peaks exactly matched with the spectrum, confirming the identity and the structure of our purified protein.
Figure 7.
2D HSQC NMR spectrum of IFNA2, and comparison with the previously published crosspeak assignments. The blue peaks correspond to IFNA2 data collected at pH 3.5. Overlaid numerical values correspond to peak assignments as determined by Klaus et al. [20].
Purified IFNA2 has functional activity comparable to International Standards
To ensure that our purified protein had functional activity, antiviral and anti-proliferation assays were conducted (Figure 8). The cytopathic effect inhibition (CPE) assay was used to verify IFNA2 antiviral activity against EMC viral challenge on human A549 cells. The experimental results showed perfect agreement between our IFNA2 and the two well-accepted standards in the interferon literature (Figure 8A and the EC50 values in Figure 8C). Purified IFNA2 has an EC50 of (1.21 ± 0.92) × 10−13 M, which matches the EC50 of (1.31 ± 1.00) × 10−13 M measured for the 2nd WHO International Standard for interferon α-2a, NIBSC 95/650, and the EC50 of (1.65 ± 0.76) × 10−13 M for the IFNA2 standard from the PBL Assay Science. Similar results were obtained with the IFNA2 anti-proliferative function assessed using human ovarian cancer cells (Figure 8B and the EC50 values in Figure 8C). Our IFNA2 has an EC50 of (1.29 ± 0.08) × 10−10 M, which matches the EC50 of (1.00 ± 0.19) × 10−10 M measured for the International Standard NIBSC 95/650 and the EC50 of (2.14 ± 0.24) × 10−10 M for the IFNA2 standard from PBL.
Figure 8.
Functional assays of IFNA2 in comparison with the two well-established standards. A) IFNA2 antiviral activity following encephalomyocarditis (EMC) viral challenge on human A549 cells. B) IFNA2 anti-proliferative function against human ovarian cancer cells. Red, blue, and green symbols represent the data on our purified IFNA2, E. coli-derived interferon α-2a standard from PBL Assay Science, and on the 2nd International WHO standard for interferon α-2a (NIBSC 95/650), respectively. C) The EC50 values determined from data fitting.
Discussion
The use of protein based pharmaceuticals is becoming increasingly important in the treatment of numerous diseases for which effective small molecule drugs are not available. The interferon family, comprising a group of immunologic factors that are up-regulated during viral or microbial onslaught, constitutes an important class of protein-based formulations. IFNA2 is one of the 13 interferon α subtypes found in humans, and is used pharmaceutically to treat various diseases including hepatitis C and leukemia [4, 8–10]. Other pharmaceutically available interferon drugs include interferon α–2b and interferon β.
Human IFNA2 is not glycosylated or otherwise modified post-translationally [12], indicating that the bacterial expression system can be utilized. However, previously published expression and purification schemes require a) purification from inclusion bodies and subsequent refolding [13–15], and b) numerous purification steps in order to obtain pure, homogenous protein [26]. A purification protocol necessitating multiple steps introduces a number of problems, such as protein stability and aggregation concerns as well as simply increasing the time required for purification. Additionally, multiple purification steps reduce the final yield [26]. Finally, refolding following purification from inclusion bodies may result in soluble protein losses due to membrane interactions, protein aggregation, and other factors. We have overcome these problems using a pET-SUMO expression system, which increased the soluble protein yield, and a single purification step. To our knowledge, ours is the first published instance of obtaining soluble IFNA2 from E. coli while still obtaining an improved overall yield of the protein.
Expression vectors using stable, soluble proteins as fusion tags have been shown to increase the soluble expression of complex, sensitive proteins [22, 27, 28]. Such technology of using solubility tags has been relatively less explored in expressing interferon proteins, except in the case of interferon α-2c [29]. Maltose binding protein (MBP) was used as a fusion tag to enhance the soluble yield of interferon α-2c. However, unlike the SUMO tag we used for IFNA2, cleaving the MBP tag left an additional methionine residue at the N-terminus of the protein. The presence of additional amino acids after proteolysis is not uncommon in recombinant protein expression. Most proteases recognize specific amino acid sequences in order to cleave the tag, and the cleavage removes majority of the tag but generally leaves a few non-native amino acids other than the protein of interest [30]. In that sense, pET-SUMO and Ulp1 expression system is unique, because cleaving the SUMO tag with Ulp1 protease leaves no residual amino acids on the protein of interest. The cleavage efficiency depends on the tertiary structure of the SUMO fusion protein in addition to the specific amino acid sequence that needs to be recognized by the Ulp1 protease [21]. The proteins obtained following the cleavage of the SUMO tag are sequentially pure. Further, using the SUMO tag has an additional advantage in terms of increasing the soluble expression of recombinant proteins in comparison with other traditional solubility tags [28]. With the addition of the SUMO tag, we obtained increased overexpression of IFNA2 in the soluble fraction.
We expressed SUMO-IFNA2 at 16°C rather than the standard temperature of 37°C commonly employed in producing recombinant proteins. Cold induction has been shown to enhance the soluble expression of recombinant proteins [31, 32]. In the case of SUMO-IFNA2, inducing at 37°C or 25°C resulted in inclusion body formation and increased fragmentation of IFNA2. Decreasing the temperature to 16°C reduced the protein fragmentation, and increased the expression of the full-length SUMO-IFNA2 as a soluble protein. Additionally, a lower concentration of IPTG was used to induce the protein expression. This allowed for slower protein expression, thus decreasing the unwanted aggregation of improperly folded protein molecules, and as a result increased the yield of soluble protein [33].
To the best of our knowledge, this is the first study where IFNA2 has been expressed in the soluble fraction of E. coli and purified in a single step to homogeneity and high purity. Additionally, in comparison with other published methods using E. coli as an expression system, we show here a reproducible and simplified method to achieve significantly improved yields of IFNA2 while maintaining the antiviral and anti-proliferative activity and the structural integrity of the protein. Combining the expression and purification protocols described here to produce IFNA2 on a small-scale with the commercial fermenter technology commonly used in pharmaceutical industry to produce recombinant proteins may further enhance the IFNA2 yields, and may promote the development of new interferon-based protein drugs to treat various disorders.
Highlights.
IFNA2 is expressed as a soluble protein in E. coli bacteria.
Improved yields of IFNA2 are obtained compared to earlier expression conditions.
A simpler purification scheme for IFNA2 is developed compared to published schemes.
Expressed IFNA2 is correctly folded, exists as a monomer, and is fully functional.
Acknowledgments
The authors thank Christopher Lima of the Sloan-Kettering Institute for his generous gift of the pET-SUMO expression system, and Deborah Wuttke and Karen Lewis for their suggestions on using pET-SUMO vector. We also thank Keith Connaghan and Michael Miura for their guidance and expertise with AUC, Joe Gomez for MS data collection and analysis, and Geoffrey Armstrong and Javier Cabello-Villegas for their help with NMR data collection and processing. This work was funded by the University of Colorado Skaggs School of Pharmacy and Pharmaceutical Sciences. Regina Bis was partially supported by a NIH Leadership training grant in Pharmaceutical Biotechnology (T32GM008732) and a predoctoral fellowship from the PhRMA Foundation (AWD-120487). Rocky Mountain Regional 900 MHz NMR Facility was funded by the National Institutes of Health instrumentation Grant P41GM068928.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Isaacs A, Lindenmann J. Virus Interference. I. The Interferon. Proceedings of the Royal Society of London. Series B - Biological Sciences. 1957;147(927):258–267. doi: 10.1098/rspb.1957.0048. [DOI] [PubMed] [Google Scholar]
- 2.Malmagaard L. Induction and regulation of IFNs during viral infections. J Interferon Cytokine Res. 2004;24:439–454. doi: 10.1089/1079990041689665. [DOI] [PubMed] [Google Scholar]
- 3.Fensterl V, Sen GC. Interferons and viral infections. Biofactors. 2009;35(1):14–20. doi: 10.1002/biof.6. [DOI] [PubMed] [Google Scholar]
- 4.Kaser A, Tilg H. Interferon-α in inflammation and immunity. Cell Mol Bio. 2000;47:609–617. [PubMed] [Google Scholar]
- 5.Theofilopoulos AN, et al. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu Rev Immunol. 2005;23:307–336. doi: 10.1146/annurev.immunol.23.021704.115843. [DOI] [PubMed] [Google Scholar]
- 6.Bekisz J, et al. Human interferons alpha, beta and omega. Growth Factors. 2004;22(4):243–251. doi: 10.1080/08977190400000833. [DOI] [PubMed] [Google Scholar]
- 7.Bergman SJ, Ferguson MC, Santanello C. Interferons as therapeutic agents for infectious diseases. Infect Dis Clin North Am. 2011;25(4):819–834. doi: 10.1016/j.idc.2011.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hiscott J, Cantell K, Weissmann C. Differential expression of human interferon genes. Nucleic Acids Res. 1984;12:3727–3746. doi: 10.1093/nar/12.9.3727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diaz M, Bohlander S, Allen G. Nomenclature of human interferon genes. J interferon Res. 1993;13:243–244. doi: 10.1089/jir.1993.13.243. [DOI] [PubMed] [Google Scholar]
- 10.Kirkwood J. Cancer immunotherapy: the interferon-alpha experience. Semin Oncol. 2002;29:18–26. doi: 10.1053/sonc.2002.33078. [DOI] [PubMed] [Google Scholar]
- 11.Rubinstein M, et al. Human leukocyte interferon: production, purification to homogeneity, and initial characterization. Proceedings of the National Academy of Sciences. 1979;76(2):640–644. doi: 10.1073/pnas.76.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pestka S. The interferons: 50 years after their discovery, there is much more to learn. J Biol Chem. 2007;282(28):20047–20051. doi: 10.1074/jbc.R700004200. [DOI] [PubMed] [Google Scholar]
- 13.Hochuli E. Large-scale recovery of interferon alpha-2 a synthesized in bacteria. Chimia. 1986;40(11):408–412. [Google Scholar]
- 14.Meng J, et al. High-yield expression, purification and characterization of tumor-targeted IFN-α2a. Cytotherapy. 2007;9(1):60–68. doi: 10.1080/14653240601094322. [DOI] [PubMed] [Google Scholar]
- 15.Piehler J, Schreiber G. Biophysical analysis of the interaction of human IFNAR2 expressed in E. coli with IFNα2. J Mol Biol. 1999;289:57–67. doi: 10.1006/jmbi.1999.2726. [DOI] [PubMed] [Google Scholar]
- 16.Champion™ pET SUMO Protein Expression System Instruction Manual, Invitrogen. 2010 [Google Scholar]
- 17.van Holde KE. Physical Biochemistry. Englewood Cliffs, California: Prentice-Hall; 1971. [Google Scholar]
- 18.Lim H, et al. Identification of 2D-gel proteins: A comparison of MALDI/TOF peptide mass mapping to μ LC-ESI tandem mass spectrometry. Journal of the American Society for Mass Spectrometry. 2003;14(9):957–970. doi: 10.1016/s1044-0305(03)00144-2. [DOI] [PubMed] [Google Scholar]
- 19.Delaglio F, et al. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. Journal of biomolecular NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 20.Klaus W, et al. The three-dimensional high resolution structure of human interferon α-2a determined by heteronuclear NMR spectroscopy in solution. J Mol Biol. 1997;274:661–675. doi: 10.1006/jmbi.1997.1396. [DOI] [PubMed] [Google Scholar]
- 21.Mossessova E, Lima CD. Ulp1-SUMO Crystal Structure and Genetic Analysis Reveal Conserved Interactions and a Regulatory Element Essential for Cell Growth in Yeast. Molecular Cell. 2000;5(5):865–876. doi: 10.1016/s1097-2765(00)80326-3. [DOI] [PubMed] [Google Scholar]
- 22.Butt TR, et al. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43(1):1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W. Protein aggregation and its inhibition in biopharmaceutics. International Journal of Pharmaceutics. 2005;289(1):1–30. doi: 10.1016/j.ijpharm.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 24.Bhatnagar BS, Bogner RH, Pikal MJ. Protein stability during freezing: separation of stresses and mechanisms of protein stabilization. Pharmaceutical development and technology. 2007;12(5):505–523. doi: 10.1080/10837450701481157. [DOI] [PubMed] [Google Scholar]
- 25.Sharma VK, Kalonia DS. Temperature-and pH-induced multiple partially unfolded states of recombinant human interferon-α2a: possible implications in protein stability. Pharmaceutical research. 2003;20(11):1721–1729. doi: 10.1023/b:pham.0000003367.62900.0f. [DOI] [PubMed] [Google Scholar]
- 26.Kim DC, Jung J. Purification of recombinant human alpha-2a interferon without using monoclonal antibodies. Journal of microbiology and biotechnology. 2002;12(6):916–920. [Google Scholar]
- 27.Esposito D, Chatterjee DK. Enhancement of soluble protein expression through the use of fusion tags. Curr Opin Biotechnol. 2006;17(4):353–358. doi: 10.1016/j.copbio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Marblestone JG, et al. Comparison of SUMO fusion technology with traditional gene fusion systems: enhanced expression and solubility with SUMO. Protein Sci. 2006;15(1):182–189. doi: 10.1110/ps.051812706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmeisser H, et al. Binding Characteristics of IFN-alpha Subvariants to IFNAR2-EC and Influence of the 6-Histidine Tag. J Interferon Cytokine Res. 2006;26(12):866–876. doi: 10.1089/jir.2006.26.866. [DOI] [PubMed] [Google Scholar]
- 30.Hefti MH, et al. A Novel Purification Method for Histidine-Tagged Proteins Containing a Thrombin Cleavage Site. Analytical Biochemistry. 2001;295(2):180–185. doi: 10.1006/abio.2001.5214. [DOI] [PubMed] [Google Scholar]
- 31.Schein CH, Noteborn MHM. Formation of Soluble Recombinant Proteins in Escherichia Coli is Favored by Lower Growth Temperature. Nat Biotech. 1988;6(3):291–294. [Google Scholar]
- 32.Qing G, et al. Cold-shock induced high-yield protein production in Escherichia coli. Nat Biotechnol. 2004;22(7):877–882. doi: 10.1038/nbt984. [DOI] [PubMed] [Google Scholar]
- 33.Tolia NH, Joshua-Tor L. Strategies for protein coexpression in Escherichia coli. Nat Meth. 2006;3(1):55–64. doi: 10.1038/nmeth0106-55. [DOI] [PubMed] [Google Scholar]