SUMMARY
Here, we show that a subset of breast cancers express high levels of the type 2 phosphatidylinositol-5-phosphate 4-kinases α and/or β (PI5P4Kα and β) and provide evidence that these kinases are essential for growth in the absence of p53. Knocking down PI5P4Kα and β in a breast cancer cell line bearing an amplification of the gene encoding PI5P4K β and deficient for p53 impaired growth on plastic and in xenografts. This growth phenotype was accompanied by enhanced levels of reactive oxygen species (ROS) leading to senescence. Mice with homozygous deletion of both TP53 and PIP4K2B were not viable, indicating a synthetic lethality for loss of these two genes. Importantly however, PIP4K2A−/−, PIP4K2B+/−, and TP53−/− mice were viable and had a dramatic reduction in tumor formation compared to TP53−/− littermates. These results indicate that inhibitors of PI5P4Ks could be effective in preventing or treating cancers with mutations in TP53.
INTRODUCTION
The phosphoinositide family of lipids includes seven derivatives of phosphatidylinositol (PI) that are formed through the phosphorylation of the 3-, 4-, and 5-positions on the inositol ring. Phosphoinositides have distinct biological roles and regulate many cellular processes, including proliferation, survival, glucose uptake, and migration. Phosphoinositide kinases, phosphatases, and phospholipases spatially and temporally regulate the generation of the different phosphoinositide species, which localize to different subcellular compartments. Phosphatidylinositol-3,4,5-trisphosphate (PI-3,4,5-P3) is synthesized by phosphoinositide 3-kinase (PI3K) and serves as the plasma membrane docking site for a subset of proteins that have pleckstrin-homology (PH) domains that bind this lipid, including the serine/threonine protein kinase AKT (also known as protein kinase B or PKB). AKT is a protooncogene that has critical regulatory roles in insulin signaling and cancer progression. Phosphatidylinositol-4,5-bisphosphate (PI-4,5-P2) is the major substrate for class I PI3Ks and has a significant role itself in mediating the localization of proteins to the plasma membrane and in nucleating cortical actin polymerization (Cantley, 2002).
Until 1997, it was thought that PI-4,5-P2 was produced exclusively by phosphorylation of phosphatidylinositol-4-phosphate (PI-4-P) at the 5 position of the inositol ring, a reaction catalyzed by the type 1 PI-4-P 5-kinases (encoded by the genes PIP5K1A, B, and C). Unexpectedly, a second highly related family of PIP kinases (called type 2) was found to produce PI-4,5-P2 by phosphorylating the 4 position of phosphatidylinositol-5-phosphate (PI-5-P), a lipid that had been previously overlooked due to its comigration with the much more abundant PI-4-P (Rameh and Cantley, 1999; Rameh et al., 1997). The type 2 PIP kinases are not present in yeast but are conserved in higher eukaryotes from worms and flies to mammals. Humans and mice have three distinct genes, PIP4K2A, B, and C encoding enzymes called PI5P4Kα, β, and γ, respectively. The bulk of PI-4,5-P2 in most tissues is almost certainly derived from the type 1 PIP5Ks, yet recent quantitative proteomic studies on cell lines have revealed a higher abundance of PI5P4Ks than PI4P5Ks (Nagaraj et al., 2011). This high abundance of the type 2 enzymes may, in part, explain why the substrate PI-5-P is present at very low levels. Although the type 1 PIP kinases generate PI-4,5-P2 at the plasma membrane, the type 2 kinases are located at internal membranes, including the endoplasmic reticulum (ER), Golgi, and nucleus and probably generate PI-4,5-P2 at those locations (Fruman et al., 1998; Sarkes and Rameh, 2010; Schaletzky et al., 2003; Walker et al., 2001). The vast majority of PI-4,5-P2 is located at the plasma membrane, and it is not clear whether the critical function of the type 2 PIP kinases is to generate PI-4,5-P2 at intracellular sites or to maintain low levels of PI-5-P (or both).
In a previous study, we generated mice in which one of the type 2 PIP kinase genes (PIP4K2B) was deleted in the germline. These mice were viable, exhibited enhanced insulin sensitivity, and enhanced insulin-dependent activation of AKT in skeletal muscle (Lamia et al., 2004). Paradoxically, despite increased AKT activation the mice were smaller and had decreased adiposity on a high-fat diet. Cell-based assays revealed that PI5P4Kβ (encoded by PIP4K2B) becomes phosphorylated by p38 at Ser326 in response to cellular stresses, such as UV and H2O2, and that this causes inhibition of the PI5P 4-kinase activity and results in increased cellular PI-5-P levels (Jones et al., 2006). These studies suggest that the type 2 PIP kinases mediate cellular stress responses downstream of p38 (presumably by altering the PI-5-P/PI-4,5-P2 ratio at intracellular locations) and that under conditions of low stress, these enzymes suppress the PI3K/AKT signaling pathway. It should be pointed out that the type 2 PIP kinases are unlikely to supply PI-4,5-P2 as a substrate for PI3K because activation of AKT correlates with loss of PI5P4K activity rather than gain.
In this study, we have interrogated the potential role of type 2 PIP kinases in cancers. We found high levels of either PI5P4Kα or PI5P4Kβ enzymes or both in a number of breast cancer cell lines and, more importantly, found amplification of the PIP4K2B gene and high levels of both the PI5P4Kα and PI5P4Kβ proteins in a subset of human breast tumors. We found that knocking down the levels of both PI5P4Kα and PI5P4Kβ in a TP53-deficient breast cancer cell line blocked growth on plastic and in xenografts. This impaired growth correlated with impaired glucose metabolism and enhanced levels of reactive oxygen species (ROS) leading to senescence. The impaired glucose metabolism, despite activation of the PI3K-AKT pathway (which typically enhances glucose metabolism) was paradoxical. The results indicate that PI3K activation is not driving the ROS production but may be an inadequate feedback attempt to restore glucose uptake and metabolism.
Finally, to assess the role of type 2 PIP kinases in tumor formation, we generated mice with germline deletions of PIP4K2A and PIP4K2B and crossed these with TP53−/− mice and evaluated tumor formation in all the viable genotypes. We found that mice with homozygous deletion of both TP53 and PIP4K2B were not viable, indicating a synthetic lethality for loss of these two genes. Importantly, mice with the genotype PIP4K2A−/−, PIP4KB+/−, TP53−/− were viable and had a dramatic reduction in tumor formation compared to siblings that were TP53−/− and wild-type for PIP4K2B and/or PIP4K2A genes. These results suggest that PI5P4Kα and PI5P4Kβ could be targets for pharmaceutical intervention in cancers that are defective in TP53.
RESULTS
Amplification of PIP4K2B in HER-2/Neu-Positive Breast Cancers and Co-Occurrence with TP53 Mutation/Deletion
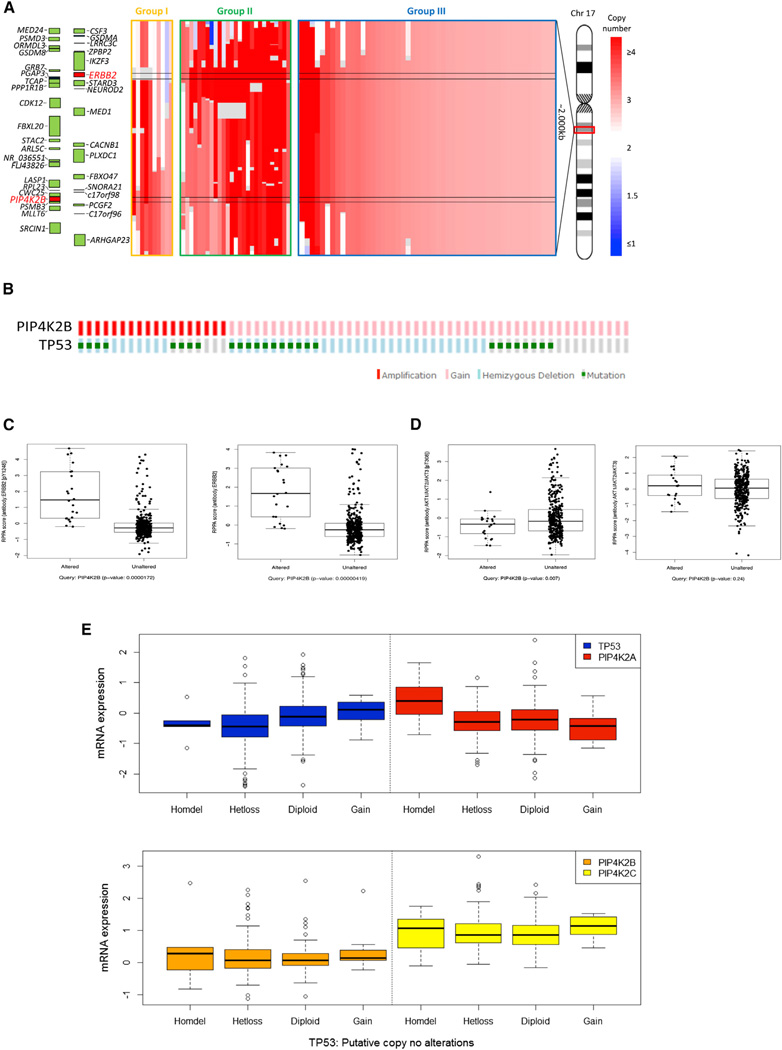
Gene amplification in breast cancer is associated with disease progression, adverse prognosis, and development of drug resistance. PIP4K2B is located in a chromosomal region (17q12) close to ERBB2 (HER2/Neu), which is amplified in about ~25% of breast cancers and in a smaller fraction of nonsmall cell lung adenocarcinomas, as well as other cancer types including colorectal and renal (Luoh et al., 2004; Slamon, 1987; Slamon et al., 1989, 2001). Approximately half of the breast tumors that exhibit ERBB2 amplification also exhibit amplification of PIP4K2B (Figure 1A). For the majority of tumors that have both ERBB2 and PIP4K2B amplified, the two genes are on the same amplicon (Figure 1B). However, for a significant fraction (27/78) these two genes appear to be on distinct amplicons. Also, tumors were identified that had relatively focal amplification of PIP4K2B without amplification of ERBB2 (Figure 1A). Amplification of PIP4K2A was only observed in a small fraction of breast cancers (data not shown). Furthermore, it is interesting to note that the genomic alteration of PIP4K2B and TP53 across 66 breast carcinomas indicate a trend of co-occurrence between PIP4K2B gain/amplification and TP53 mutation/deletion (Figure 1B). Interestingly, using reverse phase protein array (RPPA) a high-throughput proteomics technology we could correlate PIP4K2B gain/amplification with a significant increase in ERBB2 phosphorylation on tyrosine 1248 (Y1248) and of total ERBB2 levels (Figure 1C), as well as, with a small but significant decrease in the phosphorylation of AKT on threonine 308 (T308) with no change in total AKT protein levels (Figure 1D). To further address the link between PIP4K2A/B and TP53 in cancers, we more deeply interrogated the rapidly growing TCGA database of breast cancers. We found that the subgroup of breast cancers that had homozygous deletion of TP53 (analogous to the TP53-null mouse breast cancers discussed below) had significantly higher PIP4K2A mRNA compared to tumors with two alleles of TP53 or heterozygous loss of TP53 (Figure 1E). There was also a trend toward higher expression of PIP4K2B and PIP4K2C in the tumors with homozygous deletion of TP53 (Figure 1E), though these changes did not reach significance.
Figure 1. Amplification of PIP4K2B in HER-2/Neu-Positive Breast Cancers and Co-occurrence with TP53 Mutation/Deletion.
(A) Genomic landscape of PIP4K2B and ERBB2 (HER2) DNA copy number amplifications in cancer. Tumor samples are divided into three groups: group I: those with amplification in PIP4K2B but no or lower level amplification of ERBB2, group II: samples with amplification of both genes but likely to be derived from two different amplicons, and group III: samples with amplification in both genes derived from the same amplicon. Colored bar indicates degree of copy number gain (red).
(B) Oncoprints of PIP4K2B and TP53 across 66 breast carcinomas indicating a trend of co-occurrence between PIP4K2B gain/amplification and TP53 mutation/deletion (p value: 0.006306, Fisher’s exact test). Individual genes are represented as rows, and individual cases or patients are represented as columns. Genetic alterations are color-coded with red indicating amplification; pink, DNA copy number gain; light blue, hemizygous deletion and green box, point mutation. Only cases with gain/amplification of PIP4K2B are included. These oncoprints are based on data obtained from the Stand Up to Cancer cBio portal (http://cbio.mskcc.org/su2c-portal/).
(C) Box plots indicating that the levels of ERBB2 phosphorylation on tyrosine 1248 (y1248) (left) and of total ERBB2 (right) measured by RPPA are significantly higher in breast carcinomas with PIP4K2B gain/amplification (p value: 1.7 × 10−5, p value: 4.2 × 10−6, respectively).
(D) Box plots indicating that the levels of AKT phosphorylation on threonine 308 (T308) (left) measured by RPPA are significantly lower in breast carcinomas with PIP4K2B gain/amplification (p value: 0.007), whereas the total AKT levels remain unchanged (p value: 0.24) Data obtained from the Stand Up to Cancer cBio portal (http://cbio.mskcc.org/su2c-portal/).
(E) Upper: box plots indicating the expression of TP53 (blue) and PIP4K2A (red) across different subsets of breast carcinomas divided by the putative DNA copy number status of TP53. (Homdel: homozygous deletion [n = 0], Hetloss: heterozygous deletion (n = 1), diploid: n = 2, gain: n ≥ 3). TP53 mRNA expression is as expected progressively higher across the different subgroups of breast carcinomas with TP53 ploidy status ranging from n = 0 to n≥3 (one-way ANOVA, p = 4.3 × 10−6), whereas PIP4K2A expression is only significantly higher in the subgroup of breast carcinomas with homozygous deletion of TP53 (one-way ANOVA, p = 0.009). Lower: box plots indicating the expression of PIP4K2B (orange) and PIP4K2C (yellow) across different subsets of breast carcinomas divided by the putative DNA copy number status of TP53. The expression of either PIP4K2B or PIP4K2C is not significantly different across the subgroups of breast carcinomas with different ploidy status of TP53 (one-way ANOVA, p > 0.05).
PI5P4K Expression in Breast Cancer
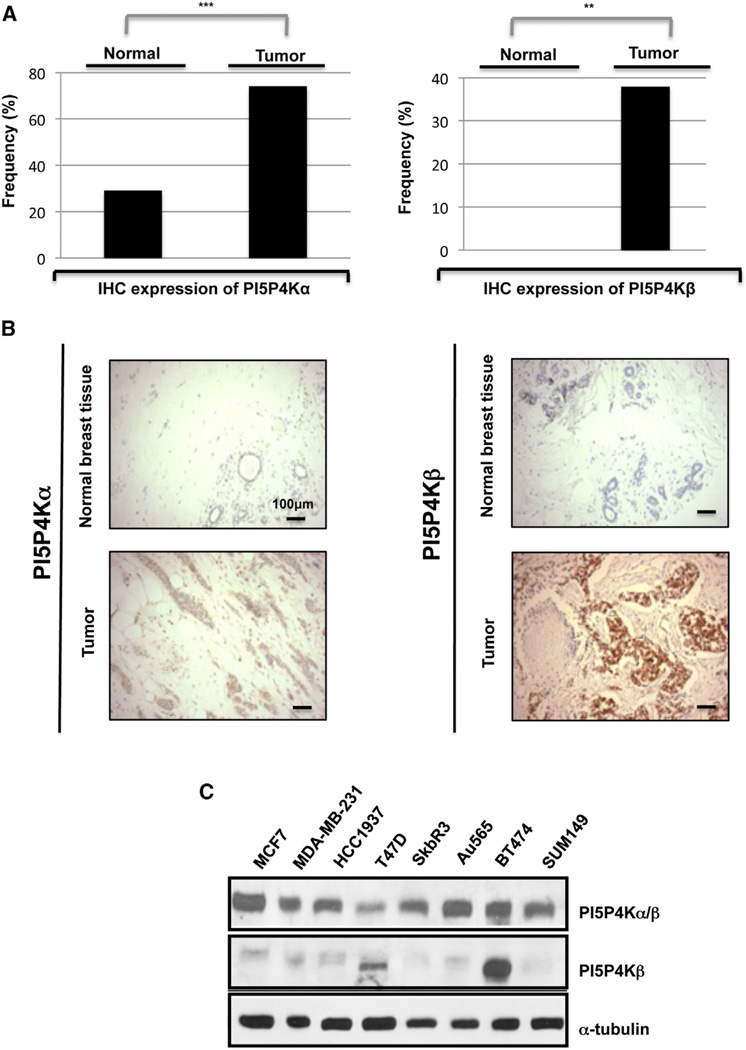
To evaluate PI5P4Kα and β protein levels in breast cancer, we utilized antibodies against these proteins for immunohistochemistry staining of a breast cancer tissue array (Figures 2A, 2B, and S1 available online; Tables S1 and S2). As shown in Figure 2, PI5P4Kα expression is detectable in both normal breast and breast cancer, but high levels of expression are found in 74% of tumors and only 29% of normal breast epithelium. This high level of expression is distributed over all the major subtypes. In contrast, PI5P4Kβ was not detected in any of the normal breast epithelial tissue but was highly expressed in 38% of the breast tumors. The subset of tumors with the highest level of expression was the HER2-positive group where 62% had high levels of PI5P4Kβ. Thus, the HER2 subtype has high protein expression, consistent with a high frequency of PI5P4Kβ gene amplification (Figure 1A).
Figure 2. PI5P4K Expression in Breast Cancer.
(A) Histograms illustrating expression levels of PI5P4Kα (***p = 0.000) or PI5P4Kβ (**p = 0.004) in breast cancer samples.
(B) Representative IHC images from breast tumor and normal samples. Scale bar, 100 µm.
(C) PI5P4Kα and PI5P4Kβ expression in panel of breast cancer cell lines.
We also evaluated the total protein expression of both isoforms in a panel of breast cancer cell lines using western blots. PI5P4Kα is expressed in all the breast cancer cell lines that we investigated, whereas PI5P4Kβ is expressed at very low levels in most breast cancer cell lines, with the exception of BT474 cells, which have both ERBB2 and PIP4K2B amplified (14 and 12 alleles, respectively) (https://cansar.icr.ac.uk/cansar/). Interestingly, the T47D cell line has four alleles of PIP4K2B and also shows some increased expression (Figure 2C). In contrast, AU565 cells have ERBB2 amplified but have only two alleles of PIP4K2B and do not express high levels of this protein (Figure 2C).
Knockdown of Both PI5P4Kα and β in BT474 Cells Abrogates Cell Proliferation and Impairs Tumor Growth in a Xenograft Model
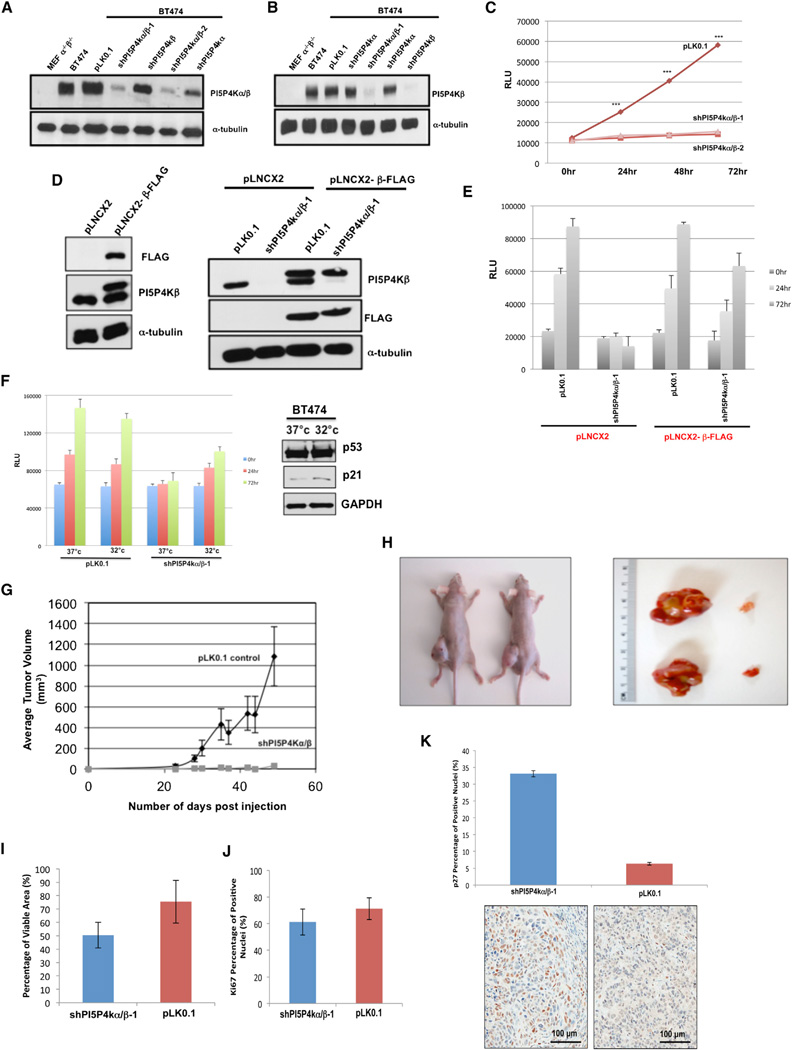
Because BT474 cells exhibit amplification of the PIP4K2B gene and express high levels of the protein encoded by this gene (PI5P4Kβ), we examined the effect of knocking down expression of either this gene alone or both PIP4K2B and PIP4K2A on cell growth. Knocking down expression of either gene alone with small hairpin RNA (shRNA) had little effect on cell growth (Figures 3 and S2; Table S3), but knocking down expression of both genes caused a dramatic inhibition of cell growth (Figure 3). Two independent sets of shRNAs targeting these proteins resulted in effective decreases in both PI5P4Kα and β (Figures 3A and 3B) and resulted in approximately 80% reduction in cell number over a 72 hr period of cell growth (Figure 3C). Importantly, to address any off target effects of the hairpins, we rescued cell proliferation of the double-knockdown line by expressing a flag-tagged version of mouse PI5P4Kβ that circumvents the shRNA directed against human PI5P4Kβ (Figures 3D and 3E).
Figure 3. Knockdown of Both PI5P4Kα and β in BT474 Cells Abrogates Cell Proliferation and Fail to Form Tumors in Xenograft Model.
(A and B) Stable knockdown of PI5P4Kα/β in BT474 cells. shPI5P4kα/β-1 (sequence 1) and shPI5P4kα/β-2 (sequence 2) are two independent hairpins targeted against PI5P4kα and PI5P4kβ. All single knockdowns are sequence 1 (see Experimental Procedures).
(C) Luminescent cell viability assay in stable PI5P4Kα/β knockdown cells. Results are from four independent experiments and are represented as the mean value ± SEM. ***p value < 0.0001 with two-tailed Student’s t test.
(D) Stable overexpression of mouse Flag-tagged PI5P4Kβ in BT474 cells and subsequent stable PI5P4Kα/β knockdown.
(E) Mouse Flag-tagged PI5P4Kβ rescues proliferation in PI5P4Kα/β knockdown cells. Results are from three independent experiments and are represented as the mean value ± SEM.
(F) Right: Luminescent cell viability assay in stable PI5P4Kα/β knockdown cells cultured at restrictive (37°C) and permissive conditions (32°C) for indicated time points. Results are from three independent experiments and are represented as the mean value ± SEM. Left: Total p53, p21, and GAPDH (loading control) protein levels in BT474 cells cultured at restrictive and permissive conditions for 24 hr.
(G) Tumor formation over time in nude mice injected with the BT474 cancer cell line expressing shRNA pLK0.1 control or shRNA PI5P4K α/β. Error bars are SEM.
(H) Images of tumors, pLK0.1 control cells (left flank), or shPI5P4Kα/β (right flank) after mice were euthanized.
(I) Histogram showing percentages of viable area in PI5P4Kα/β knockdown and control xenografts. Data from two independent experiments were considered and displayed as means ± SD.
(J) Histogram displaying expression levels of Ki67-positive cells in shPI5P4Kα/β and pLK0.1 xenografts. Quantification of Ki67-positive cells in shPI5P4Kα/β and pLK0.1 xenografts. Data from two independent experiments were considered and displayed as means ± SD.
(K) Upper: histogram illustrating expression levels of p27-positive cells in shPI5P4Kα/β and pLK0.1 xenografts. Quantification of p27-positive cells in shPI5P4Kα/β and pLK0.1 xenografts. Data from two independent experiments were considered and displayed as means ± SD (***p < 0.001). Lower: representative images of p27 immunostains are shown. Scale bar, 100 µm.
To address whether loss of p53 function contributes to the impaired growth of BT474 cells in the context of PI5P4Kα and β knockdown, we took advantage of the previous observation that the p53 E285K mutation in these cells is nonfunctional at 37°C but functional at 32°C (Dearth et al., 2007). Consistent with the previous publication, p53 activity was at least partially restored when BT474 cells were cultured at 32 degrees, as assessed by increased expression of the p53 target gene p21. Importantly, the shift to 32°C resulted in a partial rescue of proliferation (Figure 3F). As expected, the control pLKO.1 BT474 cells grew somewhat slower when shifted to 32°C. These results are consistent with a model in which knocking down PI5P4Kα and β only impairs cell growth when p53 is defective.
We also found that knocking down both PI5P4Kα and β in BT474 cells dramatically impaired tumor formation in xenografts (Figures 3G and 3H). The striking differences in tumor formation in the xenografts were due to a dramatic decrease in viable cells in the PI5P4Kα/β knockdown xenograft tumors compared to the BT474 vector control xenograft tumors (Figure 3I), along with a slight decrease in Ki67 staining (Figure 3J). Most strikingly though is the strong increase in p27 (a marker of senescence) (Young and Kaelin, 2008; Young et al., 2008) in the PI5P4K α/β knockdown xenograft tumors compared to the BT474 vector control xenograft tumors (Figure 3K). These results support a model in which the loss on PI5P4K leads to a cell-cycle-arrest phenotype in vitro and a nonviable tumor in vivo. Furthermore, these data imply that the knockdown of PI5P4K in vivo may lead to senescence.
Knockdown of Both PI5P4Kα and β in BT474 Cells Enhances PI3K Signaling, Increases ROS and Respiration, and Triggers Senescence
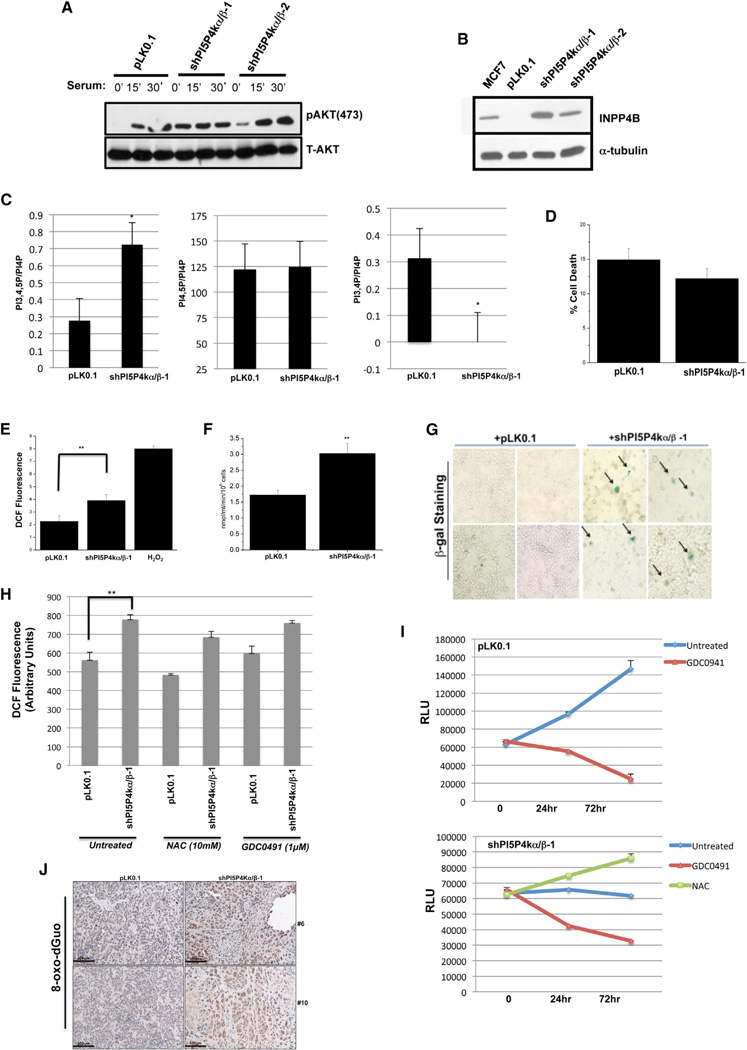
Consistent with our previous observation of increased in vivo AKT activity in response to deletion of the PIP4K2B gene in mice (Lamia et al., 2004), the knockdown of both PI5P4Kα and β resulted in increased basal AKT phosphorylation in BT474 cells (Figures 4A and S3). This increase in AKT phosphorylation could be explained by an increase in PI-3,4,5-P3 levels in response to knockdown of both PI5P4Kα and β (Figure 4C). Surprisingly, we observed a dramatic decrease in PI-3,4-P2 in the double-knockdown cells (Figure 4C). PI-3,4-P2 can be degraded to PI-3-P by the phosphatase, inositol polyphosphate 4-phosphatase-IIB (INPP4B) (Gewinner et al., 2009; Norris et al., 1995, 1997) so we investigated the level of this enzyme in the BT474 cells. The levels of INPP4B were virtually undetectable in BT474 cells prior to the knockdown of PI5P4Kα and β but were quite high following the knockdown, explaining the drop in PI-3,4-P2 levels (Figures 4B and 4C). These results suggest a negative feedback loop in which INPP4B upregulation is an attempt to suppress PI-3,4-P2 activation of AKT (Franke et al., 1997; Gewinner et al., 2009). We detected no significant change in PI-4,5-P2 levels, indicating that the majority of PI-4,5-P2 is produced by type 1 PIP kinases, as previously assumed (Figure 4C).
Figure 4. Knockdown of Both PI5P4Kα and β in BT474 Cells Enhances PI3K Signaling, Increases ROS and Respiration, and Triggers Senescence.
(A) AKT phosphorylation at serine 473(pS473) and total AKT protein levels in pLK0.1 vector control cells and in shPI5P4kα/β double-knockdown cells. Cells were serum starved overnight and then treated with media with 10% serum for the indicated time points.
(B) Total INPP4B protein levels in pLK0.1 vector control cells and in shPI5P4kα/β double-knockdown cells. The MCF7 breast cancer cell line is known to have high expression of INPP4B.
(C) BT474 pLK0.1 control vector cells or PI5P4Kα/β knockdown cells labeled with [3H]-inositol for 48 hr. Deacylated lipids were analyzed by HPLC, quantified, and normalized to PI4P levels. Results are from three independent experiments and are represented as the mean value ± SEM. *p < 0.01 with two-tailed Student’s t test.
(D) Cell death of pLK0.1 control or shRNA PI5P4K α/β double-knockdown cells was assessed by LDH release. Mean values ± SEMs from four independent experiments are shown.
(E) ROS determined by incubating pLK0.1 control or shRNA PI5P4K α/β double-knockdown cells with DCFH-DA (10 µM) for 30 min or a bolus of H2O2 (100 µM) for 15 min as positive control. n = 4 mean ± SEM. **p < 0.001 with two-tailed Student’s t test.
(F) Oxygen consumption of pLK0.1 control or shRNA PI5P4K α/β double-knockdown cells. n = 4 mean ± SEM. **p < 0.001 with two-tailed Student’s t test.
(G) pLK0.1 control or shRNA PI5P4K α/β double-knockdown cells were plated for senescence assay. Images presented from a representative experiment performed in triplicate.
(H) ROS determined by incubating pLK0.1 control or shRNA PI5P4K a/b double-knockdown cells with DCFH-DA (10 µM) for 1 hr. Cells were untreated or treated with GDC-0941 (1 µM) or N-acetylcysteine (NAC) (10 mM) for 24 hr. n = 3 mean ± SEM. **p < 0.001 with two-tailed Student’s t test.
(I) Luminescent cell viability assay in stable (upper) pLK0.1 control cells and (lower) PI5P4Kα/β knockdown cells ± GDC-0941 (1 µM) or NAC (10 mM) for indicated time points. Results are from three independent experiments and are represented as the mean value ± SEM.
(J) Immunohistochemical detection of 8-oxo-dGuo in pLK0.1 and shPI5P4Kα/β xenografts. Two representative images of 8-oxo-dGuo immunostains are shown (mouse #6 and mouse #10). Scale bar,100 µm.
See also Figure S3.
The increase in PI3K/AKT signaling, yet decrease in cell proliferation in response to knockdown of both PI5P4Kα and β, seems paradoxical given the established role of PI3K as an oncogene and in promoting cell growth and cell survival. The decrease in cell number was not due to an increased rate of cell death as judged by no change in LDH release from the cells (Figure 4D). However, we observed a significant increase in reactive oxygen species (ROS), in oxygen consumption, and in β-galactosidase staining in the BT474 cells in response to knockdown of both PI5P4Kα and β consistent with ROS-induced senescence (Figures 4E–4G).
Hyperactivation of the PI3K/AKT pathway due to loss of PTEN (and INPP4B) has previously been shown to induce senescence (Chen et al., 2005; Gewinner et al., 2009; Nogueira et al., 2008), raising the possibility that activation of AKT in response to PI5P4Kα/β knockdown is responsible for the senescence that we observed in the BT474 cells. However, BT474 cells are defective in p53, which typically mediates PI3K/AKT pathway-dependent senescence. To evaluate the possibility that the ROS and senescence observed in response to knockdown of PI5P4Kα/β is a consequence of PI3K/AKT activation, we treated these cells with the pan-PI3K inhibitor GDC-0941 to block this pathway (Figure S3). Rather than restoring growth in the PI5P4Kα/β knockdown BT474 cells, GDC-0941 caused a decrease in cell number (Figure 4I). GDC-0941 caused a small elevation in ROS in control BT474 cells and did not lower ROS levels in PI5P4Kα/β knockdown BT474 cells (Figure 4H). These data indicate that the ROS and senescence observed in PI5P4Kα/β knockdown cells is not due to activation of the PI3K/AKT pathway and raise the possibility that activation of this pathway is an attempt to rescue the cells from ROS.
In contrast to the results with BT474 cells, knocking down PI5P4Kα and β in a breast cancer cell line (MCF7 cells) that does not have PIP4K2B amplified or overexpressed and has wild-type p53 and an activating mutation in PIK3CA had no effect on growth or AKT signaling (Figure S2). Together, these data suggest that maintaining high levels of PI5P4Kα and β, in part through amplification of the PIP4K2B gene, is critical to prevent senescence in specific mutational backgrounds (e.g., HER2 amplification and p53 loss) but is not, in general, essential for the growth of cancer cell lines.
Altered Gene Expression and Metabolomic Signatures in the PI5P4Kα/β Double-Knockdown BT474 Cells
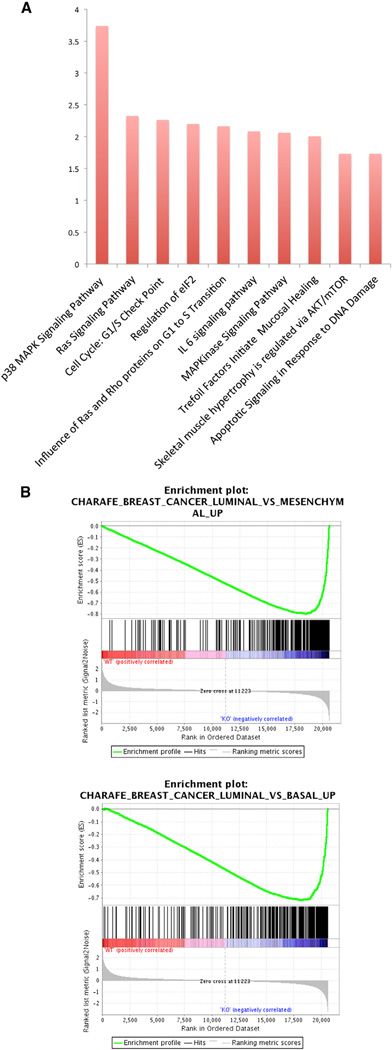
To further explore the phenotype of knocking down PI5P4Kα and β in BT474 cells, we performed a microarray analysis. As illustrated in the heatmap, there are striking differences in gene expression between the control and PI5P4Kα/β knockdown cells (Figure S4; Table S4), particularly in relation to genes involved in the p38 MAPK pathway (Figure 5A). Consistent with this, we also observe an increase in phospho-p38 MAPK in the PI5P4Kα/β knockdown cells (Figure S3). These microarray results support the idea that PI5P4Kα and β regulate cellular stress responses in the p38 MAPK pathway and corroborate previous studies indicating that p38 MAPK modulates PI-5-P levels by phosphorylating PI5P4Kβ (Jones et al., 2006). We next performed gene set enrichment analysis (GSEA) to show that the PI5P4Kα/β double-knockdown cells are significantly enriched in the luminal compared to the basal or mesenchymal-type gene expression signature (Figure 5B). We also performed gene ontology analysis of the microarray data between the control versus shPI5P4Kα/β double-knockdown cells to show that the most differentially expressed hits are genes that control the rate and direction of cellular metabolism (Figure S5; Table S5).
Figure 5. Distinct Gene Expression and Metabolomic Signatures in the PI5P4Kα/β Double-Knockdown Cells.
Expression data of PI5P4Kα/β knockdown cells (shPI5P4Kα/β) or control vector cells (pLK0.1) using Affymetrix Human Genome U133 Plus (~40,000 genes).
(A) Enrichment analysis of the curated pathways (BioCarta) following PI5P4Kα/β knockdown. The p38 MAPK and RAS signaling pathways were identified as the most significant pathways with respective p values of 1.83 × 10−4 and 0.004.
(B) Gene set enrichment analysis (GSEA) signatures highlighting coordinated differential expression of gene sets that are enriched in PI5P4Kα/β knockdown cells. The CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_UP (genes upregulated in luminal-like breast cancer cell lines compared to the mesenchymal-like ones) and CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP (genes upregulated in luminal-like breast cancer cell lines compared to the basal-like ones) were scored among the most significantly enriched gene signatures in PI5P4K knockdown cells, p < 0.001.
See also Figures S4 and S5 and Tables S4 and S5.
To better understand the effect of knocking down PI5P4Kα and β on cellular metabolism in BT474 cells, weutilized targeted mass spectrometry to examine the level of 180 metabolites. We found a significant drop in intermediates in glucose metabolism in the PI5P4Kα/β knockdown cells (Figure S5; Table S5). These results are paradoxical. In most cellular contexts, an increase in PI3K/AKT signaling results in increased glycolysis and decreased oxidative phosphorylation (the Warburg effect), but in the context of PI5P4Kα/β knockdown in BT474 cells PI3K/AKT signaling is increased but glucose metabolism is decreased and oxygen consumption is increased (reversal of Warburg effect). The increased oxygen consumption (presumably due to increased oxidative phosphorylation to maintain ATP levels at low rates of glycolysis) could explain the higher ROS levels (Figures 4E, 4F, and 4H). The metabolic imbalance and consequent high ROS levels are likely to contribute to the senescence observed.
In contrast to the effects seen in BT474 cells, knocking down PI5P4Kα/β in MCF7 cells did not cause a decrease in glucose metabolites (Figure S2; Table S3). These data suggest that it is the decreased glucose metabolism, unique to BT474 cells, that causes ROS and senescence in these cells. To further interrogate whether the ROS is responsible for the decrease in the growth of PI5P4Kα/β knockdown BT474 cells, we added the antioxidant N-acetylcysteine (NAC) to the media and found that this partially rescued the cell growth (Figure 4I). Consistent with the in vitro data, the PI5P4Kα/β knockdown xenograft tumors have higher levels of 8-hydroxydeoxyguanosine (8-oxodGuo), an indicator of ROS-dependent DNA damage, than the control BT474 tumors, indicating that oxidative stress is higher in the PI5P4Kα/β knockdown tumors (Figure 4J).
The strong correlation between high expression of PI5P4Kα or β in human breast cancers that lack both alleles of p53 and the observation that knocking down PI5P4Kα/β in a cell line that lacks p53 (BT474) results in senescence, whereas knocking down these genes in a cell line with wild-type p53 (MCF7) had no effect raises the possibility that expression of PI5P4Kα/β genes may only be essential under conditions of stress and that they may be particularly important for growth of cancers that lack p53. To explore this possibility, we generated mice with genetic deletions of PIP4K2A and crossed them into PIP4K2B−/− and TP53−/− backgrounds.
PIP4K2A−/− Mice Are Viable and Appear Normal
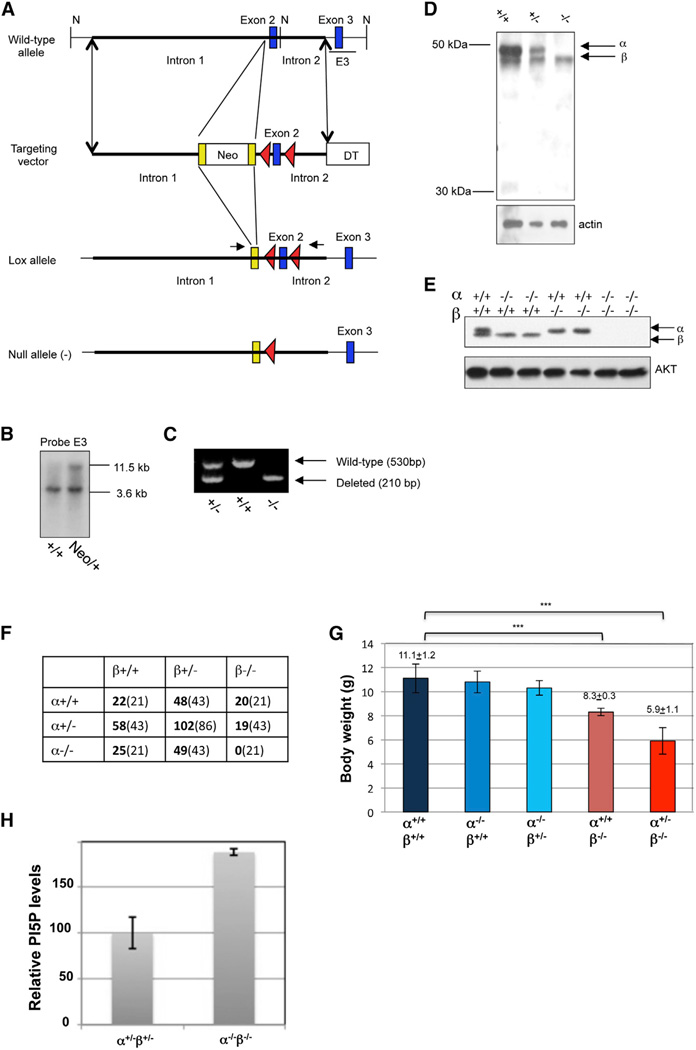
We generated mice deficient for PIP4K2A, using a conditional targeting strategy (Figure 6A). Clones were picked and analyzed for homologous recombination into the endogenous PIP4K2A locus using a combination of Southern blot (Figure 6B) and PCR analysis (Figure 6C). A representative NdeI digest, followed by Southern blot analysis of one positive and one negative neomycin resistant clone, is shown using the indicated E3 probe (Figures 6A and 6B). A representative PCR using three mice is seen in Figure 6C, indicating the expected products from germline deleted PIP4K2A−/−, PIP4K2A+/−, and PIP4K2A+/+ mice. Western blot analysis of brain tissue from PIP4K2A−/− mice using an antibody that recognizes both PI5P4Kα and β indicated that the gene product PI5P4Kα was not detected after germline deletion, nor were any truncated proteins (Figure 6D). PIP4K2A−/− mice were born in near Mendelian ratios and displayed no obvious histological, growth, or reproductive phenotypes (data not shown). Also, in contrast to PIP4K2B−/− mice that are growth retarded with decreased adiposity and increased insulin sensitivity (Lamia et al., 2004), PIP4K2A−/− mice were like wild-type littermates in regard to all these characteristics (data not shown).
Figure 6. Requirement for PIP4K2A and PIP4K2B for Survival and Growth.
(A) Schematic representation of wild-type PIP4K2A locus, targeting vector, recombined allele and FlpR/CreR-deleted null allele. Neomycin resistance cassette flanked by Frt sites (yellow boxes), exon 2 flanked by loxP sites (red arrows), and diphtheria toxin cassette located 3′ to exon 2. N, NdeI restriction site.
(B) Southern blot of embryonic stem cell clones (wild-type and targeted) using probe E3 after NdeI digestion.
(C) PCR analysis of genomic DNA derived from PIP4K2A F2 littermates.
(D) PI5P4Kα and β protein levels from brain homogenate of PIP4K2A F2 littermates.
(E) PI5P4Kα and β protein levels in primary MEFs derived from intercrossing PIP4K2A and PIP4K2B knockout mice.
(F) Genotyping results of PIP4K2A/2B double heterozygote interbreeding at weaning (observed numbers in bold, expected in parentheses).
(G) Body weight (in grams) measurements of indicated mouse genotypes. Results represented as the mean value ± SEM. ***p < 0.0001 with two-tailed Student’s t test.
(H) Quantitation of PI5P levels in PIP4K2A/2B-deficient relative to PIP4K2A/2B heterozygous MEFs. Results are from three independent experiments and are represented as the mean value ± SEM.
See also Figure S6.
PIP4K2A−/− PIP4K2B−/− Mice Develop Normally, but Exhibit Neonatal Lethality
The absence of a significant phenotype for the PIP4K2A−/− mice led us to breed these mice to the PIP4K2B−/− mice in order to generate PIP4K2A−/−, PIP4K2B−/− mice. The cross and backcross of PIP4K2A+/− mice to PIP4K2B+/− mice resulted in non-Mendelian ratios of offspring (Figure 6F). Most notably, no pups of the PIP4K2A−/− PIP4K2B−/− genotype were viable: these pups looked normal at birth and were able to suckle but died within 12 hr. There was also a sub-Mendelian ratio of viable mice of the PIP4K2A+/−, PIP4K2B−/− genotype (19 out of 43 expected). These mice were born at Mendelian ratios with normal appearance and weight, but approximately half died within 12 hr. The survivors were growth retarded with only 50% the weight of wild-type littermates at the adult stage (Figure 6G). All other genotypes appeared at Mendelian ratios. No histological abnormalities were observed in any of the genotypes. These observations indicate that type II PI5P4Kα and β are not essential for normal embryonic development but are critical for surviving the stresses of birth and contribute to growth following birth. Consistent with normal embryonic development, PIP4K2A−/− PIP4K2B−/− mouse embryonic fibroblasts (MEFs) grow at a normal rate (data not shown). Also, consistent with enhanced PI3K/AKT signaling in the PI5P4Kα/β knockdown BT474 cell lines (above) and in muscle from PIP4K2B−/− mice (Lamia et al., 2004), introduction of cre recombinase to PIP4K2Aflx/− PIP4K2B−/− MEFs to delete the second allele of PIP4K2A resulted in prolonged AKT phosphorylation at Thr308 (Figure S6). Conversely, overexpressing PIP4K2A in the same background suppresses Thr308 phosphorylation (Figure S6). Consistent with PI-5-P being the substrate of the PI5P4Ks, the PIP4K2A−/−, PIP4K2B−/− MEFs had higher levels of PI-5-P than the PIP4K2A+/−, PIP4K2B+/− MEFs derived from littermates (Figure 6H). In contrast to the BT474 cells where knockdown of these genes caused senescence, no senescence was observed in the PIP4K2A−/−, PIP4K2B−/− MEFs.
PI5P4K Deficiency Reduces Tumor-Dependent Death in TP53−/− Mice
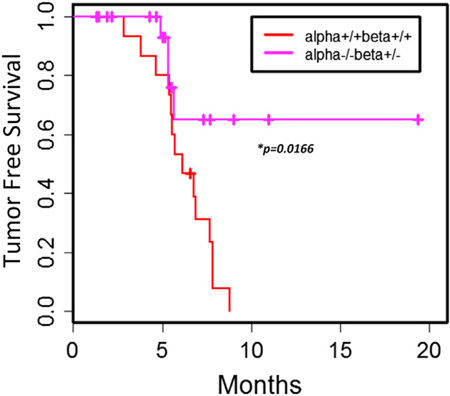
Previous studies have indicated a major role for p53 in mediating cellular responses to stress, especially metabolic stress and ROS stress (Vurusaner et al., 2012). Our observation of increased metabolic stress and ROS, leading to senescence upon knockdown of PI5P4Kα and β in a cell line lacking p53 (BT474) but no induction of senescence in MEFs in the context of deletion of PIP4K2A and PIP4K2B, raised the possibility that partial loss of PIP4K2A and/or PIP4K2B alleles, although viable in normal tissues, may result in senescence in the context of p53-deleted tumors. To test this idea, we crossed the PIP4K2A−/−, PIP4K2B+/− mice with TP53−/− mice and monitored tumor formation in all the viable mice that emerged from the backcrosses. In approximately 4–6 months, mice deficient in p53 develop spontaneous tumors, with the majority being lymphomas and soft tissue sarcomas (Jacks et al., 1994). In contrast, we find that PIP4K2A−/−, PIP4K2B+/−, TP53−/− mice that emerged from the crosses had a dramatic reduction in tumors compared to PIP4K2A+/+, PIP4K2B+/+, TP53−/− mice (Figures 7 and S7).
Figure 7. PI5P4K Deficiency Restricts Tumor Death after p53 Deletion.
Kaplan-Meier plot analysis of tumor free survival (15 PI5P4Kα+/+ PI5P4Kβ+/+ and 20 PI5P4Kα −/− PI5P4Kβ+/−). *p < 0.05 with two-tailed Student’s t test. See also Figure S7.
Deletion of PIP4K2B and TP53 Results in Synthetic Lethality
Although PIP4K2B−/− mice are viable and appear at Mendelian ratios and have a normal lifespan in TP53 wild-type backgrounds (Lamia et al., 2004), no mice with the PIP4K2B−/−, TP53−/− genotype emerged from the crosses, indicating synthetic lethality upon loss of TP53 and PIP4K2B. In order to investigate why PIP4K2B−/−, TP53−/− mice are not viable we set up timed mating pairs and at E12.5, we were able to get one double-knockout embryo out of 11 from three litters. Deletion of both alleles of the PIP4K2B and TP53 genes caused exencephaly (data not shown), a failure of neural tube closure that is occasionally observed in TP53−/− mice, but that does not usually cause early embryonic lethality (Jacks et al., 1994). In contrast, loss of only one allele of PIP4K2B and both alleles of PIP4K2A in the context of p53 deficiency results in viable mice but an apparent synthetic lethality for the TP53−/− tumors.
DISCUSSION
Here, we have shown that the PIP4K2B gene is amplified in a subset of tumors, especially in HER2-positive breast tumors, and that, although this gene is often in the same amplicon with ERBB2, in many tumors it is in a separate amplicon, and in some tumors it is amplified independent of ERBB2 amplification. Importantly, we have found that the protein product of this gene, PI5P4Kβ, is highly expressed in a majority of HER2-positive tumors. The related enzyme, PI5P4Kα, which is more broadly expressed, was also found to be elevated in many breast tumors. We showed that BT474 cells, which exhibit amplification of both ERBB2 and PIP4K2B and loss of p53, express high levels of both PI5P4Kα and β and that knocking down the expression of these proteins results in impaired glucose metabolism, increased ROS, and senescence. While this paper was in review, Jones et al. (2013) was published, demonstrating that addition of peroxide to cells increases PI-5-P levels (especially in cells lacking p53). Furthermore, overexpression of PI5P4Kα prevented the elevation in PI-5-P and also rescued cell growth in the presence of ROS providing independent evidence of a role for PI5P4Ks in rescuing cells from ROS toxicity.
The biochemical mechanism by which PI5P4Ks protect from ROS in the context of p53 deletion is not clear. Our studies suggest that high levels of PI5P4Ks are required to maintain high rates of glucose metabolism, thereby reducing rates of ROS production from oxidative phosphorylation and enhancing NADPH production via the pentose phosphate pathway. The role of PI5P4Ks in maintaining glucose metabolism appears to only be critical in the context of p53 loss. Cell lines (e.g., MCF7 cells and MEFs) with wild-type p53 do not show defects in glucose metabolism, enhanced ROS, or senescence upon loss of PI5P4Ks, and muscle tissue shows enhanced glucose uptake upon loss of PI5P4Kβ (Lamia et al., 2004). These results suggest that, in the absence of PI5P4Ks, p53 mediates an adaptive response to ROS, by maintaining glucose uptake and flux into the pentose phosphate pathway. Indeed, several studies have indicated an important role for p53 in maintaining glucose and ROS homeostasis, in part via TIGAR (Mor et al., 2011). Our studies support this concept and argue that when both p53 and PI5P4Ks are absent cells are not capable of maintaining glucose and ROS homeostasis. Interestingly, some recent studies also indicate a role for p53 in enhancing ROS in some mutational backgrounds (for review, see Vurusaner et al., 2012).
Jones et al. (2013) have proposed that the critical role of PI5P4Ks is to suppress the high levels of PI-5-P that appear in response to ROS, implying that PI-5-P is a second messenger that mediates or enhances ROS-dependent cell damage. An alternative possibility, consistent with the data presented in this article, is that PI5P4Ks, when presented with high levels of PI-5-P (due to ROS), generate PI-4,5-P2 at an intracellular location that facilitates cellular responses that enhance glucose metabolism and suppress ROS.
The fact that knocking down PI5P4Ks both activates the PI3K/AKT pathway and suppresses glucose metabolism (in p53-deficient BT474 cells) is paradoxical. In other contexts, activation of the PI3K/AKT pathway enhances glucose uptake and metabolism, and, in the context of wild-type p53, deletion of PIP4K2B in mice activates the PI3K/AKT pathway and enhances glucose uptake into muscle (Lamia et al., 2004). The data presented here, using the pan PI3K inhibitor GDC0941, show that the activation of the PI3K/AKT pathway is not responsible for the ROS and senescence in BT474 cells. It is likely that the PI5P4Ks suppress PI3K/AKT signaling because they provide an alternative mechanism for enhancing glucose metabolism in response to ROS. The activation of PI3K/AKT signaling upon suppressing PI5P4Ks is clearly not sufficient to restore glucose and ROS homeostasis when p53 is not present.
The decreased tumor incidence in the background of PIP4K2A−/−, PIP4K2B+/−, TP53−/− compared to TP53−/− alone is particularly interesting with respect to Li-Fraumeni syndrome (germline TP53 mutations). Our results indicate that expression of PI5P4Kα and/or β is critical for the growth of tumors with TP53 mutations or deletions. Thus, coamplification of PIP4K2B with ERBB2 might explain why breast cancers in patients with Li-Fraumeni syndrome show ERBB2 amplifications (HER2 positive) in over 83% of cases as opposed to 16% of age-matched patients with wild-type TP53 (Wilson et al., 2010).
The results that we present here suggest that PI5P4Kα and β play a critical role in mediating changes in metabolism in response to stress, and, in particular, ROS stress that occurs in the absence of p53. Germline deletion of either PIP4K2A or PIP4K2B alone resulted in mice with normal lifespans, and germline deletion of both PIP4K2A and PIP4K2B resulted in full-term embryos of normal size and appearance at birth, indicating that these genes do not play a major role in normal embryonic growth and development. Yet, the PIP4K2A−/−, PIP4K2B−/− pups die shortly after birth, consistent with these genes having a role in mediating stress responses known to occur following birth. Importantly, germline deletion of both PIP4K2B and TP53 resulted in lethality, whereas germline deletion of either gene alone resulted in Mendelian ratios of viable pups. Thus, the genetic studies suggest that TP53 and PIP4K2B have overlapping roles in mediating cellular responses to stress and that, whereas neither gene alone is essential, loss of both genes is not tolerated.
The most exciting observation from these studies in regard to potential new therapies for p53 mutant tumors is that germline deletion of both alleles of PIP4K2A and one allele of PIP4K2B in the context of TP53−/− results in a viable mouse with a dramatic reduction in tumor-dependent death compared to TP53−/− mice that are wild-type for PIP4K2A and B. These results (and studies of the PIP4K2A−/−, PIP4K2B+/− or PIP4K2A+/−, PIP4K2B−/− mice in the context of wild-type TP53) indicate that normal tissues tolerate well the loss of three out of four alleles of the PIP4K2A and PIP4K2B genes, but that tumors are not viable in this context. PI5P4Kα and β are kinases and pharmaceutical companies have shown that it is possible to develop highly specific inhibitors of both protein kinases and lipid kinases. The synthetic lethality that we observe between TP53 loss and loss of these kinases indicates that drugs that target either the enzyme PI5P4Kβ alone or that target both PI5P4Kα and β are likely to be well tolerated and very effective on tumors that have loss of function mutations or deletions of TP53. Our observations with BT474 cells suggest that HER2-positive tumors that have amplifications of PIP4K2B and mutations in TP53 may be particularly sensitive to PI5P4Kα,β inhibitors.
The ERBB2 (Her2) amplicon on chromosome 17 is variable in size and can contain a number of cancer-related genes in addition to the ERBB2 locus (Figure 1A). Clinically, patients who have tumors with small amplicons confined to the ERBB2 locus have the greatest benefit from ERBB2-directed therapies such as Trastuzumab, whereas tumors with wider ERBB2 amplicons have poor responses, suggesting coamplification of genes that contribute to Trastuzumab resistance (Morrison et al., 2007). PIP4K2B may be a candidate for an adjacent coamplified gene that confers Trastuzumab resistance, and, conversely, concomitant inhibition of ERBB2 and PIP4K2B could be a highly effective treatment option for ERBB2 (Her2)-positive tumors that are p53 mutant and PIP4K2B amplified.
EXPERIMENTAL PROCEDURES
Cell Culture
All cells were incubated in a 37°C or 32°C humidified incubator with 5% CO2. All breast cancer cell lines were obtained from ATCC and were cultured in Dulbecco’s modified Eagle medium (Mediatec). MEFs were cultured in Dulbecco’s modified Eagle medium (Mediatech). All the media was supplemented with 10% fetal bovine serum (Gemini Bio-Products), 100 U/ml penicillin/streptomycin (Mediatech). Mouse embryonic fibroblasts (MEFs) were isolated from E13.5 embryos as described previously (Hurov et al., 2001).
Virus Production and Infection
293T packaging cell line was used for lentiviral amplification, and all lentiviral infections were carried out as previously described (Moffat et al., 2006). In brief, viruses were collected 48 hr after infection, filtered, and used for infecting cells in the presence of 8 µg/ml polybrene prior to puromycin selection. All lentiviral vectors were obtained from Broad Institute TRC shRNA library. PI5P4Kα pLK0.1 shRNA sequence 1 is TRCN0000006009 and sequence 2 is TRCN0000006010. PI5P4Kβ pLK0.1 shRNA sequence 1 is TRCN0000006013, and sequence 2 is TRCN0000006017. The pLK0.1 vector was used as the control. To generate virus using the above-described transfer vector, we used pMD2.G (Addgene plasmid 12259) and psPAX2 (Addgene plasmid 12260). Mouse PI5P4Kβ (GenBank number BC047282) was cloned into the pLNCX2 retroviral vector along with a Kozak sequence and a FLAG tag at the N terminus of the cDNA using in-fusion cloning reagents and the manufacturer’s cloning protocol (Clontech Laboratories). BT474 were infected and stable lines were selected with 1 mg/ml of G418 for 2 weeks. Post-G418 selection, knockdown of PI5P4Kα and PI5P4Kβ were performed as above. BT474 cells were selected with 2 µg/ml of puromycin.
Immunoblot Analysis and Antibodies
Total cell lysates were prepared by washing cells with cold phosphate-buffered saline, and then the cells were lysed with buffer containing 20 mM Tris/HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, and 1% Triton as well as protease and phosphatase inhibitors. Protein was measured using the Bradford assay (Bio-Rad), and at least 50 µg of total cell lysates was run on a SDS-polyacrylamide gel electrophoresis. The proteins were transferred on to a nitrocellulose membrane, and membranes were probed overnight at 4°C with the appropriate primary antibody. Antibodies used were as follows: PI5P4Kα/β (Schulze et al., 2006), AKT (Santa Cruz Biotechnology), α-tubulin (Sigma), GAPDH (Abcam), PI5P4Kβ, phosphoAKT-473, phosphoAKT-308, phosphoP-RAS40, phosphoERK, phospho-p38MAPK, ERK, p38MAPK, INPP4B, and p21 (Cell Signaling Technology). See also Extended Experimental Procedures.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rodrick Bronson and the entire HMS Rodent Histopathology Core for technical help with the mouse histopathology and discussions concerning the project. Also, we would like to thank Jared Johnson for cloning expertise and the Nikon Imaging Center at Harvard Medical School for help with light microscopy. This work was supported by NIH grant R01 GM041890 to L.C.C., DF/HCC Career Development Award to B.M.E., and by a Stand Up to Cancer Dream Team Translational Research Grant, a Program of the Entertainment Industry Foundation (SU2C-AACR-DT0209). G.P. is a Pfizer Fellow of the Life Sciences Research Foundation. NIH DK R01-63219 supports L.E.R.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Extended Experimental Procedures, seven figures, and five tables and can be found with this article online at http://dx.doi.org/10.1016/j.cell.2013.09.057.
REFERENCES
- Bardeesy N, Sinha M, Hezel AF, Signoretti S, Hathaway NA, Sharpless NE, Loda M, Carrasco DR, DePinho RA. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature. 2002;419:162–167. doi: 10.1038/nature01045. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Chen Z, Trotman LC, Shaffer D, Lin HK, Dotan ZA, Niki M, Koutcher JA, Scher HI, Ludwig T, Gerald W, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearth LR, Qian H, Wang T, Baroni TE, Zeng J, Chen SW, Yi SY, Brachmann RK. Inactive full-length p53 mutants lacking dominant wild-type p53 inhibition highlight loss of heterozygosity as an important aspect of p53 status in human cancers. Carcinogenesis. 2007;28:289–298. doi: 10.1093/carcin/bgl132. [DOI] [PubMed] [Google Scholar]
- Franke TF, Kaplan DR, Cantley LC, Toker A. Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science. 1997;275:665–668. doi: 10.1126/science.275.5300.665. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu. Rev. Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemad-moghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, et al. Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling. Cancer Cell. 2009;16:115–125. doi: 10.1016/j.ccr.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurov JB, Stappenbeck TS, Zmasek CM, White LS, Ranganath SH, Russell JH, Chan AC, Murphy KM, Piwnica-Worms H. Immune system dysfunction and autoimmune disease in mice lacking Emk (Par-1) protein kinase. Mol. Cell. Biol. 2001;21:3206–3219. doi: 10.1128/MCB.21.9.3206-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA. Tumor spectrum analysis in p53-mutant mice. Curr. Biol. 1994;4:1–7. doi: 10.1016/s0960-9822(00)00002-6. [DOI] [PubMed] [Google Scholar]
- Jones DR, Bultsma Y, Keune WJ, Halstead JR, Elouarrat D, Mohammed S, Heck AJ, D’Santos CS, Divecha N. Nuclear PtdIns5P as a transducer of stress signaling: an in vivo role for PIP4Kbeta. Mol. Cell. 2006;23:685–695. doi: 10.1016/j.molcel.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Jones DR, Foulger R, Keune WJ, Bultsma Y, Divecha N. PtdIns5P is an oxidative stress-induced second messenger that regulates PKB activation. FASEB J. 2013;27:1644–1656. doi: 10.1096/fj.12-218842. [DOI] [PubMed] [Google Scholar]
- Lamia KA, Peroni OD, Kim YB, Rameh LE, Kahn BB, Cantley LC. Increased insulin sensitivity and reduced adiposity in phosphatidylinositol 5-phosphate 4-kinase beta−/− mice. Mol. Cell. Biol. 2004;24:5080–5087. doi: 10.1128/MCB.24.11.5080-5087.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoh SW, Venkatesan N, Tripathi R. Overexpression of the amplified Pip4k2beta gene from 17q11-12 in breast cancer cells confers proliferation advantage. Oncogene. 2004;23:1354–1363. doi: 10.1038/sj.onc.1207251. [DOI] [PubMed] [Google Scholar]
- McClintock DS, Santore MT, Lee VY, Brunelle J, Budinger GR, Zong WX, Thompson CB, Hay N, Chandel NS. Bcl-2 family members and functional electron transport chain regulate oxygen deprivation-induced cell death. Mol. Cell. Biol. 2002;22:94–104. doi: 10.1128/MCB.22.1.94-104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffat J, Grueneberg DA, Yang X, Kim SY, Kloepfer AM, Hinkle G, Piqani B, Eisenhaure TM, Luo B, Grenier JK, et al. A lentiviral RNAi library for human and mouse genes applied to an arrayed viral high-content screen. Cell. 2006;124:1283–1298. doi: 10.1016/j.cell.2006.01.040. [DOI] [PubMed] [Google Scholar]
- Mor I, Cheung EC, Vousden KH. Control of glycolysis through regulation of PFK1: old friends and recent additions. Cold Spring Harb. Symp. Quant. Biol. 2011;76:211–216. doi: 10.1101/sqb.2011.76.010868. Published online Nov 17, 2011. http://dx.doi.org/10.1101/sqb.2011.76.010868. [DOI] [PubMed] [Google Scholar]
- Morrison LE, Jewell SS, Usha L, Blondin BA, Rao RD, Tabesh B, Kemper M, Batus M, Coon JS. Effects of ERBB2 amplicon size and genomic alterations of chromosomes 1, 3, and 10 on patient response to trastuzumab in metastatic breast cancer. Genes Chromosomes Cancer. 2007;46:397–405. doi: 10.1002/gcc.20419. [DOI] [PubMed] [Google Scholar]
- Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol. Syst. Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira V, Park Y, Chen CC, Xu PZ, Chen ML, Tonic I, Unterman T, Hay N. Akt determines replicative senescence and oxidative or oncogenic premature senescence and sensitizes cells to oxidative apoptosis. Cancer Cell. 2008;14:458–470. doi: 10.1016/j.ccr.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris FA, Atkins RC, Majerus PW. The cDNA cloning and characterization of inositol polyphosphate 4-phosphatase type II. Evidence for conserved alternative splicing in the 4-phosphatase family. J. Biol. Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- Norris FA, Auethavekiat V, Majerus PW. The isolation and characterization of cDNA encoding human and rat brain inositol polyphosphate 4-phosphatase. J. Biol. Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Cantley LC. The role of phosphoinositide 3-kinase lipid products in cell function. J. Biol. Chem. 1999;274:8347–8350. doi: 10.1074/jbc.274.13.8347. [DOI] [PubMed] [Google Scholar]
- Rameh LE, Tolias KF, Duckworth BC, Cantley LC. A new pathway for synthesis of phosphatidylinositol-4,5-bisphosphate. Nature. 1997;390:192–196. doi: 10.1038/36621. [DOI] [PubMed] [Google Scholar]
- Sarkes D, Rameh LE. A novel HPLC-based approach makes possible the spatial characterization of cellular PtdIns5P and other phosphoinositides. Biochem J. 2010;428:375–384. doi: 10.1042/BJ20100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr. Biol. 2003;13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- Schulze H, Korpal M, Hurov J, Kim SW, Zhang J, Cantley LC, Graf T, Shivdasani RA. Characterization of the megakaryocyte demarcation membrane system and its role in thrombopoiesis. Blood. 2006;107:3868–3875. doi: 10.1182/blood-2005-07-2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serunian LA, Auger KR, Cantley LC. Identification and quantification of polyphosphoinositides produced in response to platelet-derived growth factor stimulation. Methods Enzymol. 1991;198:78–87. doi: 10.1016/0076-6879(91)98010-4. [DOI] [PubMed] [Google Scholar]
- Slamon DJ. Proto-oncogenes and human cancers. N. Engl. J. Med. 1987;317:955–957. doi: 10.1056/NEJM198710083171509. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Godolphin W, Jones LA, Holt JA, Wong SG, Keith DE, Levin WJ, Stuart SG, Udove J, Ullrich A, et al. Studies of the HER-2/neu proto-oncogene in human breast and ovarian cancer. Science. 1989;244:707–712. doi: 10.1126/science.2470152. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N. Engl. J. Med. 2001;344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- Vurusaner B, Poli G, Basaga H. Tumor suppressor genes and ROS: complex networks of interactions. Free Radic. Biol. Med. 2012;52:7–18. doi: 10.1016/j.freeradbiomed.2011.09.035. [DOI] [PubMed] [Google Scholar]
- Walker DM, Urbé S, Dove SK, Tenza D, Raposo G, Clague MJ. Characterization of MTMR3. an inositol lipid 3-phosphatase with novel substrate specificity. Curr. Biol. 2001;11:1600–1605. doi: 10.1016/s0960-9822(01)00501-2. [DOI] [PubMed] [Google Scholar]
- Wilson JR, Bateman AC, Hanson H, An Q, Evans G, Rahman N, Jones JL, Eccles DM. A novel HER2-positive breast cancer phenotype arising from germline TP53 mutations. J. Med. Genet. 2010;47:771–774. doi: 10.1136/jmg.2010.078113. [DOI] [PubMed] [Google Scholar]
- Young AP, Kaelin WG., Jr Senescence triggered by the loss of the VHL tumor suppressor. Cell Cycle. 2008;7:1709–1712. doi: 10.4161/cc.7.12.6124. [DOI] [PubMed] [Google Scholar]
- Young AP, Schlisio S, Minamishima YA, Zhang Q, Li L, Grisanzio C, Signoretti S, Kaelin WG., Jr VHL loss actuates a HIF-independent senescence programme mediated by Rb and p400. Nat. Cell Biol. 2008;10:361–369. doi: 10.1038/ncb1699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.