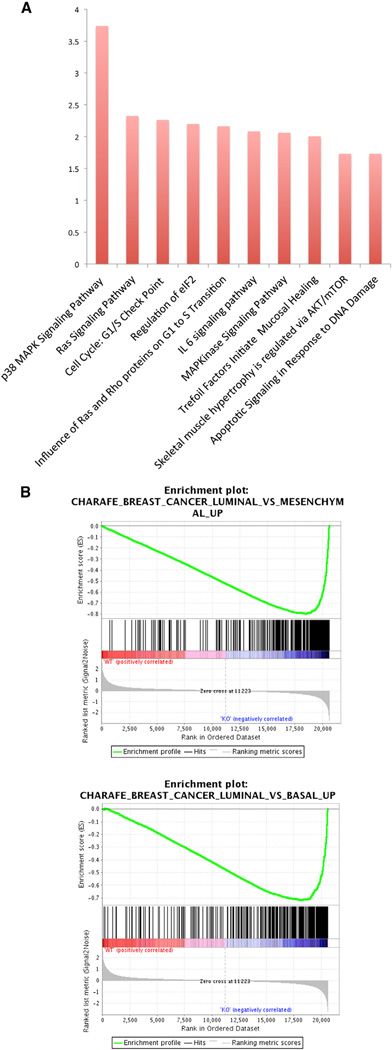
Figure 5. Distinct Gene Expression and Metabolomic Signatures in the PI5P4Kα/β Double-Knockdown Cells.
Expression data of PI5P4Kα/β knockdown cells (shPI5P4Kα/β) or control vector cells (pLK0.1) using Affymetrix Human Genome U133 Plus (~40,000 genes).
(A) Enrichment analysis of the curated pathways (BioCarta) following PI5P4Kα/β knockdown. The p38 MAPK and RAS signaling pathways were identified as the most significant pathways with respective p values of 1.83 × 10−4 and 0.004.
(B) Gene set enrichment analysis (GSEA) signatures highlighting coordinated differential expression of gene sets that are enriched in PI5P4Kα/β knockdown cells. The CHARAFE_BREAST_CANCER_LUMINAL_VS_MESENCHYMAL_UP (genes upregulated in luminal-like breast cancer cell lines compared to the mesenchymal-like ones) and CHARAFE_BREAST_CANCER_LUMINAL_VS_BASAL_UP (genes upregulated in luminal-like breast cancer cell lines compared to the basal-like ones) were scored among the most significantly enriched gene signatures in PI5P4K knockdown cells, p < 0.001.
See also Figures S4 and S5 and Tables S4 and S5.