Abstract
The first eukaryotic NER factor that recognizes NER substrates is the heterodimeric XPC-RAD23B protein. The currently accepted hypothesis is that this protein recognizes the distortions/destabilization caused by DNA lesions rather than the lesions themselves. The resulting XPC-RAD23B -DNA complexes serve as scaffolds for the recruitment of subsequent NER factors that lead to the excision of the oligonucleotide sequences containing the lesions. Based on several well-known examples of DNA lesions like the UV radiation-induced CPD and 6-4 photodimers, as well as cisplatin-derived intrastrand cross-linked lesions, it is generally believed that the differences in excision activities in human cell extracts is correlated with the binding affinities of XPC-RAD23B to these DNA lesions. However, using electrophoretic mobility shift assays, we have found that XPC-RAD23B binding affinities of certain bulky lesions derived from metabolically activated polycyclic aromatic hydrocarbon compounds such as benzo[a]pyrene and dibenzo[a,l]pyrene, are not directly, or necessarily correlated with NER excision activities observed in cell-free extracts. These findings point to features of XPC-RAD23B-bulky DNA adduct complexes that may involve the formation of NER-productive or unproductive forms of binding that depend on the structural and stereochemical properties of the DNA adducts studied. The pronounced differences in NER cleavage efficiencies observed in cell-free extracts may be due to differences in the successful recruitment of subsequent NER factors by the XPC-RAD23B-DNA adduct complexes, and/or in the verification step. These phenomena appear to depend on the structural and conformational properties of the class of bulky DNA adducts studied.
Keywords: nucleotide excision repair (NER); XPC-RAD23B binding; DNA adduct; electrophoretic mobility shift assay; benzo[a]pyrene; dibenzo[a,l]pyrene
INTRODUCTION
The nucleotide excision repair (NER) system in both prokaryotes and eukaryotes can remove a wide range of chemically and conformationally diverse DNA adducts and is a multi-step process that involves multiple proteins, nearly 30 in eukaryotes [1–3]. The DNA adducts are recognized by two different mechanisms, transcription-coupled repair and global genomic repair. In transcription-coupled repair, DNA damage stalls transcription catalyzed by RNA polymerase, which leads to the recruitment of NER factors that ultimately results in the removal of the damage and regeneration of intact DNA [4, 5].
In global genomic repair in mammals, the adducts are recognized by the heterodimeric XPC-RAD23B protein [6]. At the chromatin level in vivo, the UV-induced photolesions are first recognized by the DDB2-DDB1 heterodimer that recruits the XPC-RAD23B -centrin 2 complex to the UV photolesions [7, 8]. However, neither DDB2-DDB1 nor centrin 2 are required for the recognition of DNA adducts in cell extracts in vitro. The binding of the XPC-RAD23B heterodimer to the damaged DNA is the initial event [9, 10] that triggers the ensuing stepwise cascade of NER steps [11–15]. In order for the full processing of the DNA adducts to occur, it is believed that a two-step (bipartite) mechanism is required: (1) recognition by XPC-RAD23B and (2) the subsequent verification step that involves the helicase activity of XPD in TFIIH, the next multi-protein NER factor that binds to the XPC-RAD23B -DNA complex. Based on biochemical [16, 17] and X-ray crystallographic [18] experiments, it is now accepted that XPC-RAD23B recognizes the distortions/destabilizations of the DNA duplex caused by DNA adducts or lesions, and binds to the unmodified complementary strand rather than the lesioned strand. In this manner, the NER apparatus is capable of removing an astoundingly wide variety of DNA adducts that vary in size and shape and the site of attachment to the nucleotides[1, 2]. However, the efficiency of removing structurally different adducts can vary by two orders of magnitude, or more [15] and the origins of resistance of DNA lesions to NER are poorly understood. The relative susceptibilities of different bulky DNA adducts (termed here ‘NER cleavage efficiencies’) to excision by the human and prokaryotic NER systems in cell extracts is markedly dependent on their topological features, the nature of the adducted nucleobase, and the sequence context [19–21].
In this work, we compared the binding of XPC-RAD23B to structurally and conformationally different bulky DNA adducts and, using electrophoretic mobility shift assays (EMSA), compared the binding to the dual incisions of the same adducts catalyzed by the human NER systems in HeLa cell extracts in vitro. No direct correlation between the extent of XPC-RAD23B binding and NER cleavage efficiencies was found in the case of the six structurally or stereochemically different DNA adducts embedded in the same sequence context (Figure 1). The ramifications and significance of these findings is discussed.
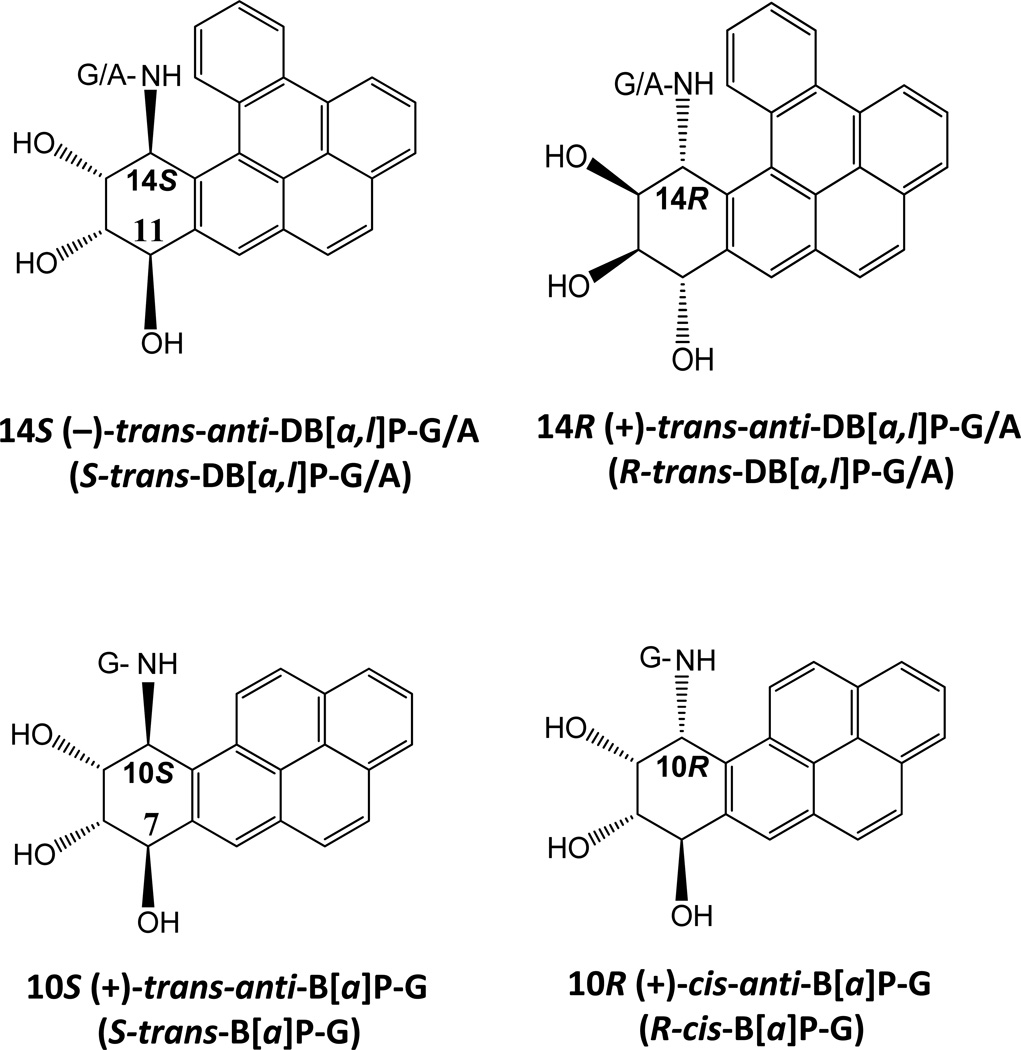
Figure 1.
Chemical structures of DNA adducts investigated.
The DNA adducts selected for study
We have previously investigated the cleavage efficiencies of DNA substrates containing different bulky adducts catalyzed by the human HeLa cell extracts in vitro [19, 21–23]. These adducts were derived from the metabolic activation of polycyclic aromatic hydrocarbons (PAH) benzo[a]pyrene (B[a]P) and dibenzo[a,l]pyrene (DB[a,l]P) to highly reactive diol epoxide intermediates that bind to the exocyclic amino groups of guanine and adenine in DNA, and form a variety of stereoisomeric adducts [24–28]. The focus of this contribution is on the stereoisomeric pairs of DNA adducts derived from the reactions of the mutagenic and tumorigenic metabolites of B[a]P and DB[a,l]P, (+)-7R,8S-dihydrodiol-9S,10R-epoxy-7,8,9,10-tetrahydro-B[a]P (B[a]PDE) and (−)-11R,12S-dihydrodiol-13S,14R-epoxy-7,8,9,10-tetrahydro-DB[a,l]P (DB[a,l]PDE) (and its mirror-image (+)-enantiomer), respectively. In general, the conformations of such DNA adducts depend markedly on their stereochemical and structural properties [29]. Here, we compared XPC-RAD23B binding to three sets of stereoisomerically different adducts, all with different DNA adduct conformations.
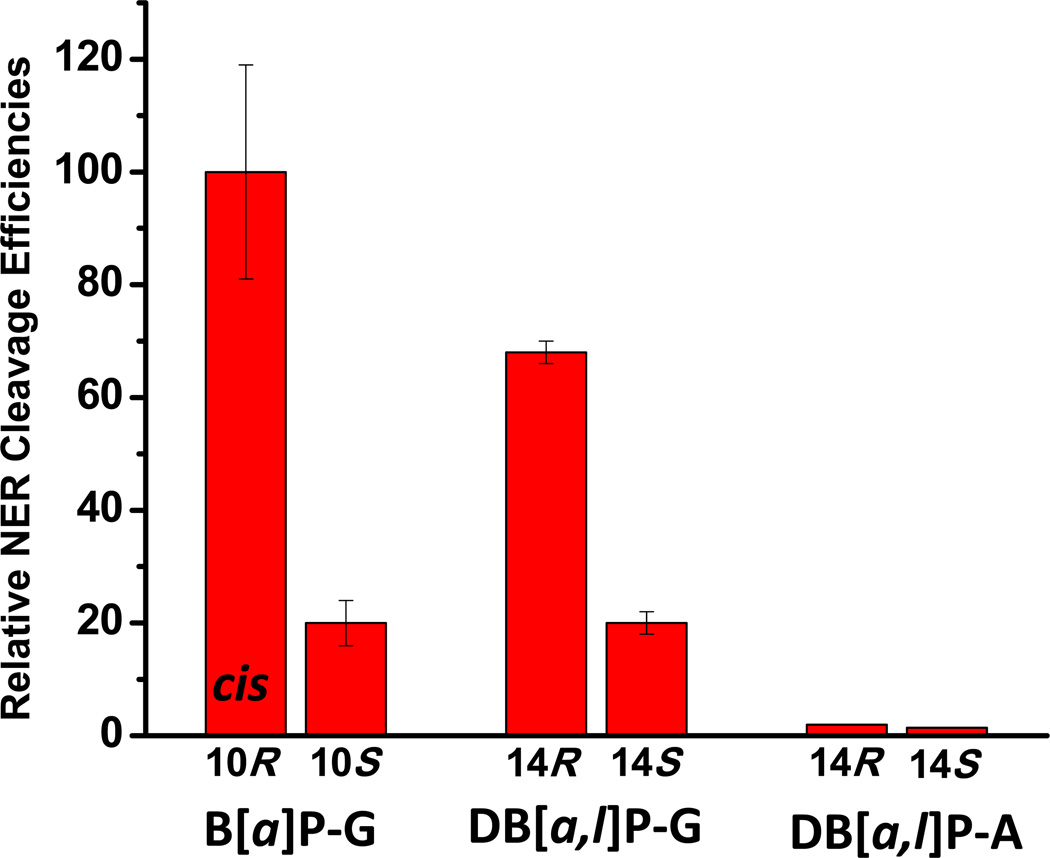
trans- and cis-B[a]P-G adducts. The cis- or trans-addition of the epoxide group of (+)-B[a]PDE at its 10-position to N2-guanine leads to the pair of stereoisomeric trans- and cis-B[a]P-G adducts shown in Figure 1. The structural properties of these two adducts in double-stranded DNA is very different. In the case of the 10S (+)-trans-anti-B[a]P-G (S-trans-B[a]P) adduct, the bulky PAH ring system is positioned in the minor groove on the 5’-side of the modified guanine residue ; however, in the 10R (+)-cis-anti-B[a]P-G (R-cis-B[a]P-G) adduct, the aromatic B[a]P residue assumes a base-displaced intercalated conformation with the modified guanine residue in the minor groove, and the partner cytosine residue displaced into the major groove [30–32]. The R-cis-B[a]P-G adduct is associated with a greater local distortion of the DNA structure (Figure 2) and the NER cleavage efficiency in HeLa cell extracts is some ~ 5 times greater than that of the stereoisomeric S-trans-B[a]P-G adduct[22, 23](Figure 3).
R- and S-trans-DB[a,l]P-G adducts. The structures of the adducts formed from the reactions of the (+)- and (−)-enantiomers of DB[a,l]PDE to the exocyclic amino groups of guanine (14R (+)- and 14S (−)-trans-anti-DB[a,l]P-G (R- and S-trans-DB[a,l]P-G)) and adenine (14R (+)- and 14S (−)-trans-anti-DB[a,l]P-A (R- and S-trans-DB[a,l]P-A)) are shown in Figure 1, and the conformations of the adducts are depicted in Figure 2. The S-trans-DB[a,l]P-G adduct is intercalatively inserted into the DNA double helix from the minor groove and causes destabilization of the duplex by disrupting hydrogen bonding at the 5’-flanking base pair [33–35]. In the case of the S-trans-DB[a,l]P-G adduct, the bulky DB[a,l]P-residue is positioned in the minor groove with the same orientation as the configurationally identical S-trans-B[a]P-G adduct whose Watson-Crick base pairing is intact (Figure 2). However, the Watson-Crick base pairing is mainly disrupted at and adjacent to the lesion site in the DB[a,l]P case, because of the greater bulk of the polycyclic DB[a,l]P residue. The minor groove in this S-trans-DB[a,l]P-G adduct is greatly widened because the large DB[a,l]P aromatic ring surface makes extensive favorable van der Waals interactions with the groove wall that help compensate for the destabilizing distortions. The NER cleavage efficiency is 3.5 times smaller in the case of the S-trans-DB[a,l]P-G adduct (Figure 3) than in the case of its R stereoisomeric counterpart [21].
R- and S-trans-DB[a,l]P-A adducts. Modeling and other evidence indicates that the stereoisomeric R-trans- and S-trans-DB[a,l]P-A adducts are intercalated between adjacent base pairs on the 5’- and the 3’-sides of the modified adenine base, respectively [36]. In contrast to the B[a]P- and DB[a,l]P-guanine adducts, both DB[a,l]P-A adducts are resistant to NER (Figure 3).
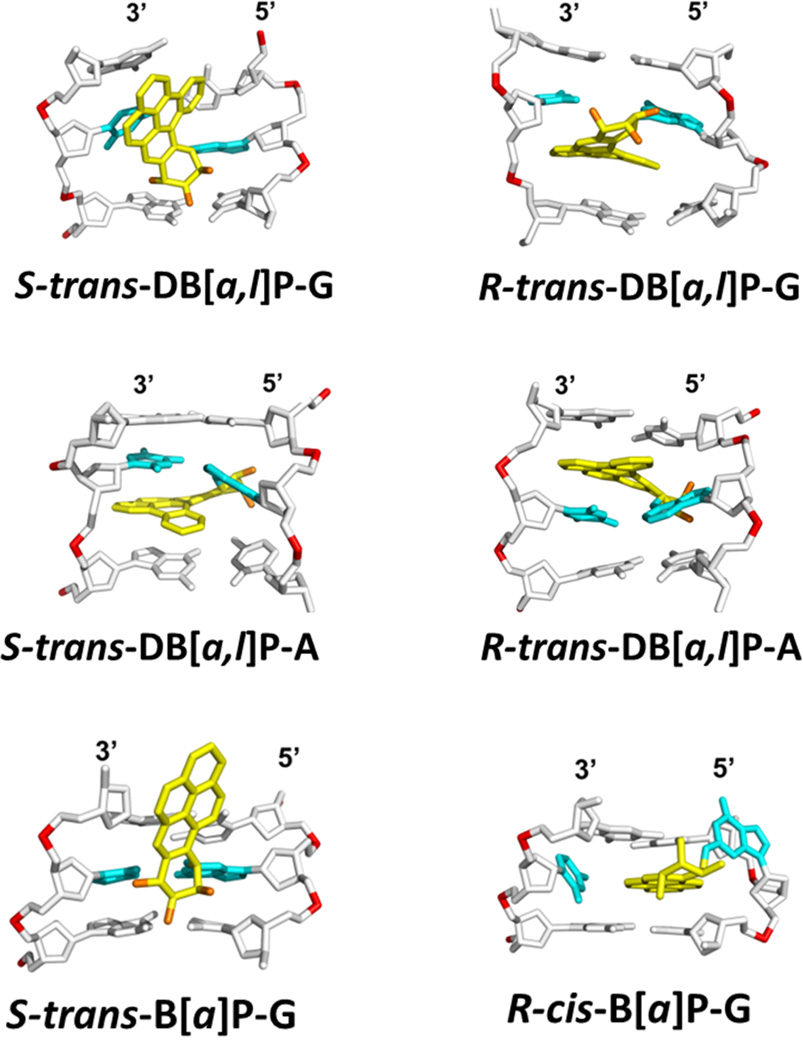
Figure 2.
Structures of lesion-containing duplexes. NMR solution structures: S-trans-B[a]P-G [31], R-cis- B[a]P-G [30], R-trans-DB[a,l]P-G [35], S-trans-DB[a,l]P-G [33, 34]. Modeled structures: R-trans-DB[a,l]P-A, and S-trans-DB[a,l]P-A [36].
Figure 3.
Relative dual incision efficiencies of 135-mer duplexes containing different DNA adducts determined in cell-free HeLa cell extracts. The data for the B[a]P-G adducts are from Hess et al. [22] and Mocquet et al [23], and for the DB[a,l]P-G/A data are from Kropachev et al. [21].
METHODS
XPC-RAD23B
Recombinant human His6-XPC-RAD23B heterodimer was overexpressed and purified from insect cells similarly as done for the yeast Rad4-Rad23 complex [18]. An N-terminal His6-tagged XPC (156-940) and RAD23B (1-409) was incorporated into a dual-expression vector (pFastBac Dual; Invitrogen) for co-expression. The residues 1-155 in XPC were highly susceptible to limited proteolysis, indicating that they are unstructured or loosely folded. Consistent with this, this region has overall low sequence complexity and is not conserved, and was omitted in order to achieve homogeneous preparation of the protein complex. The absence of the equivalent segment in yeast Rad4 (residues 1-100) does not affect the DNA binding or the lesion specificity of the complex [18]. The Hi5 insect cells co-expressing the His6-XPC-RAD23B complex were harvested 2 days after infection and were lysed in 25 mM Tris-HCl, 500 mM NaCl, 0.5 mM tris(2-carboxyethyl) phosphine hydrochloride, 0.5 mM Pefabloc SC (Roche), 1 mg/ml each of leupeptin, aprotinin and pepstatin, pH 7.5, at 4 °C, using a cell homogenizer (Avestin). The proteins were first purified using His-Select Nickel agarose resin (Sigma). They were then dialysed against 25 mM Tris-HCl, 50 mM NaCl, 4 mM dithiothreitol (DTT) and 2 mM ethylenediamine tetraacetate, pH 8.8, and were purified by anion exchange chromatography (Source Q, Amersham). The complex was further purified by cation exchange (Source S, Amersham) and gel-filtration (Superdex200, Amersham) chromatography, and it was concentrated by ultrafiltration to ~0.2 to 0.7 mg/ml in 5 mM bis-tris propane–HCl (BTP-HCl), 800 mM NaCl, 5 mM DTT, pH 6.8.
DNA substrates
The preparation of the 50-mer DNA substrate containing a single bulky DNA adduct (Figure 1) has been described elsewhere [21, 37]. Briefly, all modified bases were positioned at the 25th nucleotide counted from the 5’-end of the modified strand. The exact sequence of the 50-mer duplexes is shown in Supplementary data. For the EMSA experiments, the complementary strand was 32P-end-labeled, and was then annealed to the modified strand containing the adduct using a standard heating and slow cooling cycle. In these duplexes, all nucleotides were complementary to one another, including the adducted adenine or guanine bases.
Electrophoretic mobility shift assays
The XPC-RAD23B binding reactions were carried out in buffer solutions containing 5 mM Bis-tris propane-HCl, 75 mM NaCl, 10 mM dithioerythritol, 5% glycerol, 0.74 mM 3-[(3- cholamidopropyl)dimethylammonio]-1-propanesulfonate (CHAPS), 500 μg/ml bovine serum albumin (pH 6.8) as described [18]. The binding mixtures (8 μL aliquots) containing 0.40 nM 32P-end-labeled 50-mer DNA duplexes and various concentrations of XPC-Rad23B were incubated at 295 K for 20 mins. In the competition experiments, unlabeled and unmodified 50-mer duplexes were first incubated with 0.40 nM 32P-labeled, and various concentrations of unlabeled and unmodified DNA with the same sequence context as the 32P-labeled DNA, and then XPC-Rad23B was added and allowed to equilibrate for 20 min. The samples were then loaded onto 4.5% native polyacrylamide gels (acrylamide/bisacrylamide = 37.5:1). The sizes of the gels were 96.5 cm wide and 127 cm long containing phosphate buffer (25.5 mM KH2PO4 and 24.5 mM K2HPO4). The gel electrophesis experiments were carried out at 4 °C in a cold room at 500 V for 50 min. The gels were then dried and scanned using a Storm 860 Phosphorimager (GE Healthcare). The fractions of bound DNA molecules were evaluated using the ImageQuant 5.2 software of the phosphorimager.
RESULTS
In selecting the conditions for determining the extent of binding of XPC-RAD23B to DNA duplexes containing the single, site-specifically inserted DNA adducts (Figure 1), optimal conditions needed to be identified. The duplexes containing these lesions must be neither too short since the footprint of XPCRAD23B is about 30 base pairs [17], nor too long because this heterodimer binds non-specifically to unmodified DNA duplexes [38, 39]. Early binding experiments showed that the affinities of binding are only slightly less in 45-mer than 60-mer duplexes [38]. Thus, we selected 50-mer duplexes rather than the longer 135-mer duplexes used in the NER in vitro assays [21–23] in order to minimize the fractions of DNA-protein complexes that remain in the loading wells. We also sought to minimize the amount of radioactively labeled DNA molecules used in each experiment in order to observe saturation of XPCRAD23B binding in the < 10 nM protein range. We adopted a DNA concentration of 0.4 nM, which was consistent with this goal, yet yielded sufficiently intense bands for quantitative analysis.
General observations – single and double molecule XPC-RAD23B binding to 50-mer duplexes
Typical binding curves for all six DNA adducts (Figure 1) embedded in the identical sequence context are shown in Figures 4, 5 and 6. In the absence of the protein, a single band due to free DNA duplexes is observed at the bottom of the gel. The loading well is the weak line at the top of the gel and is due to material that did not successfully enter the gel during the electrophoresis run. The actual electrophoretic running time was limited to 20 min; the purpose was to minimize the dissociation of the protein-DNA complexes during electrophoresis that nevertheless occurred to some extent as seen by the darkness in the gels between the upper bands and the fastest moving free DNA bands. However, the short running time limited somewhat the resolution of the upper bands. Nevertheless, in the case of the unmodified DNA (UM) experiments (Figures 4, 5 and 6, panel A), a single upper band is observed at the lowest XPC concentrations, but a second, slower moving band appears at XPC-RAD23B concentrations of 2 – 3 nM, and a single band is observed at the higher XPC concentrations. The lower band is attributed to a single XPC molecule-DNA complex and the higher one is attributed to complexes with two XPC molecules bound per DNA molecule. Similar double-molecule XPC-RAD23B -DNA complexes have been reported by other workers [10, 40–42]. However, when an adduct is present, the two bands are no longer distinguishable. Only single, but broadened bands are observed that gradually exhibit lower mobilities as the protein concentrations are increased (Figures 4, 5 and 6, panel D). The broadened single bands indicate that DNA molecules form complexes with one or two protein heterodimers which exhibit variable electrophoretic mobilities, probably due to heterogeneous micro-conformations of the complexes, including the presence of both productive and non-productive binding complexes [17], discussed further below.
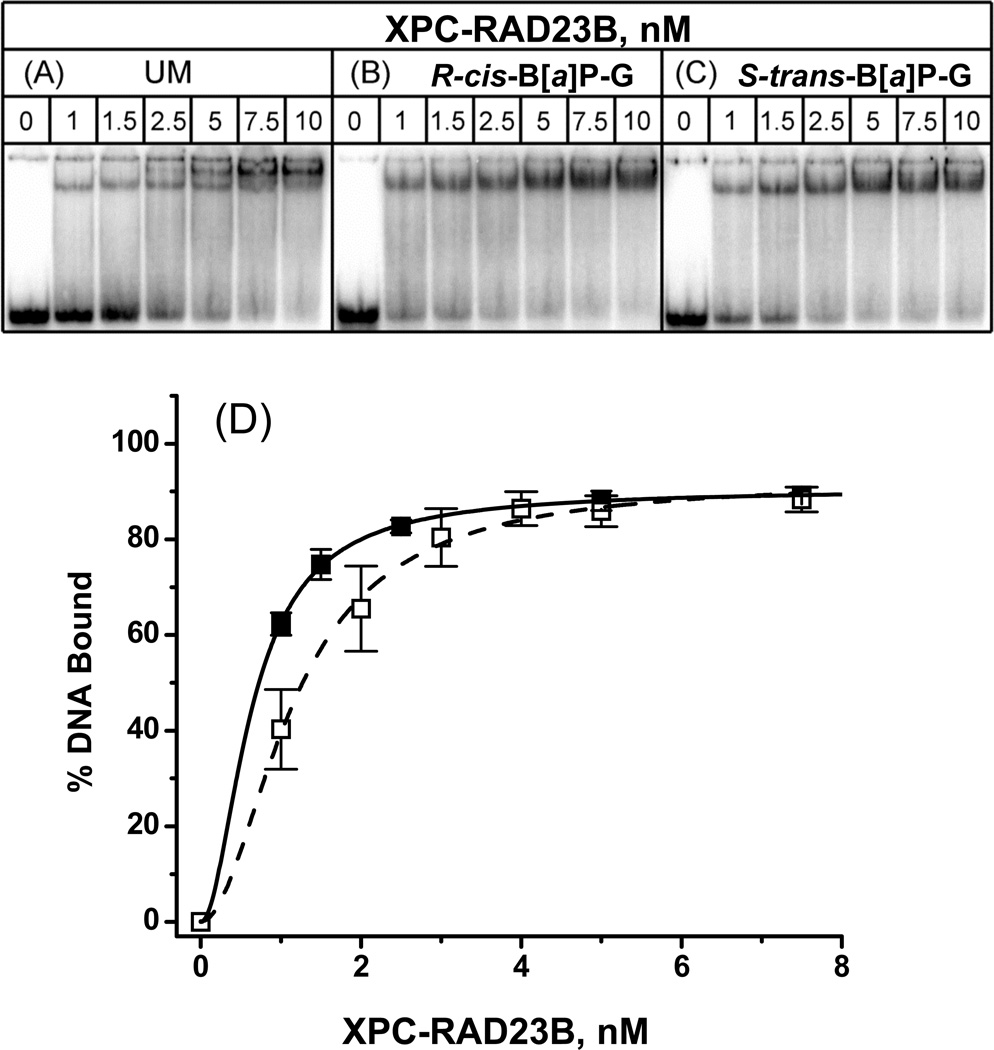
Figure 4.
Typical EMSA experiments demonstrating the binding of XPC-RAD23B to 50-mer (A) unmodified duplexes or (B) with single R-cis-B[a]P-G or (C) S-trans-B[a]P-G adducts in native 4.5% polyacrylamide gels. (D) Quantitative analysis of fractions of DNA molecules in complexes with XPC-RAD23B. The results for the cis- and trans-adducts were indistinguishable from one another within experimental error, and thus only the averages are shown (black squares). Each point and associated error bar were derived from four data points in each case (including the results for the unmodified duplexes, open circles). Solid and dashed lines are the best fits to the Hill equation based on the Hill independent bimodal binding of XPC-RAD23B to DNA (Eq. 1) with KD = (KD1KD2)1/2 = 1.2 ± 0.6 nM (unmodified duplex), 0.67±0.20 nM R-cis- and S-trans-B[a]P-G duplexes
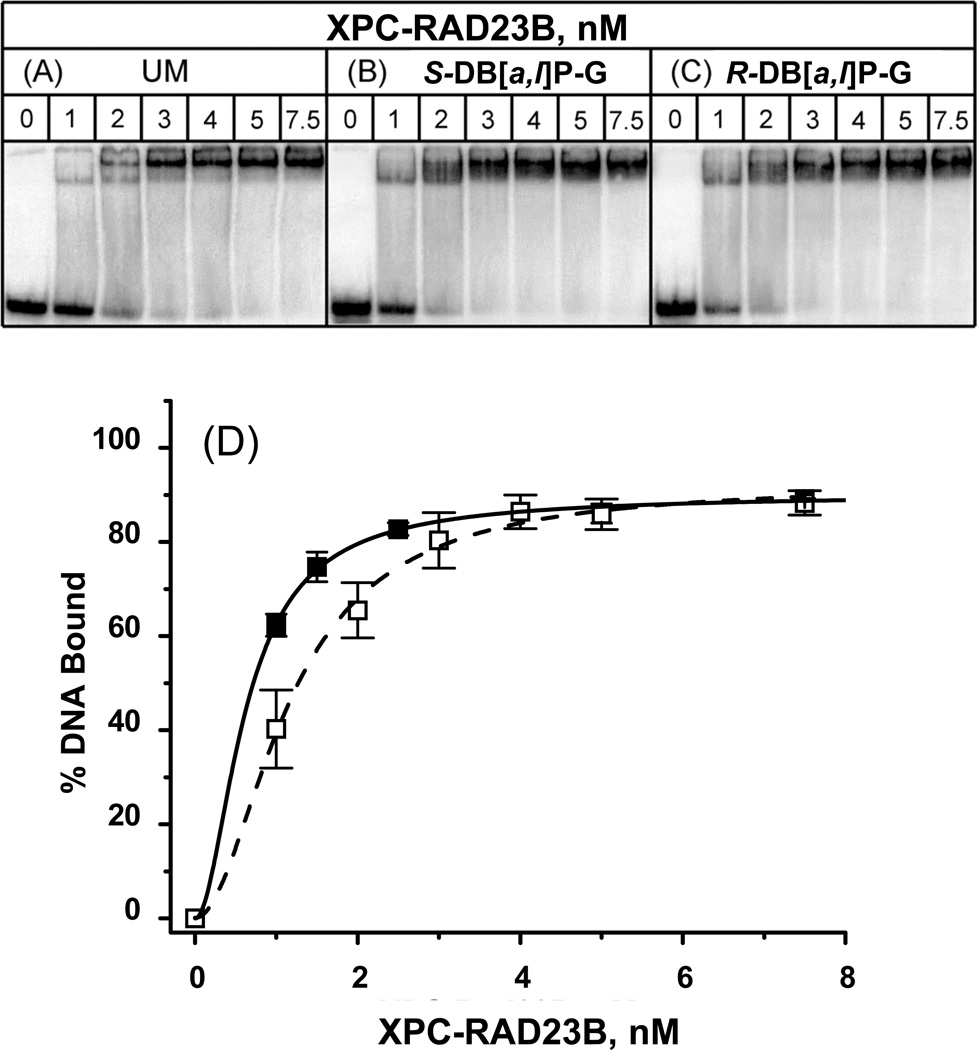
Figure 5.
Typical EMSA experiments demonstrating the binding of XPC-RAD23B to 50-mer (A) unmodified duplexes or (B) with single S-trans-DB[a,l]P-G or (C) R-trans-DB[a,l]P-G adducts in native 4.5% polyacrylamide gels. (D) Quantitative analysis of fractions of DNA molecules in complexes with XPC-RAD23B as described in the case of Figure 4, but with KD = (KD1KD2)1/2 = 0.67±0.20 nM. The results obtained with the two stereoisomeric adducts were identical to one another within experimental error.
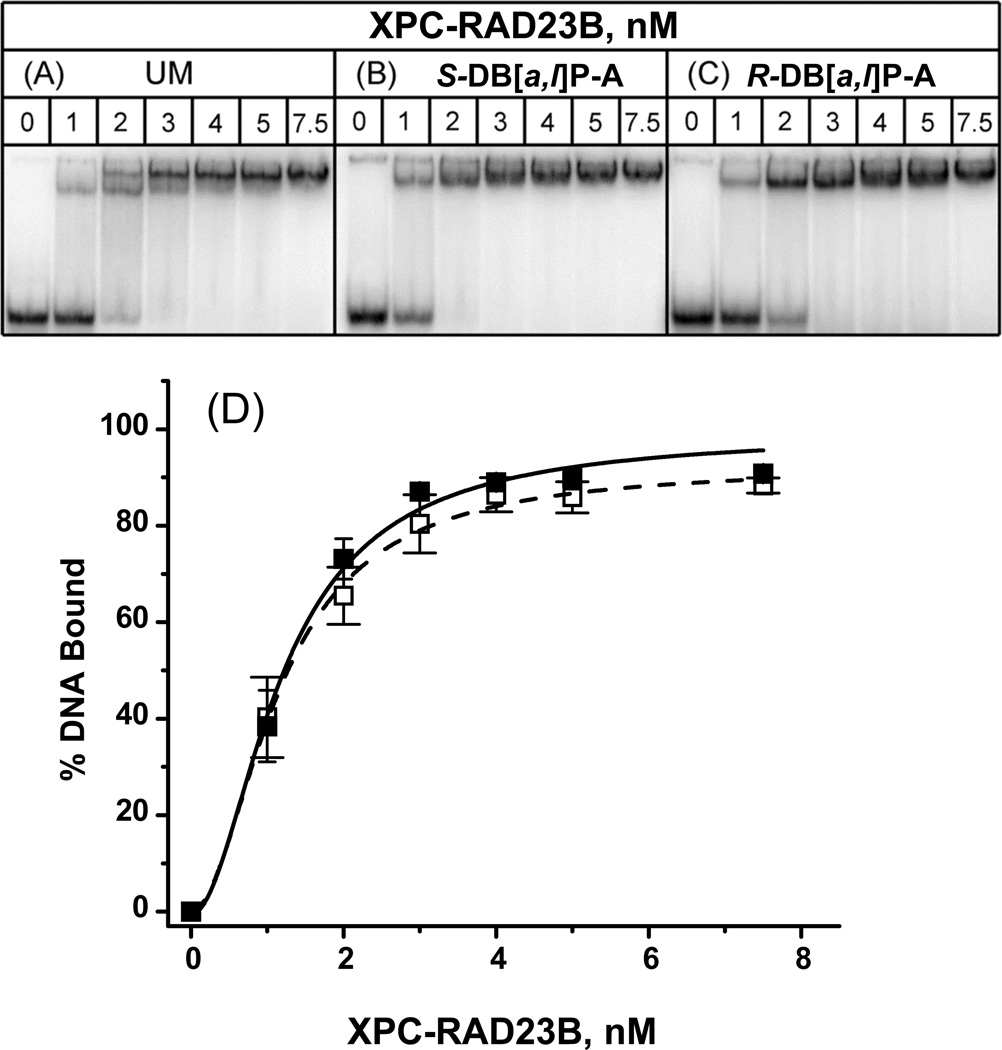
Figure 6.
Typical EMSA experiments demonstrating the binding of XPC-RAD23B to 50-mer (A) unmodified duplexes or (B) with single S-trans-DB[a,l]P-A or (C) R-trans-DB[a,l]P-A adducts in native 4.5% polyacrylamide gels. (D) Quantitative analysis of fractions of DNA molecules in complexes with XPC-RAD23B as described in the case of Figure 4, with KD = (KD1KD2)1/2 = 1.2±0.75 nM (DB[a,l]P-A) nM. The results obtained with the two stereoisomeric adducts were identical to one another within experimental error.
XPC-RAD23B binding curves
The binding curves are shown in panel D of Figures 4, 5 and 6. In the case of the DNA duplexes with R-cis and S-trans-B[a]P-G and R-trans- and S-trans-DB[a,l]P-G adducts, the binding curves are noticeably shifted to lower XPC-RAD23B concentrations relative to the unmodified duplexes. The levels of protein binding to duplexes with the stereoisomeric pairs of B[a]P-G and DB[a,l]P-G adducts are not distinguishable within experimental error. Therefore, the data points shown in panels D of Figures 4, 5 and 6 represent averages over both types of stereoisomeric DNA adducts in each case. The binding curves for the R-trans and S-trans-DB[a,l]P-A adducts are not distinguishable from the unmodified DNA binding curve within experimental error, and are thus different from those of the B[a]P-G and DB[a,l]P-G adducts.
Analysis of XPC-Rad23B binding curves
The data points in panel D of Figures 4, 5 and 6 were fit to the Hill equation describing the sequential model of binding of one and two protein molecules (P) to a single DNA (D) molecule:
| (1) |
The binding curve based on this sequential binding model can be calculated according to the Hill equation provided in Supplementary data[43]. The usual 1:1 binding model often used to derive dissociation constants using solutions of a quadratic equation [44, 45] is not appropriate here because we observe a mixture of single- and double-molecule binding. The best fits of the Hill model yield effective dissociation constants KD = (KD1KD2)1/2 = 1.2 ± 0.6 nM (unmodified duplex), 0.67±0.20 nM R-cis- and R-trans-B[a]P-G and DB[a,l]P-G), and 1.2±0.75 nM (DB[a,l]P-A). Thus, the effective dissociation constants are quite close to the values for unmodified DNA duplexes and 6-4 T^T photoproducts in 36-mer duplexes determined by the fluorescence polarization method [44].
Unmodified DNA competition binding experiments
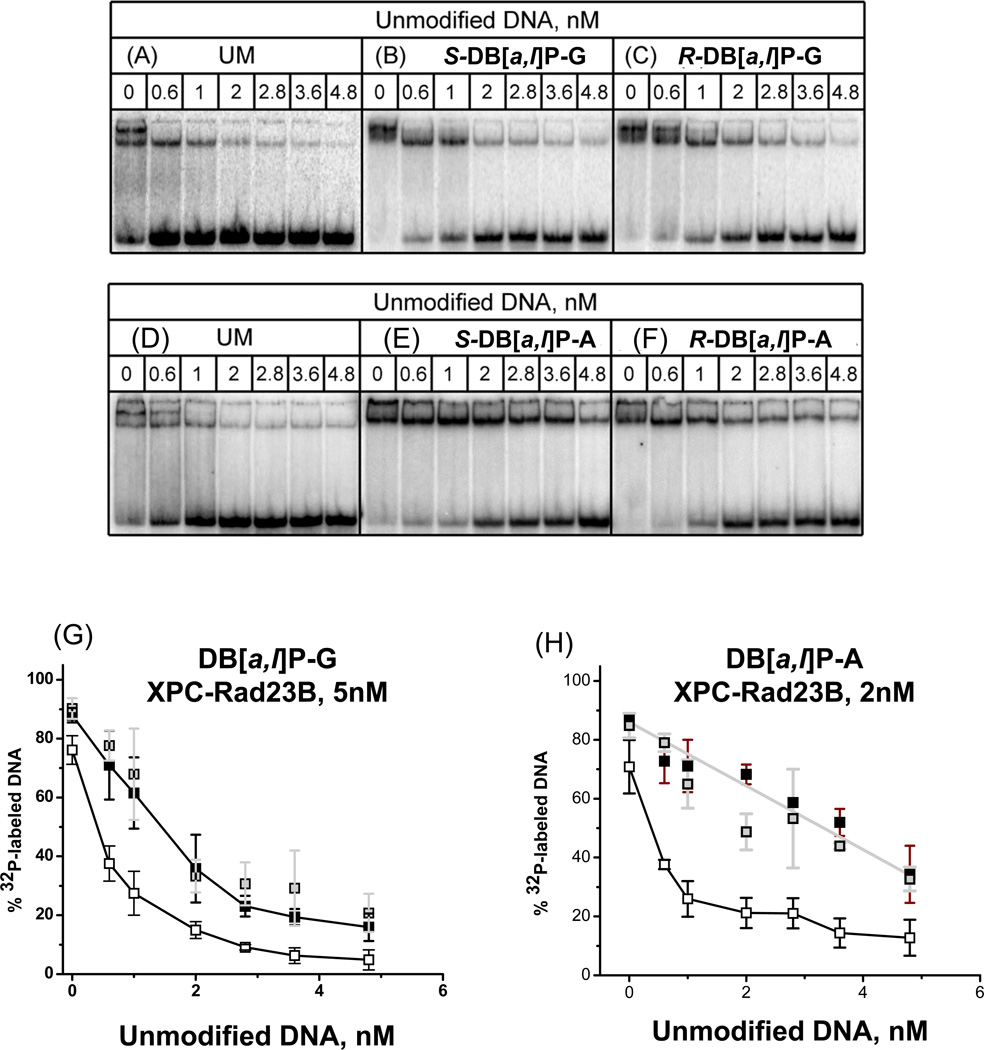
In order to further investigate whether the binding affinities of XPC-RAD23B to the pairs of stereoisomeric DB[a,l]P-G, and DB[a,l]P-A adducts are truly different, we conducted the competition experiments summarized in Figure 7. In each experiment, 0.4 nM of 32P-labeled unmodified and DB[a,l]P-modified 50-mer duplexes were mixed with unlabeled and unmodified 50-mer competitor DNA duplexes with the same sequence context as described in the Fig. 7 legends. In all cases, but to different extents, the upper bands due to the bimolecular protein complexes disappear at lower concentrations of unmodified competitor DNA. This indicates that the second XPC-RAD23B molecules bound to the same DNA molecules, have a lower binding affinity than the monomolecular complexes, and thus KD2 > KD1 (equation 1). The fraction of radioactive 32P-labeled modified duplexes as a function of unlabeled 50-mer duplex concentrations is shown in Figure 7, panels G and H. From the initial slopes of the decrease in bound, radioactively labeled DB[a,l]P-modified and unmodified DNA duplexes, the affinities of the DB[a,l]P-A duplexes are about twice the value of the DB[a,l]P-G duplexes (i.e., the rate of decrease of the fractions of 32P-labeled modified XPC-RAD23B complexes decreases more rapidly with increasing unmodified DNA competitor concentration in the case of the DB[a,l]P-G than the DB[a,l]P-A adducts (Figure 7, panels G and H, respectively)). Finally, these competition binding experiments clearly demonstrate that there is no difference in the affinities of XPC-RAD23B for duplexes containing either of the two stereoisomeric R-trans-and S-trans-DB[a,l]P-G adducts on the one hand, and the R-trans- and S-trans-DB[a,l]P-A adducts, on the other.
Figure 7.
Competition binding experiments. (A) – (H): EMSA experiments as in Figure 4, except that the unmodified and unlabeled 50-mer duplexes were first mixed with XPC-RAD23B at the concentrations indicated, allowed to equilibrate for 10 min; then the 32P-endlabeled unmodified duplexes (panels A and D), or the same duplexes bearing single stereoisomeric S-trans- or R-trans-DB[a,l]P-G adducts (B and C, respectively), or S-trans- or R-trans-DB[a,l]P-A (panels D and E, respectively) were added at the indicated concentrations.
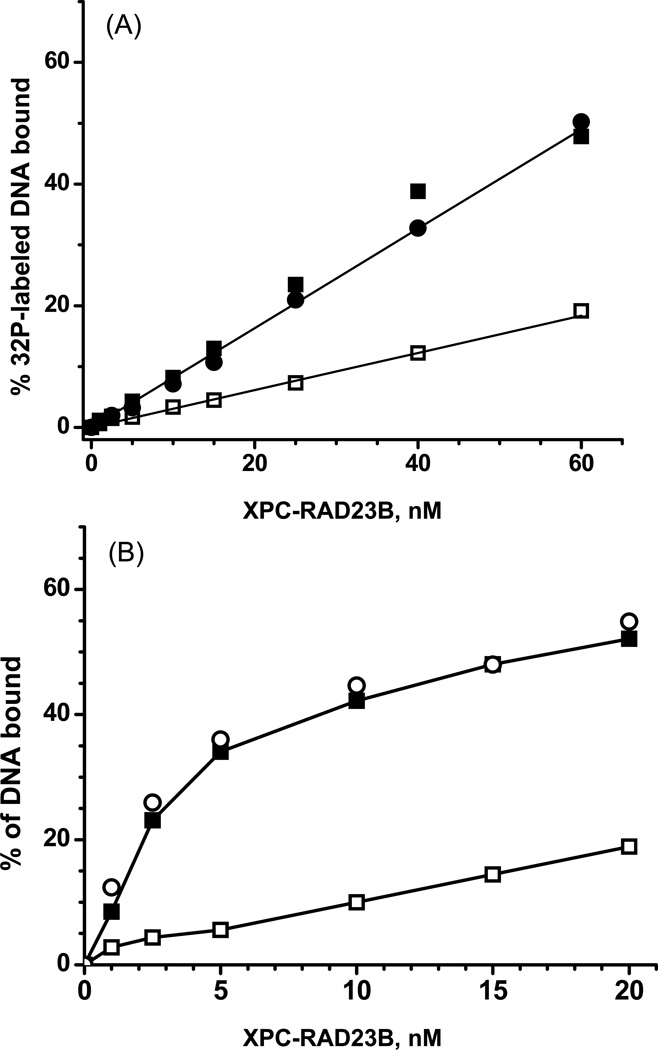
A somewhat different competition experiment in which the binding of XPC-Rad23B to 32P-labeled unmodified duplexes, or duplexes with either S-trans- or R-cis-B[a]P-G adducts (0.4 nM) was determined in the presence of an excess of 500 nM unmodified DNA duplexes with the same sequence context. The binding of XPC-RAD23B increases linearly as a function of protein concentration as expected from the relevant equation describing the fraction of DNA molecules bound to XPC-RAD23B (Supplementary data). This equation also shows that when the concentration ratio of [competitor DNA]/[unmodified DNA] >> 1, the ratio of the slopes for the modified and unmodified DNA straight lines in Figure 8A is equal to [KD (unmodified)/KD(modified DNA)] ≈ 2.7. Thus, the relative affinities of XPC-RAD23B for all six DNA adducts studied relative to unmodified DNA is only 2 – 4 times greater than for unmodified DNA.
Figure 8.
(A) Example of effects of a large excess of unlabeled and unmodified 50-mer duplexes (500 nM) on the binding of XPC-RAD23B to 32P-labeled 1 nM unmodified 50-mer duplexes (open squares), or identical duplexes but with single R-cis-B[a]P-G (black circles) or S-trans-B[a]P-G (black squares). (B) Effects of NaCl (225 mM) on the binding of XPC-RAD23B to 32P-labeled unmodified 50-mer DNA duplexes (open squares), or to 32P-labeled modified 50-mer DNA duplexes with R-cis-B[a]P-G (open circles) or with S-trans-B[a]P-G duplexes (black squares).
Effects of high salt concentration on XPC-RAD23B binding
Finally, we also investigated the effects of salt concentration on the binding of XPC-RAD23B. This experiment was prompted by the finding that electrostatic binding mechanisms contribute to the binding of RAD4-RAD23B [18], the yeast homolog of XPC, to double-stranded DNA. Such mechanisms are predicted to be sensitive to ionic strengths. An example of the binding of XPC-RAD23B to modified 50-mer duplexes and duplexes containing either the S-trans- or the R-cis-B[a]P-G adduct is shown in Figure 8B. This example shows that, at a 225 mM concentration of NaCl, non-specific binding to unmodified DNA is indeed strongly reduced, while the binding to the modified duplexes is clearly significantly higher. However, as expected from the competition experiments shown in Figure 8A, again there is no difference in binding of XPC-RAD23B to duplexes with the stereoisomeric cis- and trans- B[a]P-G adducts that are characterized by very different adduct conformations (Figure 2).
DISCUSSION
The determination of accurate binding affinities of XPC-RAD23B to DNA containing different types of bulky adducts is complicated by the strong non-specific binding of this protein heterodimer to unmodified DNA. Thus, the measured extents of overall binding are likely to depend on the lengths of the DNA duplexes, and the binding of more than one heterodimeric protein molecule to the same duplex. It is shown in Figure 8B that non-specific binding can be strongly suppressed at high salt concentrations (225 mM) which, however, is higher than physiological salt concentration. At physiological salt concentrations of 150 mM, the discrimination of XPC-RAD23B binding between unmodified and modified DNA is only somewhat better than at the 75 mM salt concentrations used in the studies described here (data not shown).
Comparisons of XPC-RAD23B binding and NER activities
The binding curves shown in Figure 4, as well as the effects of unmodified competitor DNA on XPC-RAD23B binding (Figure 8A), unambiguously demonstrate that there is no measurable difference in the binding affinities of XPC-RAD23B to 50-mer duplexes containing either the R-cis- or the S-trans-B[a]P-G adducts. Nevertheless, the NER cleavage efficiencies are greater by a factor of five in the case of the base-displaced intercalated R-cis-B[a]P-G adduct than in the case of the stereoisomeric minor groove S-trans-B[a]P-G adduct [19] (Figure 3). Thus, the XPC-RAD23B binding affinities are not correlated with the NER dual excision efficiencies in this case. Interestingly, EMSA experiments with shorter DNA duplexes have shown that the S-trans-B[a]P-G adduct causes a significant bending/flexible hinge joint in double-stranded DNA, while the R-cis-B[a]P-G adduct does not [46, 47]. Thus, in this case, bending caused by the S-trans-B[a]P-G adduct is not correlated either with XPC-RAD23B binding (as recently suggested for other forms of DNA damage [45]), nor NER cleavage efficiencies.
The binding affinities of the R-trans- and S-trans-DB[a,l]P-G adducts are also not distinguishable from one another (Figure 5 and Figure 7, panels B, C and G). However, the NER cleavage efficiencies are ~ 3.5 times greater in the case of the R-trans- than the S-trans-DB[a,l]P-G adduct [21]. In the case of the stereioisomeric R-trans- and S-trans-DB[a,l]P-A adducts, the binding curves are practically indistinguishable from the binding curve of unmodified DNA (Figure 6). These observations are consistent with the almost complete resistance of these adenine adducts to NER [21] (Figure 3). However, the competition experiments in Figure 7 demonstrate that the binding affinities of these adducts are significantly higher than in the case of unmodified DNA.
Implications
Taken together, our observations with three sets of stereoisomeric bulky DNA adducts show that XPC-RAD23B binding tends to be independent of adduct stereochemistry and conformations in double-stranded DNA, while the same features of these DNA adducts lead to distinct and remarkable differences in cleavage efficiencies. Chazin and coworkers have used high throughput fluorescence anisotropy to measure the binding affinity of XPC-RAD23B to a number of DNA lesions subject to NER and also find that the lesion identity has little effect on the XPC-RAD23B binding affinity [48].
Sugasawa has discussed two forms of binding of XPC-RAD23B; one of these is productive in terms of the subsequent strand opening catalyzed by the helicase XPD which is part of the TFIIH multiprotein complex. The other form is non-productive because the XPC molecule is bound to the lesioned rather than the complementary unmodified strand, pointing in the opposite direction from the productive complex; this binding mode is not consistent with the 5’→3’ translocation directionality of the ATP-driven strand opening helicase XPD [17, 49]. We speculate that the dominance of one type of XPC orientation over the other could play a role in governing the subsequent steps that lead to the dual incisions of the damaged strand. Among these are the recruitment of TFIIH by the XPC-damaged DNA complex, the orientation of this multi-protein complex relative to the translocation of XPD, or subsequent details of the verification step. These fascinating possibilities remain to be elucidated and point to the need for investigating the effects of DNA adduct structure and conformation on the recruitment of TFIIH by the XPC-RAD23B -DNA adduct complexes, and the mechanistic features of the subsequent verification step. These are the critical steps that govern the secrets underlying the striking differences in NER cleavage efficiencies (Figure 3), which characterize the processing of the structurally related pairs of stereoisomeric and structurally different DNA adducts by the NER apparatus.
Correlations between XPC-RAD23B binding and NER activities
While we have uncovered a set of bulky lesions that exhibit little or no correlation between NER cleavage efficiencies and XPC-RAD23B binding, there are other well-known types of DNA lesions that do exhibit a correlation between XPC-RAD23B binding and NER efficiencies. Some examples are summarized in the following paragraph.
The thymine dimer 6-4 and CPD UV photoproducts. XPC-RAD23B binds to 6-4 lesions, but weakly to CPD [39, 44], and the 6-4 photoproduct is also a much better NER substrate than CPD (e.g.,[39]). However, in duplexes with mismatched (MM) bases in the partner strand opposite to a CPD thymine dimer, the XPC-RAD23B binding is strongly enhanced and NER activities are similar to those observed in full duplexes containing the 6-4 lesions.
Acetylaminofluorene (AAF)-C8-guanine (AAF-G) and aminofluorene (AF)-C8-guanine (AF-G) adducts. The NER excision efficiencies of adducts have been found to be correlated with the binding affinities of XPC-RAD23B [10].
AAF-G adducts embedded in bubbles. Embedding AAF-G adducts within DNA duplex regions containing 3 or 5 mismatched base pairs (‘bubbles’) strongly enhances both XPC-RAD23B binding as well as the NER activity in cell-free extracts [39](relative to the adduct in normal double-stranded DNA ).
Aristolochic acid-derived DNA adducts. These adducts are NER-resistant and XPC-RAD23B binds poorly to these lesions, but in duplexes with three mismatched cytosines opposite the lesions, both XPC-RAD23B binding and the NER activities are restored [50].
cisplatin-derived lesions. The cis-Pt G*TG* intrastrand cross-linked lesion has a high affinity for XPC-RAD23B and is an excellent NER substrate [40].
Mode of binding of XPC-RAD23B to modified DNA
Min and Pavletich demonstrated that the yeast homolog of XPC-RAD23B, Rad4-Rad23, in complex with a cis-syn thymine dimer-containing duplex, forms contacts only with the unmodified strand at the site of the lesion [18]. Remarkably, this protein is positioned almost entirely on the 3’-side of the lesion. A portion of the Rad4 protein (TGD and the β-hairpin BHD1) binds non-specifically to both strands of an unmodified 11-mer region of the 24-mer duplex, while hairpins BHD2 and BHD3 bind specifically to a four base pair region around the site of the lesion; BHD3 is inserted from the major groove side between the two DNA strands and two mismatched thymine partner bases opposite the lesion are flipped into a binding pocket of the protein while the lesion is also extruded.
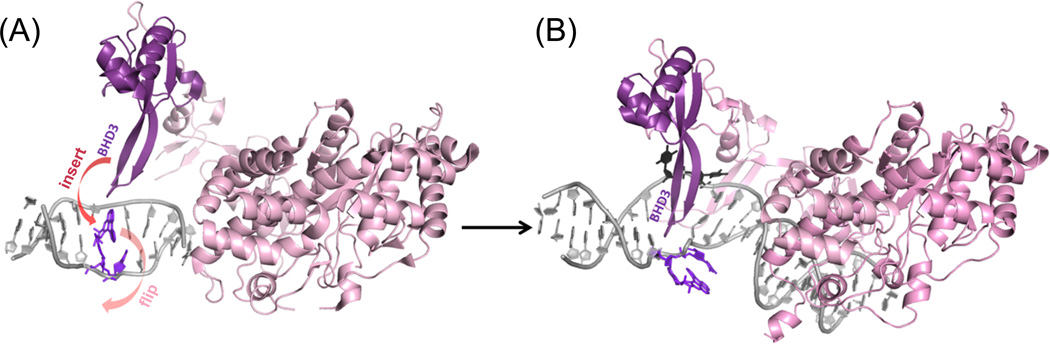
An opportunity is provided by this crystal structure to hypothesize on how productive lesion recognition may occur with intercalated adducts that are repair susceptible. With the approach of the BHD3 β-hairpin from the major groove side, it appears that partner flipping and lesion extrusion should take the minor groove route to avoid steric clashes with the incoming hairpin. In Figure 9A we have utilized the NMR solution structure of the well-repaired[21] R-trans-DB[a,l]P-G lesion [35] that is intercalated into the DNA duplex from the minor groove side, and the structure of the apo Rad4-Rad23 crystal (PDB ID: 2QSF, [18]) to illustrate the pre-bound states of the damaged duplex and the Rad4-Rad23. In Figure 9B, we have modeled (details given in Cai et al. 11]) an extruded structure for the R-trans-DB[a,l]P-G lesion based on the thymine dimer- containing crystal structure of Rad4-Rad23 (PDB ID: 2QSG, [18]). We envision that the BHD3 β-hairpin intrusion through the major groove expels the minor grove intercalated lesion via the minor groove route (Figure 9). We have speculated that the repair-resistant DB[a,l]P-A adducts that are intercalated into the DNA duplex from the major groove [36] may sterically impede BHD3 intrusion [11]. Future studies will provide essential insights on how productive recognition of bulky PAH-derived lesions by XPC-RAD23B occurs and how XPC-RAD23B interacts with such repair-resistant lesions.
Figure 9.
Structural models to illustrate the productive binding of the yeast Rad4/Rad23 apo enzyme to duplex DNA containing the R-trans-DB[a,l]P-G lesion intercalated from the minor groove; the latter is a very good NER substrate [21]. (A) NMR solution structure of the R-trans-DB[a,l]P-G lesion (PDB [53] ID: 2LZK, [35]) with the apo Rad4/Rad23 heterodimer before binding (PDB ID: 2QSF, [18]) positioned to initiate the insertion of the BHD3 β–hairpin from the major groove (red arrow), with extrusion of the lesion and its 3’ adjacent dC through the minor groove (pink arrow). The model was created by superposing, respectively, the TGD domain of the apo enzyme and the NMR structure of the R-trans-DB[a,l]P-G lesion-containing DNA, to the TGD domain and the DNA backbone of 6 base pairs around the lesion site in the complex crystal structure (PDB ID: 2QSG, [18]). (B) the BHD3 hairpin loop has inserted into the duplex from the major groove side and extruded the R-trans-DB[a,l]P-G stacked with its 3’ adjacent dC (purple in A and B) through the minor groove. The Rad4/Rad23 complex containing a cis-syn thymine dimer (PDB ID: 2QSG, [18]) was remodeled to replace the thymine dimer (whose coordinates were missing) with the R-trans-DB[a,l]P-G-C dinucleotide from the NMR structure (PDB ID:2LZK, [35]) followed by 10 ns of molecular dynamics simulation [11] The missing BHD3 hairpin loop in (A) was modeled based on its structure in (B). This model suggests how productive binding may take place for a well-repaired PAH-derived NER substrate.
Productive and unproductive modes of binding of XPC-RAD23B
Using predominantly biochemical footprinting experiments, Sugasawa et al.[17, 51] showed that the binding of XPC to lesions is asymmetric and can occur in two different ways as mentioned above. In the productive binding mode, the XPC is bound to the undamaged strand; this leads to the 5’→3’ translocation and strand opening catalyzed by XPD in TFIIH that yields the dual incisions. In the non-productive binding mode, the XPC is bound to the damaged strand, faces in the opposite direction as in productive binding, and does not lead to lesion excision. From the crystal structure of Rad4-Rad23 [18] it is clear that the productive binding mode has the N terminus of the protein facing the 3’-side of the unmodified strand; this would orient the N terminus in the non-productive binding mode to the 5’-side of the unmodified strand but bound to the damaged strand and incapable of verifying the lesion by XPD translocation and stalling [2, 52]. Our studies reveal that binding does not correlate with NER cleavage efficiencies for our series of lesions, and that there is binding of more than one XPC molecule. These experimental observations may provide rich opportunities for future elucidation of productive and nonproductive binding modes of XPC-RAD23B and their relation to cleavage efficiencies.
Supplementary Material
Acknowledgements
This research was supported by National Institutes of Health Grants 1R01-CA168469 (to N.E.G.) and R01-CA28038 (to S.B.). Computational infrastructure and systems management were partially supported by R01-CA75449 (S.B). Support to J.H.M. was provided by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust and the University of Illinois at Chicago. This work used the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation (NSF) Grant MCB060037 to S.B., and the high performance computing resources of New York University (NYU-ITS).
Abbreviations
- NER
nucleotide excision repair
- PAH
polycyclic aromatic hydrocarbon
- B[a]P
benzo[a]pyrene
- DB[a,l]P
dibenzo[a,l]pyrene
- S-trans-DB[a,l]P-G/A
14S (–)-trans-anti-DB[a,l]P-G/A
- R-trans-DB[a,l]P-G/A
14R (+)- trans-anti-DB[a,l]P-G/A
- S-trans-B[a]P-G
10S (+)-trans-anti-B[a]P-G
- R-cis-B[a]P-G
10R (+)-cis-anti-B[a]P-G
- B[a]PDE
(+)-7R,8S-dihydrodiol-9S,10R-epoxy-7,8,9,10-tetrahydro-B[a]P
- DB[a,l]PDE
(−)-11R,12S-dihydrodiol-13S,14R-epoxy-7,8,9,10-tetrahydro-DB[a,l]P
- EMSA
electrophoretic mobility shift assays
- AAF
acetylaminofluorene
- AAF-G
AAF-C8-guanine
- AF
aminofluorene
- AF-G
AF-C8-guanine
- BHD
β-hairpin domain
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 2.Scharer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. doi: 10.1101/cshperspect.a012609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kisker C, Kuper J, Van Houten B. Prokaryotic nucleotide excision repair. Cold Spring Harb Perspect Biol. 2013;5:a012591. doi: 10.1101/cshperspect.a012591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat Rev Mol Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 5.Ganesan A, Spivak G, Hanawalt PC. Transcription-coupled DNA repair in prokaryotes. Prog Mol Biol Transl Sci. 2012;110:25–40. doi: 10.1016/B978-0-12-387665-2.00002-X. [DOI] [PubMed] [Google Scholar]
- 6.Sugasawa K, Ng JM, Masutani C, Iwai S, van der Spek PJ, Eker AP, Hanaoka F, Bootsma D, Hoeijmakers JH. Xeroderma pigmentosum group C protein complex is the initiator of global genome nucleotide excision repair. Mol Cell. 1998;2:223–232. doi: 10.1016/s1097-2765(00)80132-x. [DOI] [PubMed] [Google Scholar]
- 7.Fei J, Kaczmarek N, Luch A, Glas A, Carell T, Naegeli H. Regulation of nucleotide excision repair by UV-DDB: prioritization of damage recognition to internucleosomal DNA. PLoS Biol. 2011;9:e1001183. doi: 10.1371/journal.pbio.1001183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scrima A, Konickova R, Czyzewski BK, Kawasaki Y, Jeffrey PD, Groisman R, Nakatani Y, Iwai S, Pavletich NP, Thoma NH. Structural basis of UV DNA-damage recognition by the DDB1-DDB2 complex. Cell. 2008;135:1213–1223. doi: 10.1016/j.cell.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luijsterburg MS, von Bornstaedt G, Gourdin AM, Politi AZ, Mone MJ, Warmerdam DO, Goedhart J, Vermeulen W, van Driel R, Hofer T. Stochastic and reversible assembly of a multiprotein DNA repair complex ensures accurate target site recognition and efficient repair. J Cell Biol. 2010;189:445–463. doi: 10.1083/jcb.200909175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo JE, Khoo A, Fagbemi AF, Scharer OD. The efficiencies of damage recognition and excision correlate with duplex destabilization induced by acetylaminofluorene adducts in human nucleotide excision repair. Chem Res Toxicol. 2012;25:2462–2468. doi: 10.1021/tx3003033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai Y, Zheng H, Ding S, Kropachev K, Schwaid AG, Tang Y, Mu H, Wang S, Geacintov NE, Zhang Y, Broyde S. Free energy profiles of base flipping in intercalative polycyclic aromatic hydrocarbon-damaged DNA duplexes: energetic and structural relationships to nucleotide excision repair susceptibility. Chem Res Toxicol. 2013;26:1115–1125. doi: 10.1021/tx400156a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mocquet V, Laine JP, Riedl T, Yajin Z, Lee MY, Egly JM. Sequential recruitment of the repair factors during NER: the role of XPG in initiating the resynthesis step. EMBO J. 2008;27:155–167. doi: 10.1038/sj.emboj.7601948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riedl T, Hanaoka F, Egly JM. The comings and goings of nucleotide excision repair factors on damaged DNA. EMBO J. 2003;22:5293–5303. doi: 10.1093/emboj/cdg489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Volker M, Mone MJ, Karmakar P, van Hoffen A, Schul W, Vermeulen W, Hoeijmakers JH, van Driel R, van Zeeland AA, Mullenders LH. Sequential assembly of the nucleotide excision repair factors in vivo. Mol Cell. 2001;8:213–224. doi: 10.1016/s1097-2765(01)00281-7. [DOI] [PubMed] [Google Scholar]
- 15.Wood RD. DNA damage recognition during nucleotide excision repair in mammalian cells. Biochimie. 1999;81:39–44. doi: 10.1016/s0300-9084(99)80036-4. [DOI] [PubMed] [Google Scholar]
- 16.Buterin T, Meyer C, Giese B, Naegeli H. DNA quality control by conformational readout on the undamaged strand of the double helix. Chem Biol. 2005;12:913–922. doi: 10.1016/j.chembiol.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Sugasawa K, Akagi J, Nishi R, Iwai S, Hanaoka F. Two-step recognition of DNA damage for mammalian nucleotide excision repair: Directional binding of the XPC complex and DNA strand scanning. Mol Cell. 2009;36:642–653. doi: 10.1016/j.molcel.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 18.Min JH, Pavletich NP. Recognition of DNA damage by the Rad4 nucleotide excision repair protein. Nature. 2007;449:570–575. doi: 10.1038/nature06155. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Reeves D, Kropachev K, Cai Y, Ding S, Kolbanovskiy M, Kolbanovskiy A, Bolton JL, Broyde S, Van Houten B, Geacintov NE. Probing for DNA damage with beta-hairpins: similarities in incision efficiencies of bulky DNA adducts by prokaryotic and human nucleotide excision repair systems in vitro. DNA Repair (Amst) 2011;10:684–696. doi: 10.1016/j.dnarep.2011.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai Y, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Broyde S, Patel DJ, Geacintov NE. Recognition and removal of bulky DNA lesiosn by the nucleotide excision repair system. In: Geacintov NE, Broyde S, editors. The Chemical Biology of DNA Damage. Weinheim, Germany: Wiley-VCH; 2010. pp. 261–298. Chapter 12. [Google Scholar]
- 21.Kropachev K, Kolbanovskiy M, Liu Z, Cai Y, Zhang L, Schwaid AG, Kolbanovskiy A, Ding S, Amin S, Broyde S, Geacintov NE. Adenine-DNA adducts derived from the highly tumorigenic dibenzo[a,l]pyrene are resistant to nucleotide excision repair while guanine adducts are not. Chem. Res. Toxicol. 2013;26:783–793. doi: 10.1021/tx400080k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hess MT, Gunz D, Luneva N, Geacintov NE, Naegeli H. Base pair conformation-dependent excision of benzo[a]pyrene diol epoxide-guanine adducts by human nucleotide excision repair enzymes. Mol Cell Biol. 1997;17:7069–7076. doi: 10.1128/mcb.17.12.7069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mocquet V, Kropachev K, Kolbanovskiy M, Kolbanovskiy A, Tapias A, Cai Y, Broyde S, Geacintov NE, Egly JM. The human DNA repair factor XPC-HR23B distinguishes stereoisomeric benzo[a]pyrenyl-DNA lesions. EMBO J. 2007;26:2923–2932. doi: 10.1038/sj.emboj.7601730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng SC, Hilton BD, Roman JM, Dipple A. DNA adducts from carcinogenic and noncarcinogenic enantiomers of benzo[a]pyrene dihydrodiol epoxide. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 25.Conney AH. Induction of microsomal enzymes by foreign chemicals and carcinogenesis by polycyclic aromatic hydrocarbons: G. H. A. Clowes Memorial Lecture. Cancer Res. 1982;42:4875–4917. [PubMed] [Google Scholar]
- 26.Cosman M, Hingerty BE, Luneva N, Amin S, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (-)-cis-anti-benzo[a]pyrenyl-dG adduct opposite dC in a DNA duplex: intercalation of the covalently attached BP ring into the helix with base displacement of the modified deoxyguanosine into the major groove. Biochemistry. 1996;35:9850–9863. doi: 10.1021/bi9605346. [DOI] [PubMed] [Google Scholar]
- 27.Li KM, George M, Gross ML, Lin CH, Jankowiak R, Small GJ, Seidel A, Kroth H, Rogan EG, Cavalieri EL. Structure elucidation of the adducts formed by fjord region Dibenzo[a,l]pyrene-11,12-dihydrodiol 13,14-epoxides with deoxyguanosine. Chem Res Toxicol. 1999;12:778–788. doi: 10.1021/tx980234k. [DOI] [PubMed] [Google Scholar]
- 28.Yagi H, Frank H, Seidel A, Jerina DM. Revised assignment of absolute configuration of the cisand trans-N6-deoxyadenosine adducts at C14 of (+/-)-11beta,12alpha-dihydroxy-13alpha,14alpha-epoxy-11,12,13,14-tetrahydr odibenzo[a,l]pyrene by stereoselective synthesis. Chem Res Toxicol. 2008;21:2379–2392. doi: 10.1021/tx800268f. [DOI] [PubMed] [Google Scholar]
- 29.Geacintov NE, Cosman M, Hingerty BE, Amin S, Broyde S, Patel DJ. NMR solution structures of stereoisometric covalent polycyclic aromatic carcinogen-DNA adduct: principles, patterns, and diversity. Chem Res Toxicol. 1997;10:111–146. doi: 10.1021/tx9601418. [DOI] [PubMed] [Google Scholar]
- 30.Cosman M, de los Santos C, Fiala R, Hingerty BE, Ibanez V, Luna E, Harvey R, Geacintov NE, Broyde S, Patel DJ. Solution conformation of the (+)-cis-anti-[BP]dG adduct in a DNA duplex: intercalation of the covalently attached benzo[a]pyrenyl ring into the helix and displacement of the modified deoxyguanosine. Biochemistry. 1993;32:4145–4155. doi: 10.1021/bi00067a001. [DOI] [PubMed] [Google Scholar]
- 31.Cosman M, de los Santos C, Fiala R, Hingerty BE, Singh SB, Ibanez V, Margulis LA, Live D, Geacintov NE, Broyde S, et al. Solution conformation of the major adduct between the carcinogen (+)-anti-benzo[a]pyrene diol epoxide and DNA. Proc Natl Acad Sci U S A. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.de los Santos C, Cosman M, Hingerty BE, Ibanez V, Margulis LA, Geacintov NE, Broyde S, Patel DJ. Influence of benzo[a]pyrene diol epoxide chirality on solution conformations of DNA covalent adducts: the (-)-trans-anti-[BP]G.C adduct structure and comparison with the (+)-trans-anti-[BP]G.C enantiomer. Biochemistry. 1992;31:5245–5252. doi: 10.1021/bi00138a002. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez F. Chemistry Department. New York, NY: New York University; 2007. Nuclear Magnetic Resonance Solution Structure of Covalent Polycyclic Aromatic Carcinogen–DNA adducts: Influence of Base Sequence Contexts and Carcinogen Topology. [Google Scholar]
- 34.Rodríguez FA, Liu Z, Lin CH, Ding S, Cai Y, Kolbanovskiy A, Kolbanovskiy M, Amin S, Broyde S, Geacintov NE. Nuclear magnetic resonance studies of N2-guanine adducts derived from the tumorigen dibenzo[a,l]pyrene in DNA: Impact of adduct stereochemistry, size, and local DNA structure on solution conformations. Biochemistry. 2014;53:1827–1841. doi: 10.1021/bi4017044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tang Y, Liu Z, Ding S, Lin CH, Cai Y, Rodriguez FA, Sayer JM, Jerina DM, Amin S, Broyde S, Geacintov NE. Nuclear magnetic resonance solution structure of an N(2)-guanine DNA adduct derived from the potent tumorigen dibenzo[a,l]pyrene: intercalation from the minor groove with ruptured Watson-Crick base pairing. Biochemistry. 2012;51:9751–9762. doi: 10.1021/bi3013577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai Y, Ding S, Geacintov NE, Broyde S. Intercalative conformations of the 14R(+)- and 14S(-)-trans-anti-DB[a,l]P-N(6)-dA adducts: Molecular modeling and MD simulations. Chem Res Toxicol. 2011;24:522–531. doi: 10.1021/tx1004002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kropachev K, Kolbanovskii M, Cai Y, Rodriguez F, Kolbanovskii A, Liu Y, Zhang L, Amin S, Patel D, Broyde S, Geacintov NE. The sequence dependence of human nucleotide excision repair efficiencies of benzo[a]pyrene-derived DNA lesions: insights into the structural factors that favor dual incisions. J Mol Biol. 2009;386:1193–1203. doi: 10.1016/j.jmb.2008.12.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reardon JT, Mu D, Sancar A. Overproduction, purification, and characterization of the XPC subunit of the human DNA repair excision nuclease. J Biol Chem. 1996;271:19451–19456. doi: 10.1074/jbc.271.32.19451. [DOI] [PubMed] [Google Scholar]
- 39.Sugasawa K, Okamoto T, Shimizu Y, Masutani C, Iwai S, Hanaoka F. A multistep damage recognition mechanism for global genomic nucleotide excision repair. Genes Dev. 2001;15:507–521. doi: 10.1101/gad.866301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Batty D, Rapic'-Otrin V, Levine AS, Wood RD. Stable binding of human XPC complex to irradiated DNA confers strong discrimination for damaged sites. J Mol Biol. 2000;300:275–290. doi: 10.1006/jmbi.2000.3857. [DOI] [PubMed] [Google Scholar]
- 41.Trego KS, Turchi JJ. Pre-steady-state binding of damaged DNA by XPC-hHR23B reveals a kinetic mechanism for damage discrimination. Biochemistry. 2006;45:1961–1969. doi: 10.1021/bi05196t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krasikova YS, Rechkunova NI, Maltseva EA, Pestryakov PE, Petruseva IO, Sugasawa K, Chen X, Min JH, Lavrik OI. Comparative analysis of interaction of human and yeast DNA damage recognition complexes with damaged DNA in nucleotide excision repair. J Biol Chem. 2013;288:10936–10947. doi: 10.1074/jbc.M112.444026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weiss JN. The Hill equation revisited: uses and misuses. FASEBJ. 1997;11:835–841. [PubMed] [Google Scholar]
- 44.Hey T, Lipps G, Sugasawa K, Iwai S, Hanaoka F, Krauss G. The XPC-HR23B complex displays high affinity and specificity for damaged DNA in a true-equilibrium fluorescence assay. Biochemistry. 2002;41:6583–6587. doi: 10.1021/bi012202t. [DOI] [PubMed] [Google Scholar]
- 45.Krasikova YS, Rechkunova NI, Maltseva EA, Anarbaev RO, Pestryakov PE, Sugasawa K, Min JH, Lavrik OI. Human and yeast DNA damage recognition complexes bind with high affinity DNA structures mimicking in size transcription bubble. J Mol Recognit. 2013;26:653–661. doi: 10.1002/jmr.2308. [DOI] [PubMed] [Google Scholar]
- 46.Tsao H, Mao B, Zhuang P, Xu R, Amin S, Geacintov NE. Sequence dependence and characteristics of bends induced by site-specific polynuclear aromatic carcinogen-deoxyguanosine lesions in oligonucleotides. Biochemistry. 1998;37:4993–5000. doi: 10.1021/bi980291c. [DOI] [PubMed] [Google Scholar]
- 47.Xu R, Mao B, Xu J, Li B, Birke S, Swenberg CE, Geacintov NE. Stereochemistry-dependent bending in oligonucleotide duplexes induced by site-specific covalent benzo[a]pyrene diol epoxideguanine lesions. Nucleic Acids Res. 1995;23:2314–2319. doi: 10.1093/nar/23.12.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shell SM, Hawkins EK, Tsai MS, Hlaing AS, Rizzo CJ, Chazin WJ. Xeroderma pigmentosum complementation group C protein (XPC) serves as a general sensor of damaged DNA. DNA Repair (Amst) 2013;12:947–953. doi: 10.1016/j.dnarep.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naegeli H, Sugasawa K. The xeroderma pigmentosum pathway: decision tree analysis of DNA quality. DNA Repair (Amst) 2011;10:673–683. doi: 10.1016/j.dnarep.2011.04.019. [DOI] [PubMed] [Google Scholar]
- 50.Sidorenko VS, Yeo JE, Bonala RR, Johnson F, Scharer OD, Grollman AP. Lack of recognition by global-genome nucleotide excision repair accounts for the high mutagenicity and persistence of aristolactam-DNA adducts. Nucleic Acids Res. 2012;40:2494–2505. doi: 10.1093/nar/gkr1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sugasawa K, Shimizu Y, Iwai S, Hanaoka F. A molecular mechanism for DNA damage recognition by the xeroderma pigmentosum group C protein complex. DNA Repair (Amst) 2002;1:95–107. doi: 10.1016/s1568-7864(01)00008-8. [DOI] [PubMed] [Google Scholar]
- 52.Naegeli H, Bardwell L, Friedberg EC. The DNA helicase and adenosine triphosphatase activities of yeast Rad3 protein are inhibited by DNA damage. A potential mechanism for damage-specific recognition. J Biol Chem. 1992;267:392–398. [PubMed] [Google Scholar]
- 53.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.