Summary
Membrane phospholipids typically contain fatty acids (FAs) of 16 and 18 carbon atoms. This particular chain length is evolutionarily highly conserved and presumably provides maximum stability and dynamic properties to biological membranes in response to nutritional or environmental cues. Here, we show that the relative proportion of C16 versus C18 FAs is regulated by the activity of acetyl-CoA carboxylase (Acc1), the first and rate-limiting enzyme of FA de novo synthesis. Acc1 activity is attenuated by AMPK/Snf1-dependent phosphorylation, which is required to maintain an appropriate acyl-chain length distribution. Moreover, we find that the transcriptional repressor Opi1 preferentially binds to C16 over C18 phosphatidic acid (PA) species: thus, C16-chain containing PA sequesters Opi1 more effectively to the ER, enabling AMPK/Snf1 control of PA acyl-chain length to determine the degree of derepression of Opi1 target genes. These findings reveal an unexpected regulatory link between the major energy-sensing kinase, membrane lipid composition, and transcription.
Highlights
-
•
AMPK/Snf1 inhibition of acetyl-CoA carboxylase controls fatty acyl-chain length
-
•
Opi1 repressor preferentially binds to C16 rather than C18 acyl-chains in PA
-
•
Acyl-chain length tunes Opi1 sequestration to the ER and target gene derepression
-
•
AMPK/Snf1 thus uses its effect on acyl-chain length to control Opi1 target genes
Hofbauer et al. find that yeast Snf1/AMPK regulation of acetyl-CoA carboxylase affects acyl-chain length (C16 versus C18) in membrane phospholipids. Moreover, preferential binding of the transcriptional repressor Opi1 to C16-containing phosphatidic acid controls Opi1 sequestration and target gene derepression. Snf1/AMPK regulation of membrane-lipid composition thus determines a key transcriptional output.
Introduction
The biogenesis of biological membranes that delineate the cell surface or act as intracellular space partition is a fundamental biological process and a remarkable evolutionary achievement. The hydrophobic barrier of membranes is comprised of fatty acid (FA)-derived acyl-chains, typically 16 or 18 carbon atoms in length; these FAs are synthesized in a cyclic series of reactions by the multifunctional FA synthase (FAS) complex by the addition of C2 units that are derived from malonyl-coenzyme A (CoA) (Smith et al., 2003). Remarkably, the FA chain length in membrane phospholipids (PLs) is evolutionarily highly conserved (Daum et al., 1999; Schneiter et al., 1999; Alex Brown, 2012; Han et al., 2012) and apparently provides maximum stability to the otherwise fluid bilayer structure through their hydrophobic interactions. At the same time, this acyl-chain composition permits rapid and significant adjustments by a single elongation step (C16→C18) and/or desaturation (C16:0→C16:1; C18:0→C18:1) in response to alterations in environmental conditions such as temperature. Furthermore, gradual increase in chain length and, subsequently, membrane thickness along the secretory pathway is an important determinant and provides sufficient differentiation for protein sorting (Bretscher and Munro, 1993). Similar to mammalian cells, yeast membranes predominantly consist of mono- and di-unsaturated PLs containing C16 and C18 acyl-chains to maintain the liquid crystalline state at physiological temperature (de Kroon et al., 2013). A decrease in the ambient temperature primarily triggers an increase in the C16/C18 ratio rather than a change in the degree of acyl-chain desaturation (Martin et al., 2007).
The mechanisms that control the ratio between C16 versus C18 FAs in cellular lipids are unknown. FAs are synthesized de novo by the FAS complex, which typically limits acyl-chain length extension to C16 and C18 carbon atoms, both in yeast and in mammals (Okuyama et al., 1979; Wakil et al., 1983); FAs are subsequently channeled through the common precursor phosphatidic acid (PA) into the synthesis of neutral and polar lipid species, i.e., storage triacylglycerols (TAG) and membrane PLs, respectively. The substrate for the FAS complex, malonyl-CoA, is synthesized by the multifunctional enzyme, acetyl-CoA carboxylase (Acc1), the initial and rate-limiting step in cellular FA de novo synthesis in yeast (Al-Feel et al., 1992; Chirala et al., 1994; Hasslacher et al., 1993; Woods et al., 1994) and in mammals (for a review, see Wakil and Abu-Elheiga, 2009).
Acc1 activity is tightly regulated at multiple levels. Nutritional and metabolic signals, such as glucose limitation and salt stress, are transduced to Acc1 by the Snf1 kinase, the yeast ortholog of mammalian AMP-activated protein kinase, AMPK (Carling et al., 1994; Hardie et al., 1998; Hedbacker and Carlson, 2008). Snf1 is the major energy-sensing kinase that phosphorylates and inactivates Acc1 in vivo under conditions of low energy load (Woods et al., 1994). Thus, yeast mutants lacking Snf1 kinase display 3-fold elevated Acc1 activity; in addition, snf1 mutants are defective in transcription of multiple genes including INO1, which encodes inositol-3-phosphate synthase and represents the most highly regulated gene of PL biosynthesis (Galdieri and Vancura, 2012; Jesch et al., 2005; Santiago and Mamoun, 2003; Shirra et al., 2001). Inositol-3-phosphate synthase is indispensable for the endogenous production of inositol, which makes snf1 mutants dependent on inositol supplementation for growth (Ino− phenotype).
Notably, transcription of ACC1 and PL biosynthetic genes, including INO1, is strongly regulated by inositol in the growth media, which is mediated by the UASINO element in the promoter of these genes (Carman and Han, 2009; Henry et al., 2012, 2014). Full activation of INO1 transcription occurs in the absence of inositol and requires the Ino2/Ino4 transcriptional activator complex that binds to the UASINO element (Ambroziak and Henry, 1994; Lopes and Henry, 1991). UASINO-containing genes are repressed by Opi1 (White et al., 1991) by its interaction with Ino2 in the nucleus (Wagner et al., 2001). Opi1 is a PA-binding protein and shuttles between the endoplasmic reticulum (ER) and the nucleus, dependent on the level of PA and the presence of the yeast VAMP-associated protein (VAP) homolog Scs2, in the ER. In the absence of inositol, PA levels are high, Opi1 is bound to the ER, and transcription of INO1 is enabled, leading to de novo production of inositol. In contrast, supplementation of inositol to the growth medium depletes PA by draining it into phosphatidylinositol (PI) synthesis, which promotes translocation of Opi1 into the nucleus and repression of the INO1 gene (Loewen et al., 2004; Young et al., 2010). PA is a central lipid intermediate and precursor both for membrane PL and storage TAG synthesis; however, it is currently unclear how the metabolic flux either way is regulated in growing cells. Defects in PL metabolism that lead to an increased steady-state concentration of PA in the ER cause sequestration of the Opi1 repressor to the ER and elevated transcription of UASINO-dependent genes. The cho2 and opi3 mutants, for instance, are defective in PL methylation and, thus, accumulate PA and overexpress INO1 leading to overproduction and excretion of inositol into the growth medium (Opi− phenotype; Greenberg et al., 1982; McGraw and Henry, 1989; Summers et al., 1988).
We previously showed that the inositol requirement of the snf1 mutant is suppressed by downregulation of Acc1, suggesting that its activity might be connected to the Ino2/Ino4/Opi1 regulatory circuit (Shirra et al., 2001). However, due to the pleiotropic defects of the snf1 mutant, other regulatory mechanisms could not be ruled out. For instance, lack of INO1 expression in the snf1 mutant was suggested to be a result of defective chromatin modification (Lo et al., 2001). To uncouple Acc1 activity from Snf1-dependent phosphorylation, we have now characterized and mutated the specific phosphorylation site at position serine 1157 of Acc1. We provide evidence for a direct regulatory function of FA de novo synthesis in the transcriptional regulation of PL biosynthesis through the Ino2/Ino4/Opi1 regulatory circuit. We show that Acc1 activity is a critical determinant of the C16/C18 acyl-chain ratio in cellular lipids; in conjunction with the specific binding preference of the Opi1 repressor for C16-PA species, this describes a regulatory mechanism of gene transcription in response to altered acyl-chain length in cellular lipids and unveils a function of the Snf1/AMP-activated protein kinase in regulating lipid homeostasis.
Results
Acc1 Is Phosphorylated by Snf1 Kinase at Serine 1157
Acc1 is one of the few direct enzymatic targets of Snf1 kinase. A large-scale phosphoproteome analysis identified a single phosphorylation site at serine 1157 of Acc1 that is embedded in a “MNRAVSVSDLS” sequence and matches the AMPK consensus phosphorylation motif, R/K-X-X-S (Brinkworth et al., 2006; Dale et al., 1995). Using avidin affinity-purified Acc1 protein, we characterized and confirmed the phosphorylation site by qualitative and quantitative mass spectrometry analyses (Figure 1; Figure S1 available online; see Supplemental Experimental Procedures for a detailed description). In wild-type, about 63% of the Acc1 protein was phosphorylated at serine 1157, suggesting that the majority of the enzyme is rather inactive in logarithmically growing yeast cells. However, in snf1 cells, the phosphorylation level at this residue in the Acc1 protein decreased by half to about 35%, confirming serine 1157 as the target site of Snf1 phosphorylation. Alternative phosphorylation of serine 1159, which was recently predicted to be a potential phosphorylation site (Oliveira et al., 2012), or phosphorylation of any other sites was not detected in our phosphoproteome analyses of log-phase yeast cells.
Figure 1.
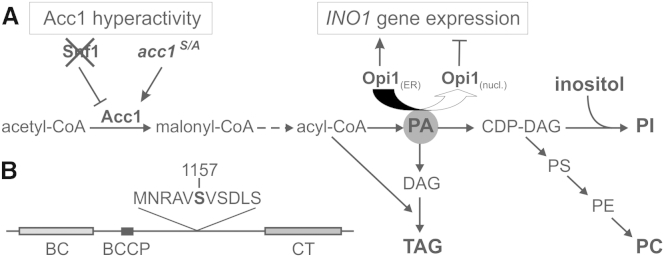
Role of Acc1 in Lipid Metabolism
(A) Metabolic pathway for FA and lipid synthesis. Acc1, which is negatively regulated by Snf1 kinase, catalyzes the rate-limiting step in FA de novo production. The level of PA in the ER is responsible for transcriptional regulation of many genes involved in PL synthesis, in particular INO1, by binding and sequestering the repressor Opi1 at the nuclear ER.
(B) Schematic view of the yeast Acc1 protein (2,233 amino acids). The enzyme consists of three domains: the biotin carboxylase domain (BC, amino acids [aa] 58–567), the biotin-carboxyl carrier protein (BCCP, aa 701–767) and the carboxyltransferase domain (CT, aa 1603–2101). The serine residue at position 1157 is the phosphorylation site for Snf1 kinase, which is located in an R/K-X-X-S consensus motif that is present in many Snf1 substrates.
See also Figure S1.
The acc1S/A Mutant Exhibits Inositol Auxotrophy and TAG Accumulation
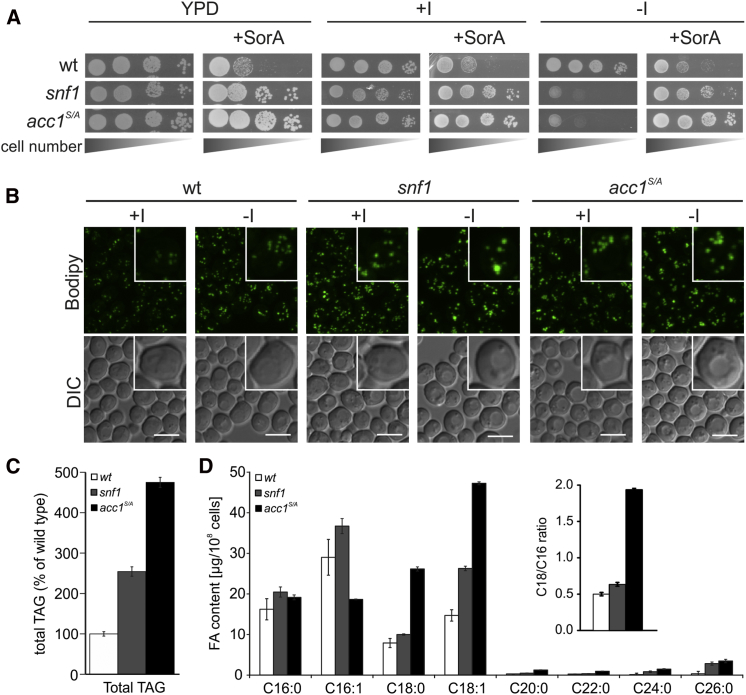
To assess its in vivo function, we mutated serine 1157 to an alanine residue, thus locking the Acc1 enzyme in a nonphosphorylatable state. As shown in Figure 2A, the acc1S/A mutant, generated in this fashion, grew like wild-type on complete yeast extract peptone dextrose (YPD) media, but similarly to the snf1 mutant, it was resistant to the Acc1-specific inhibitor, Soraphen A (SorA) (Shen et al., 2004; Vahlensieck et al., 1994). Increased resistance to SorA indicates hyperactivity of Acc1 in these strains. In addition, acc1S/A mutants, like the snf1 mutant strain, were unable to grow on media lacking inositol (−I medium). This inositol auxotrophy (Ino− phenotype) could be suppressed either by decreasing Acc1 activity with SorA (Figure 2A) or in the snf1 mutant by downregulation of ACC1 transcription under control of the heterologous doxycycline-repressible tet07 promoter (tetO7-ACC1; Figure S2A). Yeast snf1 mutants were originally identified by their inability to utilize exogenous sucrose (Snf−, sucrose nonfermenting phenotype), resulting from their failure to express the SUC2 gene (encoding invertase) on glucose depletion (Carlson et al., 1981). Notably, the acc1S/A mutant was capable of growing on sucrose as the sole carbon source or on nonfermentable carbon sources like glycerol or ethanol, which distinguishes its phenotype from that of snf1 mutants with respect to carbon source utilization (Figures S2B and S2C). Acc1 hyperactivity in both snf1 and acc1S/A mutants leads to highly elevated FA production and massive accumulation of TAG and lipid droplets during logarithmic growth (Figures 2B and 2C); these phenotypes are even more pronounced in stationary phase (Figure S2D). Most notably, as a result of elevated malonyl-CoA production by hyperactive Acc1 in the acc1S/A mutant, FA acyl-chain distribution was significantly shifted toward longer acyl-chain lengths, increasing the C18:C16 acyl-chain ratio from ∼0.5 in wild-type to ∼2.0 in the acc1S/A mutant (Figure 2D). This shift is also reflected in the altered acyl-chain composition of virtually all glycerolipid classes—for instance, TAG (Figure S2E). Taken together, the acc1S/A mutant displays similar phenotypes as the snf1 mutant with respect to FA overproduction, a shift to longer acyl-chains, and TAG accumulation, as well as inositol auxotrophy.
Figure 2.
Increased Acc1 Activity Leads to Inositol Auxotrophy and Accumulation of TAG and Lipid Droplets
(A) Growth of wild-type (WT), snf1, and acc1S/A strains was monitored on rich media plates (YPD) and inositol-containing (+I) or inositol-free (−I) media plates. Cells were spotted on to the indicated media and grown for 2 days at 30°C. The concentration of Soraphen A (SorA) was 1 μg/ml.
(B–D) Cells were precultured logarithmically for 20 hr in +I medium.
(B) Visualization of lipid droplets with Bodipy in cells grown logarithmically for 3 hr in fresh +I or −I medium. Scale bar, 5 μm.
(C) Total TAG levels were measured in cells grown logarithmically for 3 hr on inositol-containing (+I) medium and normalized to wild-type levels. Results are presented as means ± SD; n = 3.
(D) Total FA content and composition was measured in cells grown as in (C). Results are presented as mean ± SD; n = 3.
See also Figure S2.
Most notably, growth on nonfermentable carbon sources clearly distinguishes the acc1S/A mutant from the pleiotropic phenotypes that are associated with deletion of SNF1 and uncouples Acc1 activity from other roles of Snf1 kinase in energy and carbon metabolism.
Acc1 Activity Regulates Transcription of INO1
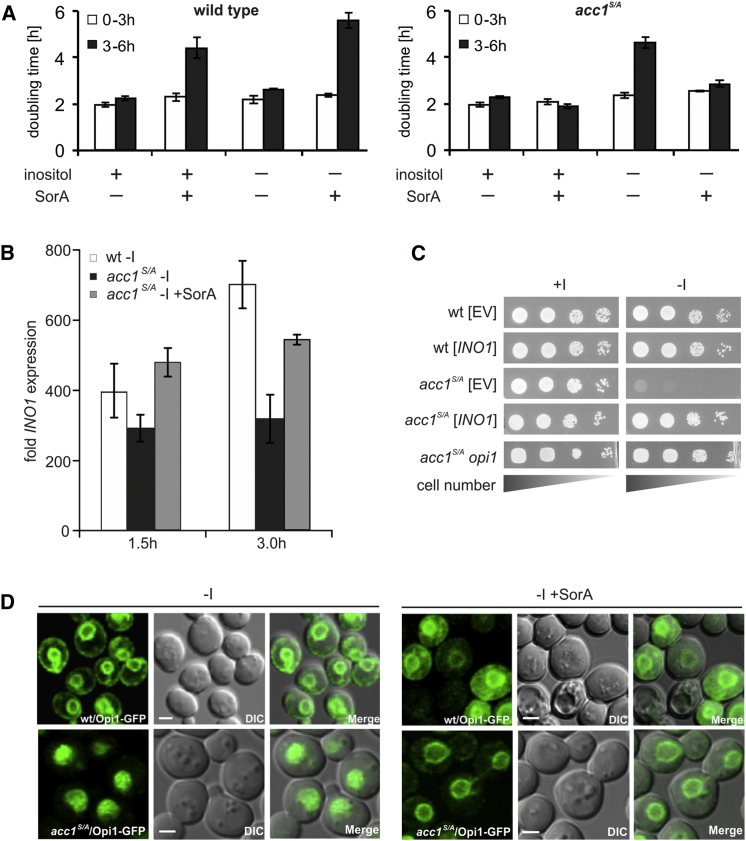
The observed growth defect of acc1S/A mutants on solid media lacking inositol was reproduced in liquid cultures. When cultivated in the presence of inositol (+I medium), the acc1S/A mutant grew at a rate identical to that of wild-type cells (Figure 3A; Figure S3A). After the shift to inositol-free medium, only a slightly reduced growth rate of the acc1S/A mutant strain was observed during the first 3 hr compared to wild-type, most likely due to intracellular inositol pools that are consumed during that period (Figure 3A; see also Figure S3B for detailed inositol titration experiment). However, a dramatic increase in doubling time of the acc1S/A mutant occurred between 3 and 6 hr after the shift to inositol-free medium (Figure 3A). Notably, cells eventually reached wild-type-like final cell density after 20 hr, suggesting that cells are impaired in growth but remain viable (Figure S3A). This reduced growth of the acc1S/A mutant in the absence of inositol was cured by inhibition of Acc1 by SorA, clearly linking the requirement for inositol supplementation to Acc1 activity. Wild-type cells treated with the same concentration of SorA, on the other hand, exhibited a severe growth defect in both the presence and absence of inositol, which is consistent with the essential role of Acc1 in cellular metabolism (Figure 3A).
Figure 3.
Inositol Auxotrophy of acc1S/A Is Linked to the Ino2/Ino4/Opi1 Regulatory Circuit
In each experiment, cells were precultured logarithmically for 20 hr in inositol-containing (+I) medium and shifted to fresh medium, as indicated. The concentration of Soraphen A (SorA) was 1 μg/ml.
(A) Doubling times of wild-type and the acc1S/A mutant between 0 and 3 and from 3 to 6 hr of growth. Results are presented as mean ± SD; n = 3.
(B) INO1 expression kinetics of wild-type and acc1S/A strains were measured at indicated time points, and values were normalized to wild-type grown for 1.5 hr on medium containing inositol (INO1-repressing conditions). ACT1 gene served as internal control. Results are presented as mean ± SD; n = 3.
(C) Cells were grown overnight in YPD medium at 30°C and washed twice with sterile water. Five-microliter aliquots of dilutions of OD600 = 1, 0.1, 0.01, and 0.001/ml were spotted on to the indicated plates and incubated at 30°C for 2 days. [EV] – cells transformed with empty vector, [INO1] – cells transformed with INO1 wild-type gene under the regulation of the TEF1 promoter.
(D) Localization of Opi1-GFP fusion protein was determined in wild-type and acc1S/A strains shifted for 3 hr to inositol-free (−I) medium in the presence or absence of Soraphen A (SorA). Scale bar, 2 μm.
See also Figure S3.
We next asked whether the poor growth rate of the acc1S/A mutant in the absence of exogenous inositol was indeed associated with a reduced de novo production of that essential metabolite, i.e., at the level of transcription of the INO1 gene. In wild-type, as expected, INO1 transcription was highly induced 3 hr following the shift to inositol-free medium; in contrast, expression of INO1 was significantly attenuated in the acc1S/A mutant strain. Consistent with the observed restoration of growth in the absence of inositol, inhibition of Acc1 in the acc1S/A mutant by SorA also restored wild-type-like INO1 expression kinetics (Figure 3B). These data support the notion that lack of growth of the acc1S/A (and snf1) mutant on media lacking inositol is linked to a defect in INO1 expression as a consequence of Acc1 hyperactivity. Indeed, overexpression of INO1 from a high-copy-number plasmid or deletion of the OPI1 gene fully restored growth of the acc1S/A strain in the absence of inositol (Figure 3C and Figure S3C). In addition, downregulation of ACC1 gene expression in the tetO7ACC1 strain by 0.5–3.0 μg/ml doxycycline led to overproduction and secretion of inositol (Opi− phenotype; Figure S4A), clearly demonstrating the inverse correlation between Acc1 activity and the ability to express INO1 (Figure S4B). Full repression of ACC1 transcription (>5.0 μg/ml doxycycline) led to cell death of the tetO7-ACC1 strain, further supporting the essential function of Acc1.
From previous studies, it is known that INO1 expression responds to the localization of Opi1, the transcription factor that senses the levels of the central lipid intermediate, PA, in the ER (Loewen et al., 2004). Consistent with the current model, Opi1-green fluorescent protein (GFP) translocated in wild-type cells from the nucleus to the nuclear and peripheral ER within 3 hr in inositol-free medium, leading to the observed rapid INO1 induction. In marked contrast, the acc1S/A mutant retained nuclear localization of Opi1-GFP after the shift to inositol-free medium (Figure 3D); this defect in Opi1-GFP export from the nucleus parallels the reduced INO1 expression kinetics. Notably, inhibition of Acc1 activity in the acc1S/A mutant readily promoted export of Opi1-GFP from the nucleus, leading to the observed relief of INO1 expression (Figures 3B and 3D). Taken together, these analyses demonstrate that this growth deficit of the acc1S/A mutant is due to impaired INO1 expression through the Ino2/Ino4/Opi1 regulatory circuit.
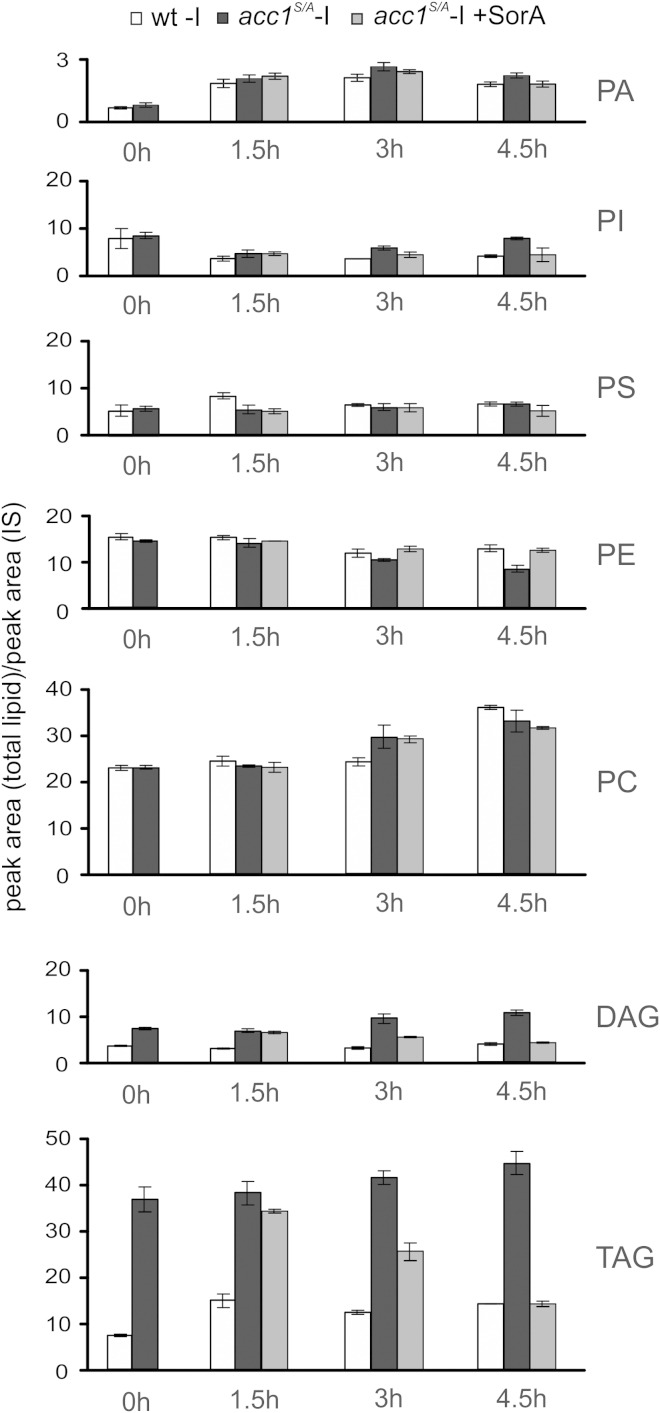
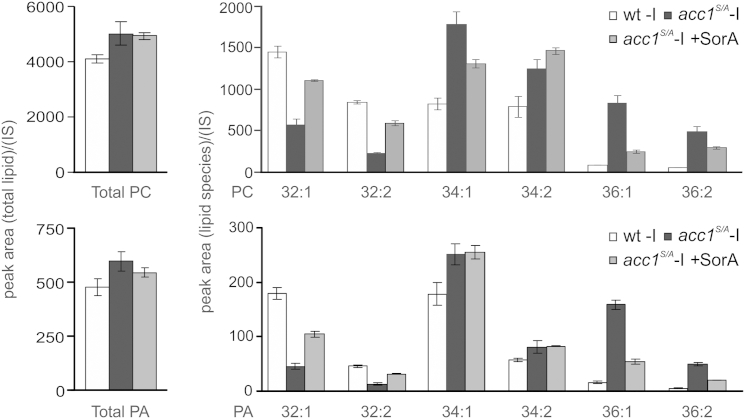
Since Opi1 localization responds to the amount of PA in the ER (Loewen et al., 2004), we hypothesized that overproduction of FA might lead to reduced cellular PA levels, perhaps due to its increased channeling into TAG synthesis. This, however, was not the case: analysis of the lipid profile in the acc1S/A mutant showed that total glycerophospholipid content remained almost unaltered compared to wild-type. Diacylglycerol (DAG) and TAG levels were raised about 3- to 4-fold, which was reversed by SorA treatment within 4.5 hr (Figure 4). Thus, elevated de novo synthesized FAs are preferentially channeled into TAG synthesis, without affecting total PL content. This observation is surprising since both pathways share the common intermediate, PA. We next analyzed to what extent cellular PA content was altered as a result of hyperactive Acc1 in the acc1S/A mutant. Surprisingly, the total PA pool was not decreased but rather slightly increased compared to wild-type (Figure 5). This finding is inconsistent with the proposed mechanism by Loewen et al., that reduced PA levels are responsible for translocation of Opi1 from the ER into the nucleus to repress Ino2/Ino4-dependent INO1 transcription (Loewen et al., 2004). Since the molecular species distribution in all glycerolipids displayed a characteristic shift toward increased acyl-chain length in acc1S/A, we hypothesized that not the total amount of PA but rather its specific molecular acyl-chain composition may define its binding affinity to Opi1. Thus, we next analyzed the molecular species composition of PA (and of the major cellular PL phosphatidylcholine [PC] as a reference) in wild-type and acc1S/A strains in greater detail. As shown in Figure 5, PA 32:1 (containing C16:0 and C16:1 acyl-chains) and PA 32:2 (containing two C16:1 acyl-chains) species were indeed greatly reduced in the acc1S/A mutant, whereas PA 36:1 (containing C18:0 and C18:1 acyl-chains) and PA 36:2 (containing two C18:1 acyl-chains) species were increased. This shift in acyl-chain composition was reversed by the addition of SorA, a condition that also led to inositol prototrophy (Figures 2A and 3A), derepression of INO1 (Figure 3B), and nuclear export of Opi1-GFP (Figure 3D) in the acc1S/A strain.
Figure 4.
Modulation of Acc1 Activity Affects Neutral Lipid Levels but Renders Membrane PL Content Unaltered
Cells were precultured logarithmically for 20 hr in inositol-containing (+I) medium and shifted to medium lacking inositol (−I). The concentration of Soraphen A (SorA) was 1 μg/ml. Total lipid content and composition were analyzed at indicated time points. Lipid amounts were normalized to an internal standard (IS) as described in the Experimental Procedures. PS, phosphatidylserine; PE, phosphatidylethanolamine. Results are presented as mean ± SD; n = 3.
See also Figure S4.
Figure 5.
Acc1 Hyperactivity Affects Acyl-Chain Composition of PL Classes
Cells were precultured logarithmically for 20 hr in inositol-containing (+I) medium. Total content and species composition of PC and PA were determined in cells shifted for 3 hr to fresh inositol-free (−I) medium with or without SorA (1 μg/ml) supplementation. Lipid amounts were normalized to an internal standard (IS) as described in the Experimental Procedures. Results are presented as mean ± SD; n = 3.
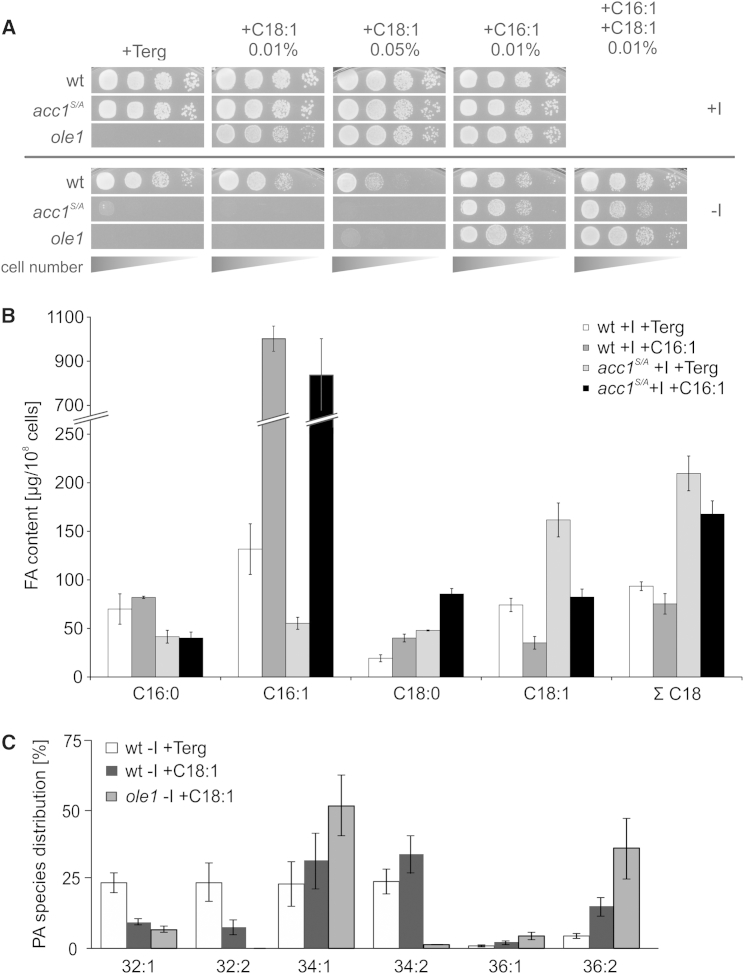
Oleic Acid Supplementation Induces Inositol Auxotrophy in Wild-Type and ole1 Strains
Based on the hypothesis that an increase in the proportion of C18 versus C16 acyl-chains in PA is responsible for nuclear localization of Opi1, we next tested whether exogenous C18:1 supply and concomitant changes in the acyl-chain composition would phenocopy the effects of the hyperactive acc1S/A allele also in wild-type cells. As shown in Figure 6A, addition of C18:1 indeed induced inositol auxotrophy in wild-type cells. This phenotype was not observed in the presence of palmitoleic acid (C16:1) and, thus, cannot be due to a general “lipotoxic” effect of supplied FA on inositol-free media. Similarly, the ole1 mutant, which lacks the sole FA desaturase in yeast (Stukey et al., 1989), and which is dependent on unsaturated FA supplementation, displayed a strict inositol auxotrophy in the presence of C18:1. Notably, growth of wild-type, ole1, and acc1S/A strains on media lacking inositol was fully rescued by (co)supplementation with C16:1, indicating that this FA reversed the inositol auxotrophy induced by excess C18:1 FA. This rescue of the acc1S/A mutant growth was not due to repression of Acc1 activity by C16:1, since total C18 FAs remained at a high level, characteristic for Acc1 hyperactivity; rather, the ratio between C16 and C18 FAs was restored by C16:1 supplementation (Figure 6B). The analysis of PA species in wild-type and ole1 strains supplemented with C18:1 showed a similar shift toward longer acyl-chain lengths as in the acc1S/A strain as compared to wild-type grown without C18:1 (Figure 6C): PA 32:1 and PA 32:2 species were significantly decreased, whereas PA species with longer FA chain length increased, especially 36:2.
Figure 6.
Impact of FA Supplementation on Inositol Auxotrophy
(A) Growth of wild-type, acc1S/A, and ole1 strains was monitored on media plates with (+I) or without (−I) inositol plus indicted FAs. Oleic acid (C18:1) and palmitoleic acid (C16:1) were dissolved in tergitol and supplemented as indicated. The final concentration of tergitol was 1%. Dilution series of cells were spotted on to the indicated plates and grown for 2 days at 30°C.
(B) Effect of C16:1 supplementation on the FA composition of wild-type and acc1S/A strains. Cells were precultured logarithmically for 20 hr in +I medium. FA content was measured after growing cells for 3 hr in fresh +I medium with or without 0.05% C16:1 supplementation. The final concentration of tergitol was 1%. Results are presented as mean ± SD; n = 3.
(C) Impact of C18:1 supplementation on PA species composition. Cells were precultured logarithmically for 20 hr in MES-buffered +I medium with 0.01% C18:1. PA species were measured after growing cells for 6 hr in MES-buffered −I medium with or without 0.01% C18:1 supplementation. The final concentration of tergitol was 1%. Results are presented as mean ± SD; n = 3.
The Opi1 Repressor Preferentially Binds Shorter-Chain and More-Saturated PA Species
To further confirm whether the acyl-chain composition of PA affects the interaction with Opi1, we performed in vitro binding assays with PC 36:2 liposomes as a matrix, containing 20 mol% of different PA species. Indeed and in line with our hypothesis, we found significantly different binding affinities of Opi1 for PA species dependent on the acyl-chain composition. Liposomes containing saturated PA species (PA 32:0 and PA 36:0) showed the highest affinity, followed by liposomes containing 32:1 PA; liposomes containing PA 36:2 showed the lowest binding affinity for purified Opi1. Liposomes, which consisted solely of PC 36:2, showed no Opi1 binding at all (Figure 7). These results clearly demonstrate that the acyl-chain composition of PA is a major determinant of its interaction with the transcriptional repressor Opi1. This preferred binding affinity explains the observed phenotypes and significantly extends the current model of the transcriptional regulation of PL biosynthesis in yeast in response to deregulated FA de novo synthesis.
Figure 7.
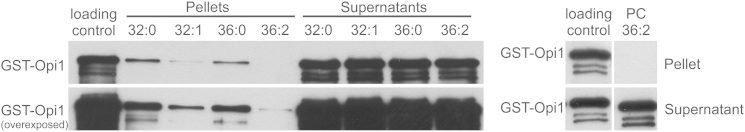
Opi1 Shows Preferred Binding toward Shorter-Chain and More-Saturated PA Species
Binding assays were performed with purified GST-tagged Opi1 toward different PA species embedded in liposomes consisting of 80 mol% di-oleoyl-PC (PC 36:2) and 20 mol% PA as indicated (left panel). Pellets represent the bound GST-Opi1 fraction and the supernatants represent the unbound GST-Opi1 fraction. An identical aliquot of GST-Opi1 as for the binding assays was used as loading control (100% input), and a liposome consisting solely of PC 36:2 served as negative control for Opi1 binding (right panel). The western blot shows a representative result of three independent experiments.
Discussion
Throughout the eukaryotic kingdom, FAs of 16 or 18 carbon atoms in length are the preferred acyl components in all glycerolipids that serve as bulk lipids in biological membranes or as storage lipids. Whereas mechanisms controlling the level of FA desaturation in response to ambient temperature and membrane fluidity have been described in detail, much less is known about the mechanisms regulating chain length distribution (de Kroon et al., 2013). In addition to their fundamental roles as components of biological membranes, FAs also serve essential functions as energy substrates, signaling molecules, or protein modifiers. Cellular FA synthesis is also coordinated with nutritional supply, activation, and incorporation into complex lipids to support proper cell function and to prevent potentially detrimental FA-induced lipotoxic effects, which are believed to be causative for lipid-associated disorders, such as obesity and type 2 diabetes in humans (Kim and Ye, 2013; Unger, 2002; Vidal-Puig and Unger, 2010). Thus, endogenous de novo synthesis of particular FAs also plays an important role to counterbalance the physiological impact of nutritional FAs that may be incorporated into cellular lipids. According to their central role in cellular lipid synthesis, both Acc1 and FAS have emerged as potential drug targets in obesity-related diseases, as well as in cancer, which are characterized by deregulated FA and lipid metabolism (Wakil and Abu-Elheiga, 2009; Furuta et al., 2010; Pandey et al., 2012; Wang et al., 2010; Liu et al., 2010; Natter and Kohlwein, 2013).
In yeast and mammalian cells, acyl-chain length is largely determined by the FAS complex that limits acyl-chain length to 16 and 18 carbon atoms (Okuyama et al., 1979). We now show that the activity of Acc1, the initial and rate-limiting enzyme of de novo FA synthesis, is a major determinant of total cellular FA production and also has a fundamental impact on the relative distribution of C16 and C18 acyl-chains in cellular lipids. Overproduction of FAs leads to the accumulation of TAG and proliferation of cytosolic lipid droplets, whereas membrane PL content remains unaltered. On the other hand, the shift in acyl-chains toward longer residues is reflected in all cellular glycerolipids, with consequences for the binding and sequestration of the transcriptional repressor Opi1 to the ER.
The synthesis of FAs is a major energy- and carbon-consuming pathway and, therefore, under several levels of control. One level involves the expression of the ACC1 gene, which is coordinately regulated with PL synthesis by the PL precursors, inositol and choline, and the positive and negative transcription factors Ino2/Ino4 and Opi1, respectively (Hasslacher et al., 1993; Henry et al., 2012). The regulation of ACC1 expression in coordination with PL biosynthetic genes (for review, see Tehlivets et al., 2007) suggests that FA production must also be balanced with TAG formation in growing cells in order to sustain requirements for PL and membrane biogenesis. In addition, the activity of Acc1 is strongly regulated by Snf1-dependent phosphorylation (Mitchelhill et al., 1994; Woods et al., 1994), which, in turn, depends on the energy status of the cell (Wilson et al., 1996).
Here, we made use of a yeast mutant that lacks the single Snf1 phosphorylation site in Acc1 at serine 1157 (acc1S/A) and exhibits phenotypes very similar to snf1 mutants with respect to TAG and lipid droplet accumulation as well as inositol auxotrophy, demonstrating that these snf1 phenotypes are caused by Acc1 hyperactivity. The evidence we present here strongly supports the hypothesis that the inositol-requiring phenotype of the acc1S/A strain is due to decreased INO1 expression that is controlled by the well-characterized Ino2/Ino4/Opi1regulatory circuit (Henry et al., 2012; Loewen et al., 2004), as both deletion of OPI1, which uncouples transcriptional repression of INO1 from cellular PA levels, and overexpression of INO1 fully restored inositol prototrophy to the acc1S/A mutant strain. We further suggest that PL synthesis may be limited by the coordinated repression of the UASINO-containing PL biosynthetic genes in the presence of excess FA. The observed Opi1 translocation to the nucleus concomitantly slows cell growth as PL and inositol production become limiting due to PA diversion into TAG accumulation. Most notably, neither increased endogenous FA production nor exogenous FA supply led to increased PL accumulation, despite the fact that an altered acyl-chain composition is reflected in all PL classes; instead, excess FAs are preferentially channeled into TAG. In addition, the similarity of the cellular response to these two situations suggests that functional TAG synthesis is crucial for accommodating these FAs, thus avoiding the buildup of potential signal molecules, such as PA or DAG, or free FAs (Garbarino and Sturley, 2009; Kohlwein, 2010; Kohlwein and Petschnigg, 2007; Petschnigg et al., 2009; Fakas et al., 2011).
Importantly, our evidence significantly extends the current model that indeed the acyl-chain composition of PA is the critical determinant of Opi1 sequestration to the ER. We show that alterations in the acyl-chain composition of the acc1S/A strain or of wild-type and the ole1 mutant in the presence of oleic acid lead to inositol auxotrophy due to the reduced binding affinity of the Opi1 repressor to PA molecular species containing longer acyl residues. We hypothesize that the recognition of the negatively charged head group of PA by the basic tract of Opi1 might only represent the first level of interaction, whereas the second level is presumably determined by sensing the nature of the acyl-chains of PA within the ER membrane. Several other studies also support the importance of acyl-chain composition of PA for interaction with proteins (Kooijman and Burger, 2009); for example, the binding specificity of human protein phosphatase 1 (PP1cγ) to PA species harboring a higher degree of acyl-chain unsaturation (Jones and Hannun, 2002). However, no consensus sequence motif for PA binding proteins has been identified yet (Stace and Ktistakis, 2006). It should be noted that the acidic PLs PA and PI are exceptional regarding their membrane activity compared to other PLs with respect to impact on membrane packing, electrostatics, and membrane curvature (Bigay and Antonny, 2012). PA is the only anionic PL with a pronounced cone shape, facilitating protein penetration into the membrane bilayer and recognition of the acyl moieties (Kooijman et al., 2007; Shin and Loewen, 2011).
The actual concentration of PA in the yeast ER membrane is not well defined. Previous studies reported PA levels between 0.5% and 3% of total PLs in isolated microsomal fractions (Zinser et al., 1991; Pichler et al., 2001), whereas a recent shotgun lipidomics study found about 10% PA of total lipids in whole cell extracts (Klose et al., 2012). These findings indicate that the short-lived intermediate PA may be metabolized during cell fractionation and may indeed be present in higher amounts than previously suggested. On the other hand, sole variation of the acyl-chain composition of PA in the liposome binding assay eliminated influences of membrane electrostatics and curvature as well as of proteins, such as the yeast VAP Scs2, which is required for binding of Opi1 to PA in vivo (Loewen et al., 2004).
The results obtained in this study have a number of implications. (1) Currently, the mechanisms that regulate the total amount of cellular PLs and their specific acyl-chain composition in response to altered FA supply are largely unclear. A mechanism, as described here, that responds not only to the content of critical lipid intermediates such as PA but also to its specific molecular composition provides an attractive regulatory circuit to integrate metabolic input parameters, such as availability of nutritional FAs. (2) FA synthesis itself is under several levels of control and has a profound impact on metabolic and transcriptional regulation. (3) The major and highly conserved energy-sensing Snf1 kinase operates through additional levels of regulation, namely, its metabolic downstream target Acc1. In conclusion, we demonstrate that Acc1 hyperactivity leads to overproduction of FAs and a shift in acyl-chain length distribution toward longer chains in all PL classes as well as in TAG. The acyl-chain distribution of PA is critical for its interaction with the Opi1 repressor, which displays reduced affinity for PA molecular species containing longer and more unsaturated acyl-chains. In the context of the snf1 mutant scenario, Acc1 hyperactivity is a potent player in sequestering carbon into fat production and regulating cell growth and proliferation. This unveils an impact of the major energy-sensing kinase on membrane lipid composition and function and may explain some of the seemingly unrelated pleiotropic consequences of its deficiency.
Experimental Procedures
Strains and Media
S. cerevisiae strains used in this study, except for the AID strain, are congenic with BY, a derivative of S288C, and are listed in the Supplemental Experimental Procedures. For liquid media experiments, cells were cultivated on a rotary shaker at 30°C and 180 rpm using threonine-free minimal synthetic defined media with (+I) or without (−I) 75 μM inositol (Villa-García et al., 2011). Solid YPD (1% yeast extract, 2% peptone and 2% glucose, 2% agar) and yeast extract peptone (1% yeast extract, 2% peptone, 2% agar) with 2% of the nonfermentable carbon sources sucrose, glycerol, ethanol, or 1% glycerol and 1% ethanol were additionally used for plate tests.
Culture Conditions and Calculation of Doubling Times
Cell cultivation—if not described differently—was performed as follows; a single colony of the desired strains was taken from a YPD plate and grown for 16 hr in +I medium (50 ml) to midlogarithmic growth phase, rediluted to optical density 600 (OD600) = 0.1/ml in 100 ml of fresh +I medium, and grown for an additional 4.5 hr maintaining midlogarithmical growth. A cell aliquot of OD600 = 20 was then shifted by filtration (pore size, 0.45 μm) to 40 ml of fresh +I and −I media, respectively, resulting in an initial OD600 = 0.5/ml (0 hr time point). Cell growth was monitored during growth, and doubling times were determined from the OD600 readings as described elsewhere (Gaspar et al., 2011).
For cultivation of the FA auxotrophic ole1 strain, the media for the 16 hr precultivation and the 4.5 hr growth after redilution contained 0.01% oleic acid and 1% tergitol. Additionally, 20 ml of a 1 M stock solution of MES, pH 6.0, was added per liter medium, resulting in an initial pH of 5.7. Wild-type cells were also precultivated the same way in this experiment. Cells were then shifted to fresh −I media containing 20 mM MES with 0.01% oleic acid and 1% tergitol or with solely 1% tergitol, as a control, and grown for 6 hr.
Acc1 activity was inhibited in vivo by adding 1 μg/ml SorA to the culture medium where indicated: SorA is highly specific for the eukaryotic form of acetyl-CoA carboxylase, and by its binding to the biotin carboxylase dimer interface, it inhibits oligomerization, which is required for enzyme activity (Shen et al., 2004).
Phenotypic Characterization
For plate tests, a single colony of the desired strains was taken from a YPD plate and grown overnight in YPD medium (for the ole1 strain, 0.01% oleic acid was supplemented) to reach late-logarithmic growth phase. A cell aliquot of OD600 = 10 was harvested and washed twice with sterile water. Five microliters of serial 1:10 dilutions starting with OD600 = 1/ml were spotted on to the indicated media plates. Plates were analyzed after 2 days of growth at 30°C.
To determine overproduction and excretion of inositol (Opi− phenotype), a sensitive bioassay was used (Greenberg et al., 1982): 5 μl OD600 (1 aliquot of wild-type, tetO7ACC1, and the control strains opi1Δ and cho2Δ) were spotted on −I medium, containing the indicated concentration of doxycycline. Cells were grown for 2 days at 30°C and sprayed with a suspension of the inositol auxotrophic (Ino−) tester strain (AID), which grows as a halo only around strains excreting inositol. Plates were analyzed after an additional 2 days of growth at 30°C.
For microscopic analysis, cultures were grown as described above and analyzed 3 hr after the shift or in stationary phase. Labeling of neutral lipids with BODIPY 493/503 or Nile Red was performed by adding the fluorescence dye directly to the cell culture for 10 min at room temperature in the dark (final concentration, 1 μg/ml). Refer to Supplemental Experimental Procedures for detailed microscope settings.
Lipid Analysis
Cells were grown as described above, and a cell aliquot of OD600 = 20 was harvested at the indicated time points after the media shift. Lipids were extracted according to a slightly modified Folch method (Folch et al., 1957; Schneiter and Daum, 2006) and analyzed on a UPLC-Synapt qTOF HDMS system (Waters), as described by Knittelfelder et al. (2014). For total FA analysis, an aliquot of the lipid extract was used for methyl ester production and gas chromatography-mass spectrometry measurements. Refer to Supplemental Experimental Procedures for lipid extraction, chromatography, and mass spectrometry parameters.
Relative Quantification of INO1 Gene Expression
Cultures were grown as described above and harvested 1.5 hr and 3 hr after the shift. Real-time PCR was conducted as described elsewhere (Gaspar et al., 2011). INO1 values were normalized to wild-type grown on +I media for 1.5 hr following the shift (INO1-repressing conditions). ACT1 levels served as an internal standard for normalization. Refer to the Supplemental Experimental Procedures for RNA isolation, reverse transcription, and real-time PCR parameters.
Opi1 In Vitro Binding Assays
For Opi1 binding assays, 150 μl of liposome suspension and 150 μl glutathione S-transferase (GST)-Opi1 were incubated for 30 min at 22°C and centrifuged at 30,000 rpm for 10 min at 22°C in an Optima TLX Ultracentrifuge using a TLA-100.3 fixed angle rotor (Beckmann). Supernatants were precipitated with 1 ml acetone at −20°C overnight for nonbinding controls. Pellets were washed once with elution buffer without glutathione, resuspended in 150 μl elution buffer without glutathione, and precipitated with 600 μl acetone at −20°C overnight. Additionally, 150 μl GST-Opi1 were precipitated with 600 μl acetone at −20°C overnight as a loading control. Precipitated GST-Opi1 protein was pelleted at 13,200 rpm for 10 min at 4°C in an Eppendorf centrifuge (Type 5415R). Pellets were washed once in 600 μl acetone and dissolved in protein loading buffer for SDS-PAGE. For detection of GST-Opi1 fusion protein, western blot analysis was performed using an anti-GST antibody from goat (Sigma-Aldrich, 1:10.000) and an anti-goat antibody conjugated with horseradish peroxidase (Pierce, 1:7.500) for detection with SuperSignal West Pico Chemiluminescent Substrate (Thermo Scientific).
Additional experimental information in expanded form on ACC1 in vitro mutagenesis, phospho-mass spectrometry, fluorescence microscopy, lipid analytics, INO1 expression, Opi1 overexpression and purification, liposome studies, and PA 32:1 synthesis is available in the Supplemental Experimental Procedures.
Acknowledgments
We thank Christoph Kurat, Olaf Merkel, Andreas Ulrich, and Julia Petschnigg for contributions in the early phase of the project; members of the Kohlwein and Henry laboratories for discussions; Dr. David L. Silver for critically reading the manuscript and helpful suggestions; Dr. Klaus Gerth (Helmholtz-Zentrum für Infektionsforschung, Braunschweig, Germany) for the gift of Soraphen A; and Dr. Jens Nielsen (Chalmers University of Technology, Gothenburg, Sweden) for providing the plasmid pSP-G1. H.F.H. was a member of the PhD program Molecular Enzymology, funded by the Austrian Science Funds FWF (project W901-B12). This work was supported by grants from NIH (project GM19629) to S.A.H.; the Austrian Federal Ministry for Science and Research, project GOLD: Genomics of Lipid-Associated Disorders, to S.D.K. and H.W.; the European Commission via the FP7 projects MEIOsys and Prime-XS to K.M.; and the Austrian Science Funds FWF, projects SFB F3402-B3, P2465-B24, and TRP 308-N15 to K.M. and project F3005-B12 LIPOTOX and NAWI Graz to S.D.K.
Footnotes
This is an open access article under the CC BY-NC-SA license (http://creativecommons.org/licenses/by-nc-sa/3.0/).
Supplemental Information
References
- Al-Feel W., Chirala S.S., Wakil S.J. Cloning of the yeast FAS3 gene and primary structure of yeast acetyl-CoA carboxylase. Proc. Natl. Acad. Sci. USA. 1992;89:4534–4538. doi: 10.1073/pnas.89.10.4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alex Brown H. Lipidomics: when apocrypha becomes canonical. Curr. Opin. Chem. Biol. 2012;16:221–226. doi: 10.1016/j.cbpa.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambroziak J., Henry S.A. INO2 and INO4 gene products, positive regulators of phospholipid biosynthesis in Saccharomyces cerevisiae, form a complex that binds to the INO1 promoter. J. Biol. Chem. 1994;269:15344–15349. [PubMed] [Google Scholar]
- Bigay J., Antonny B. Curvature, lipid packing, and electrostatics of membrane organelles: defining cellular territories in determining specificity. Dev. Cell. 2012;23:886–895. doi: 10.1016/j.devcel.2012.10.009. [DOI] [PubMed] [Google Scholar]
- Bretscher M.S., Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- Brinkworth R.I., Munn A.L., Kobe B. Protein kinases associated with the yeast phosphoproteome. BMC Bioinformatics. 2006;7:47. doi: 10.1186/1471-2105-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carling D., Aguan K., Woods A., Verhoeven A.J., Beri R.K., Brennan C.H., Sidebottom C., Davison M.D., Scott J. Mammalian AMP-activated protein kinase is homologous to yeast and plant protein kinases involved in the regulation of carbon metabolism. J. Biol. Chem. 1994;269:11442–11448. [PubMed] [Google Scholar]
- Carlson M., Osmond B.C., Botstein D. Mutants of yeast defective in sucrose utilization. Genetics. 1981;98:25–40. doi: 10.1093/genetics/98.1.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carman G.M., Han G.-S. Regulation of phospholipid synthesis in yeast. J. Lipid Res. 2009;50(Suppl):S69–S73. doi: 10.1194/jlr.R800043-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chirala S.S., Zhong Q., Huang W., al-Feel W. Analysis of FAS3/ACC regulatory region of Saccharomyces cerevisiae: identification of a functional UASINO and sequences responsible for fatty acid mediated repression. Nucleic Acids Res. 1994;22:412–418. doi: 10.1093/nar/22.3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dale S., Wilson W.A., Edelman A.M., Hardie D.G. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361:191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- Daum G., Tuller G., Nemec T., Hrastnik C., Balliano G., Cattel L., Milla P., Rocco F., Conzelmann A., Vionnet C. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- de Kroon A.I., Rijken P.J., De Smet C.H. Checks and balances in membrane phospholipid class and acyl chain homeostasis, the yeast perspective. Prog. Lipid Res. 2013;52:374–394. doi: 10.1016/j.plipres.2013.04.006. [DOI] [PubMed] [Google Scholar]
- Fakas S., Qiu Y., Dixon J.L., Han G.S., Ruggles K.V., Garbarino J., Sturley S.L., Carman G.M. Phosphatidate phosphatase activity plays key role in protection against fatty acid-induced toxicity in yeast. J. Biol. Chem. 2011;286:29074–29085. doi: 10.1074/jbc.M111.258798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J., Lees M., Sloane Stanley G.H. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Furuta E., Okuda H., Kobayashi A., Watabe K. Metabolic genes in cancer: their roles in tumor progression and clinical implications. Biochim. Biophys. Acta. 2010;1805:141–152. doi: 10.1016/j.bbcan.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdieri L., Vancura A. Acetyl-CoA carboxylase regulates global histone acetylation. J. Biol. Chem. 2012;287:23865–23876. doi: 10.1074/jbc.M112.380519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbarino J., Sturley S.L. Saturated with fat: new perspectives on lipotoxicity. Curr. Opin. Clin. Nutr. Metab. Care. 2009;12:110–116. doi: 10.1097/MCO.0b013e32832182ee. [DOI] [PubMed] [Google Scholar]
- Gaspar M.L., Hofbauer H.F., Kohlwein S.D., Henry S.A. Coordination of storage lipid synthesis and membrane biogenesis: evidence for cross-talk between triacylglycerol metabolism and phosphatidylinositol synthesis. J. Biol. Chem. 2011;286:1696–1708. doi: 10.1074/jbc.M110.172296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg M.L., Reiner B., Henry S.A. Regulatory mutations of inositol biosynthesis in yeast: isolation of inositol-excreting mutants. Genetics. 1982;100:19–33. doi: 10.1093/genetics/100.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Yang K., Gross R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie D.G., Carling D., Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- Hasslacher M., Ivessa A.S., Paltauf F., Kohlwein S.D. Acetyl-CoA carboxylase from yeast is an essential enzyme and is regulated by factors that control phospholipid metabolism. J. Biol. Chem. 1993;268:10946–10952. [PubMed] [Google Scholar]
- Hedbacker K., Carlson M. SNF1/AMPK pathways in yeast. Front. Biosci. 2008;13:2408–2420. doi: 10.2741/2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S.A., Kohlwein S.D., Carman G.M. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry S.A., Gaspar M.L., Jesch S.A. The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast. Chem. Phys. Lipids. 2014;180:23–43. doi: 10.1016/j.chemphyslip.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jesch S.A., Zhao X., Wells M.T., Henry S.A. Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 2005;280:9106–9118. doi: 10.1074/jbc.M411770200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J.A., Hannun Y.A. Tight binding inhibition of protein phosphatase-1 by phosphatidic acid. Specificity of inhibition by the phospholipid. J. Biol. Chem. 2002;277:15530–15538. doi: 10.1074/jbc.M111555200. [DOI] [PubMed] [Google Scholar]
- Kim H., Ye J. Cellular responses to excess fatty acids: focus on ubiquitin regulatory X domain-containing protein 8. Curr. Opin. Lipidol. 2013;(Dec):29. doi: 10.1097/MOL.0000000000000048. [DOI] [PubMed] [Google Scholar]
- Klose C., Surma M.A., Gerl M.J., Meyenhofer F., Shevchenko A., Simons K. Flexibility of a eukaryotic lipidome—insights from yeast lipidomics. PLoS ONE. 2012;7:e35063. doi: 10.1371/journal.pone.0035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittelfelder O.L., Weberhofer B.P., Eichmann T.O., Kohlwein S.D., Rechberger G.N. A versatile ultra-high performance LC-MS method for lipid profiling. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014;951-952:119–128. doi: 10.1016/j.jchromb.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohlwein S.D. Obese and anorexic yeasts: experimental models to understand the metabolic syndrome and lipotoxicity. Biochim. Biophys. Acta. 2010;1801:222–229. doi: 10.1016/j.bbalip.2009.12.016. [DOI] [PubMed] [Google Scholar]
- Kohlwein S.D., Petschnigg J. SLipid-induced cell dysfunction and cell death: lessons from yeast. Curr. Hypertens. Rep. 2007;9:455–461. doi: 10.1007/s11906-007-0084-5. [DOI] [PubMed] [Google Scholar]
- Kooijman E.E., Burger K.N. Biophysics and function of phosphatidic acid: a molecular perspective. Biochim. Biophys. Acta. 2009;1791:881–888. doi: 10.1016/j.bbalip.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Kooijman E.E., Tieleman D.P., Testerink C., Munnik T., Rijkers D.T., Burger K.N., de Kruijff B. An electrostatic/hydrogen bond switch as the basis for the specific interaction of phosphatidic acid with proteins. J. Biol. Chem. 2007;282:11356–11364. doi: 10.1074/jbc.M609737200. [DOI] [PubMed] [Google Scholar]
- Liu H., Liu J.Y., Wu X., Zhang J.T. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int. J. Biochem. Mol. Biol. 2010;1:69–89. [PMC free article] [PubMed] [Google Scholar]
- Lo W.S., Duggan L., Emre N.C., Belotserkovskya R., Lane W.S., Shiekhattar R., Berger S.L. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Loewen C.J.R., Gaspar M.L., Jesch S.A., Delon C., Ktistakis N.T., Henry S.A., Levine T.P. Phospholipid metabolism regulated by a transcription factor sensing phosphatidic acid. Science. 2004;304:1644–1647. doi: 10.1126/science.1096083. [DOI] [PubMed] [Google Scholar]
- Lopes J.M., Henry S.A. Interaction of trans and cis regulatory elements in the INO1 promoter of Saccharomyces cerevisiae. Nucleic Acids Res. 1991;19:3987–3994. doi: 10.1093/nar/19.14.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C.E., Oh C.S., Jiang Y. Regulation of long chain unsaturated fatty acid synthesis in yeast. Biochim. Biophys. Acta. 2007;1771:271–285. doi: 10.1016/j.bbalip.2006.06.010. [DOI] [PubMed] [Google Scholar]
- McGraw P., Henry S.A. Mutations in the Saccharomyces cerevisiae opi3 gene: effects on phospholipid methylation, growth and cross-pathway regulation of inositol synthesis. Genetics. 1989;122:317–330. doi: 10.1093/genetics/122.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchelhill K.I., Stapleton D., Gao G., House C., Michell B., Katsis F., Witters L.A., Kemp B.E. Mammalian AMP-activated protein kinase shares structural and functional homology with the catalytic domain of yeast Snf1 protein kinase. J. Biol. Chem. 1994;269:2361–2364. [PubMed] [Google Scholar]
- Natter K., Kohlwein S.D. Yeast and cancer cells—common principles in lipid metabolism. Biochim. Biophys. Acta. 2013;1831:314–326. doi: 10.1016/j.bbalip.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuyama H., Saito M., Joshi V.C., Gunsberg S., Wakil S.J. Regulation by temperature of the chain length of fatty acids in yeast. J. Biol. Chem. 1979;254:12281–12284. [PubMed] [Google Scholar]
- Oliveira A.P., Ludwig C., Picotti P., Kogadeeva M., Aebersold R., Sauer U. Regulation of yeast central metabolism by enzyme phosphorylation. Mol. Syst. Biol. 2012;8:623. doi: 10.1038/msb.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P.R., Liu W., Xing F., Fukuda K., Watabe K. Anti-cancer drugs targeting fatty acid synthase (FAS) Recent Patents Anticancer. Drug Discov. 2012;7:185–197. doi: 10.2174/157489212799972891. [DOI] [PubMed] [Google Scholar]
- Petschnigg J., Wolinski H., Kolb D., Zellnig G., Kurat C.F., Natter K., Kohlwein S.D. Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J. Biol. Chem. 2009;284:30981–30993. doi: 10.1074/jbc.M109.024752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H., Gaigg B., Hrastnik C., Achleitner G., Kohlwein S.D., Zellnig G., Perktold A., Daum G. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 2001;268:2351–2361. doi: 10.1046/j.1432-1327.2001.02116.x. [DOI] [PubMed] [Google Scholar]
- Santiago T.C., Mamoun C.B. Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 2003;278:38723–38730. doi: 10.1074/jbc.M303008200. [DOI] [PubMed] [Google Scholar]
- Schneiter R., Daum G. Extraction of yeast lipids. Methods Mol. Biol. 2006;313:41–45. doi: 10.1385/1-59259-958-3:041. [DOI] [PubMed] [Google Scholar]
- Schneiter R., Brügger B., Sandhoff R., Zellnig G., Leber A., Lampl M., Athenstaedt K., Hrastnik C., Eder S., Daum G. Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J. Cell Biol. 1999;146:741–754. doi: 10.1083/jcb.146.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Volrath S.L., Weatherly S.C., Elich T.D., Tong L. A mechanism for the potent inhibition of eukaryotic acetyl-coenzyme A carboxylase by soraphen A, a macrocyclic polyketide natural product. Mol. Cell. 2004;16:881–891. doi: 10.1016/j.molcel.2004.11.034. [DOI] [PubMed] [Google Scholar]
- Shin J.J., Loewen C.J. Putting the pH into phosphatidic acid signaling. BMC Biol. 2011;9:85. doi: 10.1186/1741-7007-9-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirra M.K., Patton-Vogt J., Ulrich A., Liuta-Tehlivets O., Kohlwein S.D., Henry S.A., Arndt K.M. Inhibition of acetyl coenzyme A carboxylase activity restores expression of the INO1 gene in a snf1 mutant strain of Saccharomyces cerevisiae. Mol. Cell. Biol. 2001;21:5710–5722. doi: 10.1128/MCB.21.17.5710-5722.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S., Witkowski A., Joshi A.K. Structural and functional organization of the animal fatty acid synthase. Prog. Lipid Res. 2003;42:289–317. doi: 10.1016/s0163-7827(02)00067-x. [DOI] [PubMed] [Google Scholar]
- Stace C.L., Ktistakis N.T. Phosphatidic acid- and phosphatidylserine-binding proteins. Biochim. Biophys. Acta. 2006;1761:913–926. doi: 10.1016/j.bbalip.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Stukey J.E., McDonough V.M., Martin C.E. Isolation and characterization of OLE1, a gene affecting fatty acid desaturation from Saccharomyces cerevisiae. J. Biol. Chem. 1989;264:16537–16544. [PubMed] [Google Scholar]
- Summers E.F., Letts V.A., McGraw P., Henry S.A. Saccharomyces cerevisiae cho2 mutants are deficient in phospholipid methylation and cross-pathway regulation of inositol synthesis. Genetics. 1988;120:909–922. doi: 10.1093/genetics/120.4.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tehlivets O., Scheuringer K., Kohlwein S.D. Fatty acid synthesis and elongation in yeast. Biochim. Biophys. Acta. 2007;1771:255–270. doi: 10.1016/j.bbalip.2006.07.004. [DOI] [PubMed] [Google Scholar]
- Unger R.H. Lipotoxic diseases. Annu. Rev. Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- Vahlensieck H.F., Pridzun L., Reichenbach H., Hinnen A. Identification of the yeast ACC1 gene product (acetyl-CoA carboxylase) as the target of the polyketide fungicide soraphen A. Curr. Genet. 1994;25:95–100. doi: 10.1007/BF00309532. [DOI] [PubMed] [Google Scholar]
- Vidal-Puig A., Unger R.H. Special issue on lipotoxicity. Biochim. Biophys. Acta. 2010;1801:207–208. doi: 10.1016/j.bbalip.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Villa-García M.J., Choi M.S., Hinz F.I., Gaspar M.L., Jesch S.A., Henry S.A. Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics. 2011;285:125–149. doi: 10.1007/s00438-010-0592-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner C., Dietz M., Wittmann J., Albrecht A., Schüller H.J. The negative regulator Opi1 of phospholipid biosynthesis in yeast contacts the pleiotropic repressor Sin3 and the transcriptional activator Ino2. Mol. Microbiol. 2001;41:155–166. doi: 10.1046/j.1365-2958.2001.02495.x. [DOI] [PubMed] [Google Scholar]
- Wakil S.J., Abu-Elheiga L.A. Fatty acid metabolism: target for metabolic syndrome. J. Lipid Res. 2009;50(Suppl):S138–S143. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakil S.J., Stoops J.K., Joshi V.C. Fatty acid synthesis and its regulation. Annu. Rev. Biochem. 1983;52:537–579. doi: 10.1146/annurev.bi.52.070183.002541. [DOI] [PubMed] [Google Scholar]
- Wang C., Rajput S., Watabe K., Liao D.F., Cao D. Acetyl-CoA carboxylase-a as a novel target for cancer therapy. Front. Biosci. (Schol. Ed.) 2010;2:515–526. doi: 10.2741/s82. [DOI] [PubMed] [Google Scholar]
- White M.J., Hirsch J.P., Henry S.A. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J. Biol. Chem. 1991;266:863–872. [PubMed] [Google Scholar]
- Wilson W.A., Hawley S.A., Hardie D.G. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr. Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Woods A., Munday M.R., Scott J., Yang X., Carlson M., Carling D., Mundays M.R. Yeast SNF1 is functionally related to mammalian AMP-activated protein kinase and regulates acetyl-CoA carboxylase in vivo. J. Biol. Chem. 1994;269:19509–19515. [PubMed] [Google Scholar]
- Young B.P., Shin J.J., Orij R., Chao J.T., Li S.C., Guan X.L., Khong A., Jan E., Wenk M.R., Prinz W.A. Phosphatidic acid is a pH biosensor that links membrane biogenesis to metabolism. Science. 2010;329:1085–1088. doi: 10.1126/science.1191026. [DOI] [PubMed] [Google Scholar]
- Zinser E., Sperka-Gottlieb C.D., Fasch E.V., Kohlwein S.D., Paltauf F., Daum G. Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 1991;173:2026–2034. doi: 10.1128/jb.173.6.2026-2034.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.