Abstract
Background
Abuse of heroin and prescription opiate medications has grown to disturbing levels. Opioids mediate their effects through mu opioid receptors (MOR), but minimal information exists regarding MOR-related striatal signaling relevant to the human condition. The striatum is a structure central to reward and habitual behavior and neurobiological changes in this region are thought to underlie the pathophysiology of addiction disorders.
Methods
We examined molecular mechanisms related to MOR in postmortem human brain striatal specimens from a homogenous European Caucasian population of heroin abusers and control subjects and in an animal model of heroin self-administration. Expression of ets-like kinase 1 (ELK1) was examined in relation to polymorphism of the MOR gene OPRM1 and drug history.
Results
A characteristic feature of heroin abusers was decreased expression of MOR and extracellular regulated kinase (ERK) signaling networks, concomitant with dysregulation of the downstream transcription factor ELK1. Striatal ELK1 in heroin abusers associated with the polymorphism rs2075572 in OPRM1 in a genotype dose-dependent manner and correlated with documented history of heroin use, an effect reproduced in an animal model that emphasizes a direct relationship between repeated heroin exposure and ELK1 dysregulation. A central role of ELK1 was evidenced by an unbiased whole transcriptome microarray that revealed ~20% of downregulated genes in human heroin abusers are ELK1 targets. Using chromatin immuneprecipitation, we confirmed decreased ELK1 promoter occupancy of the target gene Use1.
Conclusions
ELK1 is a potential key transcriptional regulatory factor in striatal disturbances associated with heroin abuse and relevant to genetic mutation of OPRM1.
Keywords: opioid, self-administration, MAPK, transcriptome, addiction, rat
Introduction
Opioids have become the second most abused class of drugs in the United States and are prevalent among teens and young adults.(1) Given that opioid abuse is associated with high rates of drug-related mortality, morbidity and criminality, the ramifications of this trend are cause for public health concern.(2) The rewarding effects of heroin are mediated by the activation of the G-protein coupled μ-opioid receptor (MOR).(3, 4) MOR stimulation leads to modulation of multiple effectors, including the inhibition of adenylyl cyclase,(5) regulation of voltage-gated calcium (Ca2+) channels,(6, 7) and control of the canonical mitogen-activated protein kinases (MAPK) extracellular regulated kinase (ERK)-1 and -2.(8) Active ERK1/2 phosphorylates the transcription factors ELK1 and cyclic AMP responsive element binding protein (CREB), which translocate to the nucleus to regulate expression of target genes,(9) the physiological consequences of which result in synaptic plasticity and aberrant behaviors that maintain the cycle of drug abuse.(10) Experimental models have led to the postulation that long-term repeated exposure to opioid drugs results in receptor desensitization and/or down-regulation events linked to opioid tolerance and dependence.(11, 12) However, the signaling and transcriptional events that underlie adaptation within the human brain as a result of repeated MOR activation remain poorly understood.
An important consideration in human studies is genetics which confer an increased vulnerability to heroin addiction (13, 14) and gene variants encoding opioid receptors have been shown to be associated with this disorder.(15–18) Numerous single nucleotide polymorphisms (SNPs) have been identified in the human MOR gene (OPRM1) and while the functional relevance of some variants to MOR signaling(19–21) and expression(22) has been described, the downstream transcriptional events have not been well characterized.
In the present study, we investigated signaling molecules directly associated with MOR function and downstream transcription factors in the striatum, a central structure involved in mediating goal-directed behavior, habit formation and reward.(23–25) We report dysregulation of striatal MOR and ERK signaling pathways in human heroin abusers and identify ELK1 as a key transcriptional regulator for differentially expressed genes in the striatum of heroin abusers and in association with OPRM1 variants. These findings highlight ELK1 as a potentially new molecular target in the cellular dysregulation of opioid addiction.
Materials and Methods
Human brain specimens
Postmortem brains samples from heroin abusers and control subjects were collected at the Department of Forensic Medicine at Semmelweis University, Hungary, and the National Institute of Forensic Medicine at Karolinska Institutet, Stockholm, Sweden under the ethics guidelines approved by each institution. Heroin subjects died from heroin intoxication, were predominantly heroin users not receiving methadone or buprenorphine treatment, and were negative for human immunodeficiency virus infection. No information was known about their physical dependence. The control group had negative blood levels of opiates and other drugs of abuse. Demographic information can be found in Supplemental Table 1. Striatal tissue was dissected as previously described.(18) Due to the low abundance of nucleus accumbens (NAc) tissue, putamen tissue was used for the majority of protein experiments. Putamen punches (~200 mg) were pulverized on dry ice and stored at −80°C. For laser captured samples, striatal regions were dissected from 20μm sections from human subjects using an Arcturus XT Laser Capture Microdissection Instrument (Molecular Devices, Sunnyvale, CA).
Rodent heroin self-administration
A heroin self-administration paradigm was performed in adult male Long Evans rats (Charles River, Wilmington, MA) for two weeks as previously described.(26–28) This dosing paradigm does not induce physical dependence.(29) Animals were sacrificed either 1 hour or 24 hours after the last self-administration session by CO2 followed by decapitation, brains frozen and striata dissected using a 2mm micro-punch.
SNP genotyping
Genomic DNA was purified from human cerebellar tissue using DNeasy columns (Qiagen, Valencia, CA). Genotyping for the OPRM1 rs1799971 (Taqman Assay ID: C_8950074_1_) and rs2075572 (Taqman Assay ID: C_1691815_1_, Applied Biosystems, Foster City, CA) SNPs was performed as previously described.(18, 30) The PLINK 1.07 genetic association analysis program(31) was used to verify SNP data quality, test for departure from Hardy-Weinberg equilibrium, and test individual SNPs for statistical association with significance set at p<0.05.
Western blotting
Western blots were performed on a subset of heroin (N=32) and control (N=15) human subjects, and on dorsal striata from the rodent self-administration study, as previously described(32) and probed with primary antibodies directed against the following proteins: MOR (GeneTex, Inc., Irvine, CA); ERK1/2, ELK1 and phospho-ELK1(Ser383)(Cell Signaling Technology, Danvers, MA for human samples and Santa Cruz Biotechnology, Santa Cruz, CA for rat); βarrestin2 (Santa Cruz Biotechnology); and MEK1 (Invitrogen, Carlsbad, CA). Proteins were analyzed using ImageJ software(33, 34) and normalized to total protein levels(35) obtained with the Memcode Reversible Protein Stain Kit (Thermo Scientific). Logarithmic transformations were performed to render normal data distribution. A general linear stepwise regression was used to calculate statistical significance and identify covariates (brain pH, gender, age) using JMP software (SAS Institute, Cary, NC). Two-tailed t-tests were used when no covariates were found. Spearman correlations were calculated to assess the relationship between protein levels and heroin toxicology.
Microarray analysis
mRNA microarray experiments were performed on NAc tissue from a subset of heroin (N=22; 19 males and 3 females) and control (N=27; 22 males and 5 females) subjects at the Purdue Pharma, L.P. (Cranbury, NJ). Total RNA isolation, assessment of RNA integrity, and microarray hybridization were performed as previously described.(36) Expression data were collected using HG-U133A Affymetrix Chips (Affymetrix, Santa Clara, CA). Raw data were normalized using the Robust Multichip Average (RMA)(37) from the Affymetrix Expression Console. To control for RNA quality, actin 3′/5′ ratios of less than 3 and % present calls of 50% or greater were used. To identify Differentially Expressed Genes (DEGs), two-tailed t-tests were performed with a multiple-test correction (p<0.05) (38). Hierarchical clustering was performed on log2 expression levels of DEGs with Euclidian distance and average-linkage for constructing dendograms. Common transcription factors for DEGs were identified with Lists2Networks, a web-based software system for performing gene list enrichment analysis.(39) The transcription factor gene set library in List2Networks was created from putative targets of transcription factors.(40) All other computational analyses were performed in MATLAB. (Mathworks, Natick, MA)
Gene expression verification
Microarray findings were verified with Nanostring Technologies (Nanostring, Seattle, WA) using 100ng NAc RNA from human heroin (N=20) and control (N=16) subjects. Design and synthesis of probes was performed according to company protocol and hybridization reactions were carried out as previously described.(41) Expression values were normalized to ACTB and GAPDH, which were stable in the human population. Probe sequences are listed in supplementary table 2. Statistical analyses were performed as above using JMP software.
Chromatin immuneprecipitation
Chromatin immuneprecipitation (ChIP) was performed as previously described (42) using the NAc from rats that underwent the self-administration paradigm. 30ug of purified chromatin per animal was immuneprecipitated with 1.25ug of ELK1 antibody (Epitomics, Burlingame, CA) or nonspecific rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA). Chromatin was analyzed by QPCR at genomic regions of BAG1 and USE1. Primer sequences are listed in supplementary table 3. Statistical analyses as described above were performed using JMP software.
Results
Dysregulation of MOR signaling molecules in the putamen of human heroin abusers
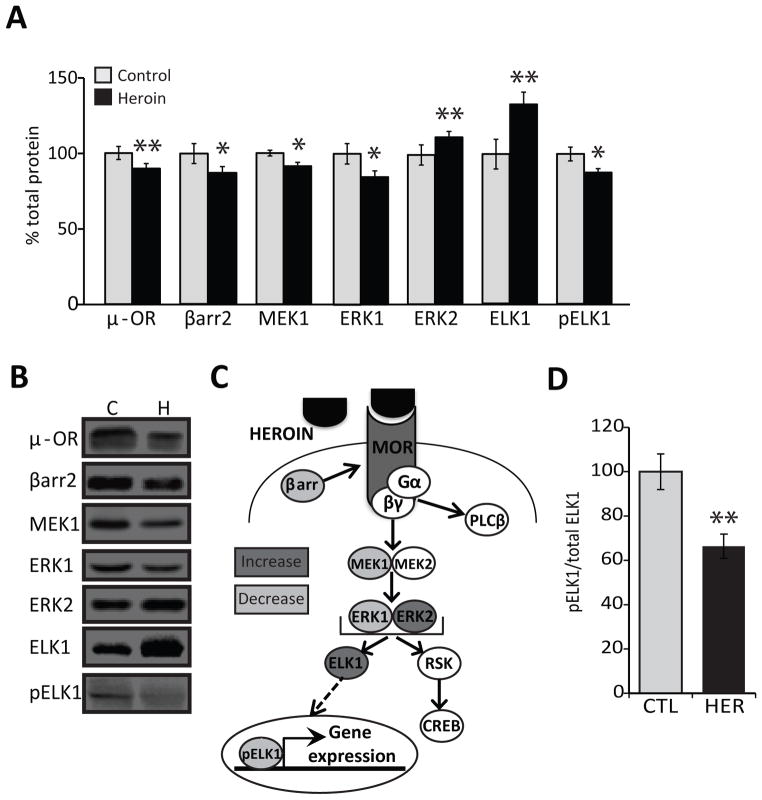
Protein expression levels of MOR (89.55±3.42%, p<0.005; F2,43=6.73) and β-arrestin2 (87.11±4.26%, p<0.05; F3,41=8.1), which regulates MOR desensitization and internalization,(43) were significantly reduced in heroin abusers compared to controls (Fig 1A–B), indicating that the tone of MOR-signaling cascades may be altered. One of the key intracellular pathways activated upon MOR stimulation is the MAPK pathway, depicted in Figure 1C. We focused on the core components of the pathway including MAPK ERK kinases (MEK)1/2 and ERK1/2 because they regulate phosphorylation status and activity of transcription factors. Levels of both MEK1 (91.28±2.55%, p<0.05; F1,41=4.33) and its substrate ERK1 (84.78 ± 3.83%, p<0.05; F1,46= 4.40) were significantly reduced. We observed an opposing increase in the protein levels of ERK2 (111.78±2.32%. p<0.005; F1,44=9.33). The levels of phosphorylated ERK1 (105.67 ± 13.01%) and phosphorylated ERK2 (120.1 ± 11.4%) were not different from controls due to the strong confound of brain pH which significantly impacted pERK1 (p<0.0005) and pERK2 (p< 0.00001) levels (data not shown), suggesting that phosphorylation of ERK1/2 is sensitive to the perimortem state (data not shown).
Figure 1. MOR signaling networks and the MAPK pathway are dysregulated in human heroin abusers.
(A,B) Protein levels of MOR, β-arrestin2, MEK1, ERK1, ERK2, ELK1, and pELK1 in the putamen of human control subjects and heroin abusers. N=15 control, N=32 heroin. Representative bands are shown next to the histograms; Mean ± SEM. * p < 0.05, ** p < 0.01. (C) Schematic depiction of the MOR signaling networks. (D) Ratio of phosphorylated versus total ELK1 protein in human subjects. Abbreviations: C, CTL, control; H, HER, heroin.
Dysregulation of transcription factors in the putamen of heroin abusers
To assess further downstream events of MAPK signaling, we examined expression of transcription factors ELK1 and CREB, which are nuclear targets for ERK1/2.(44, 45) CREB is activated in response to ERK1/2 through signaling by kinases such as p90RSK.(46–48) Both p90RSK (82.01±10.2% control) and CREB (79.07±4.69% control) were reduced in the putamen, but this was confounded by sensitivity to pH (p<0.0001). However, heroin abusers had significantly higher expression of ELK1 (133.26±8.21%, p<0.01; F1,46=7.38). In order to gain entry into the nucleus, ELK1 must be phosphorylated by ERK1/2(49) and the levels of activated pELK1 were significantly reduced (86.92±3.35%, p<0.05; F1,41=5.21) in heroin subjects. Furthermore, the ratio of pELK1 to total ELK1 was significantly decreased in heroin abusers (66.37±5.46, p<.001; F1,46=12.01; Fig 1D), indicating that repeated heroin use leads to an imbalance in total versus phosphorylated ELK1 protein.
Elevation of ELK1 protein levels in the NAc of heroin abusers
The NAc plays a key role in heroin abuse vulnerability,(50, 51) thus we next examined protein levels of ELK1 in the NAc to determine whether alterations in the putamen were also evident in the mesolimbic component of the striatum. Laser capture microdissected samples were taken from the NAc, putamen and caudate nucleus from glass-mounted striatal sections from the same subjects studied above (Fig. 2A). Similar to the observations in putamen tissue punches, ELK1 expression was significantly elevated in all striatal regions of the heroin abusers, but was most pronounced in the NAc (p<0.05; F2,41=4.923, Fig. 2B).
Figure 2. ELK1 expression levels are increased in striatal regions in heroin abusers.
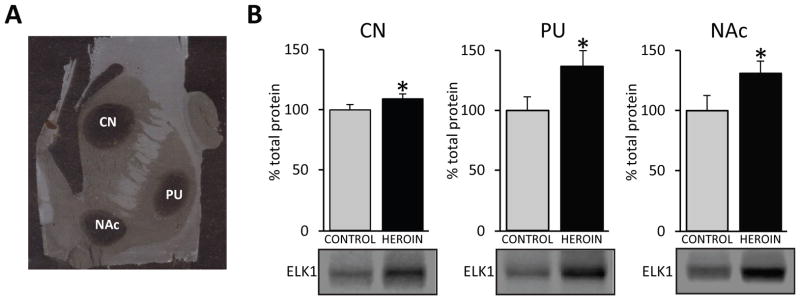
(A) A representative photomicrograph of striatal-mounted sections (20μm) after laser capture microdissection. Circular dark areas indicate striatal regions that were isolated. (B) ELK1 protein levels were significantly increased in all striatal regions in heroin abusers. Representative bands are shown below histograms; mean ± SEM. Abbreviations: CN, caudate; NAc, nucleus accumbens; PU, putamen. For Western analyses, N=15 control, N=32 heroin. * p < 0.05.
Striatal ELK1 protein expression is associated with an OPRM1 polymorphism
Given the contribution of genetics to opiate abuse, we determined if individual polymorphisms of OPRM1 are related to ELK1 expression. The rs1799971 SNP (A118G variant) in exon 1 is the most common mutation in the coding region as well as the most studied OPRM1 polymorphism.(52) Unfortunately, only two control subjects carried the minor G allele which rendered it challenging to interpret the results in regard to non-drug protein expression. Nevertheless, there was an overall group-genotype effect (F2,42=4.789, p<0.05) with ELK1 levels significantly higher in heroin subjects than controls for the A/A genotype (p<.001) and heroin subjects carrying the G allele had the highest ELK1 levels (p<0.01; Fig. 3A).
Figure 3. ELK1 expression levels are associated with polymorphism of the OPRM1 gene.
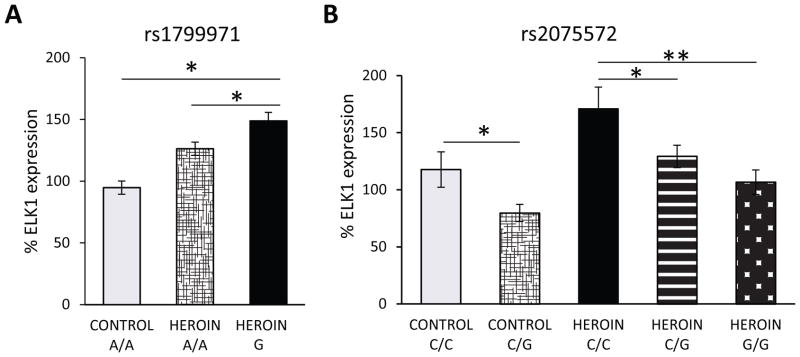
(A) Heroin subjects carrying the G allele of the rs1799971 SNP have highest the ELK1 expression. Control A/A, N=13; Heroin A/A, N=22, Heroin G, N=10. (B) Genotype-dose effect evident for the rs2075572 SNP in controls and heroin subjects with ELK1 expression levels highest in C/C subjects in each respective group. Control: C/C and C/G, N= 8 and 5, respectively; Heroin: C/C, C/G and G/G, N=8, 10 and 15, respectively. Mean ± SEM. * p < 0.05, ** p < 0.01.
Considering the low frequency of rs1799971, we also evaluated the rs2075572 SNP, which is located in intron 2(53) and has been associated with initiation of smoking.(54) There was not an overall significant association of rs2075572 with heroin abuse, but a greater number of G/G subjects tended to be heroin abusers, while the control group tended to have higher numbers of C/C individuals (Supplementary Table 4). We observed a pronounced genotype-dose effect association of rs2075572 with striatal ELK1 protein levels (F4,46=4.338, p<0.01;Fig. 3B). Heroin carriers of the minor C allele had significantly higher ELK1 levels than heroin subjects homozygous for the G allele (Fig 3B). A similar trend was observed for changes in ELK1 protein expression between control subjects, indicating that individual variation in the OPRM1 gene may result in heterogeneous levels of ELK1. Importantly, the mean value of ELK1 expression for each individual heroin genotype (C/C- 170.8%; C/G- 129.2%; G/G- 106.6%) was higher than the mean value for all control subjects combined (100%).
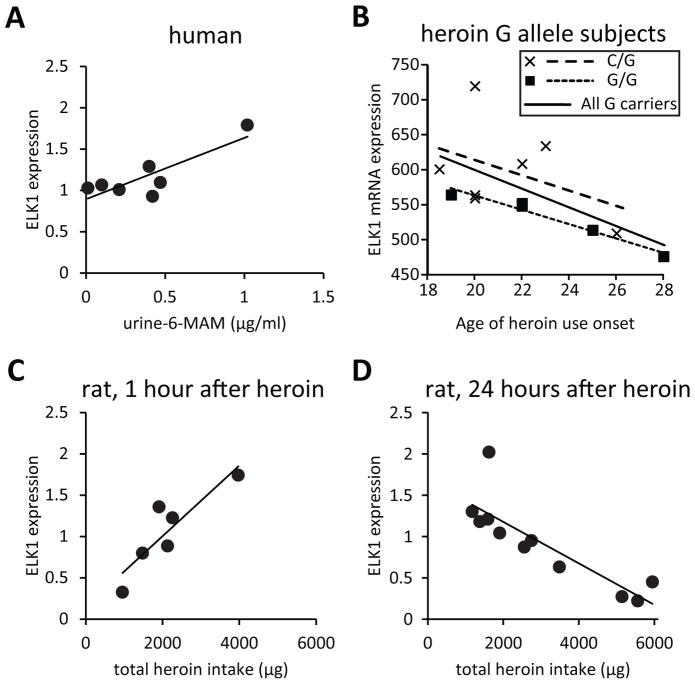
Striatal ELK1 levels relate to heroin use history
Assessment of toxicology in heroin abusers revealed a strong positive correlation between ELK1 protein expression in the putamen and urine concentrations of 6-MAM, the rapid metabolite of heroin (r2=0.70, p<0.05), but this was strongly influenced by one subject (Figure 4A). Interestingly, documentation of drug use history available for some subjects showed that levels of ELK1 mRNA tended to be negatively correlated with age of onset of heroin use for subjects carrying the G allele of rs2075572 (r2=0.37, p<0.05), suggesting elevated ELK1 with long-term heroin use, but this was predominantly driven by those with the G/G genotype (r2=0.95, p<0.01; Fig 4B). Importantly, this subset of G allele heroin abusers was representative of the larger population of subjects as their ELK1 mRNA expression levels (124.00±4.04% of controls, p<0.01; F1,28=10.328, data not shown) were comparable to that in the larger cohort.
Figure 4. Striatal ELK1 protein levels correlate with heroin use.
(A) A strong positive correlation (r2=0.70) was observed between ELK1 protein expression in the putamen and urine levels of the rapid heroin metabolite 6-MAM in human heroin abusers; seven subjects had detectable 6-MAM levels. (B) ELK1 mRNA levels negatively correlate with the age of onset of heroin use in human subjects that carry the G allele of rs2075572 (r2=0.37), with the strongest correlation in G/G subjects (r2=0.95). (C) Total ELK1 protein levels in the dorsal striatum are positively correlated (r2=0.79) to history of heroin intake in a rat heroin self-administration model 1 hour after the last drug session, while a negative correlation (r2=0.72) was observed 24 hours later (D). N= 5–11/group.
Given the challenge of human postmortem studies that lack a complete drug history about the subjects, we examined the regulation of ELK1 in a rat model of addiction using a heroin self-administration paradigm where animals controlled their individual drug intake.(26–28) Since the post-mortem interval for many of the human heroin subjects is relatively short, we examined ELK1 protein 1 hour after the last self-administration session as well as 24 hours later to obtain insights regarding the stability of the observations. ELK1 expression was tightly correlated with each animal’s history of heroin intake. We observed a positive correlation 1 hour after the final heroin self-administration session (r2=0.79, p<0.05; Fig 4C) that was reversed 24 hours later with a negative correlation to ELK1 levels (r2=0.72, p<0.001; Fig 4D).
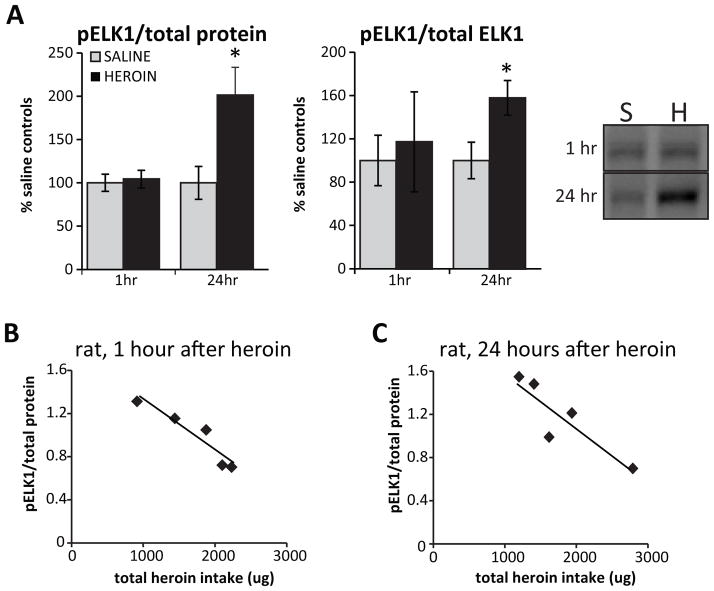
To assess the functional state of ELK1 in relation to heroin exposure, we also measured levels of pELK1 in animals that self-administered the drug. While no changes in pELK1 were observed at the 1 hour time-point, we observed a significant increase in pELK1 relative to both total protein and total ELK1 protein at 24 hours after the last heroin session (pELK1/total protein: 201.31 ± 32.13%, p<0.05; F1,10= 6.03; pELK1/total ELK1: 158.14 ± 16.10%, p<0.05; F1,10= 6.50; Figure 5A.). Correlations to heroin intake revealed that at both time points, pELK1 negatively correlated to heroin intake (1 hour: r2=0.86, p<0.05; Figure 5B; 24 hours: r2=0.79, p<0.05; Figure 5C), indicating that high levels of heroin were associated with decreased active ELK1. These data support the notion that repeated heroin use dysregulates ELK1 expression relative to the overall history of drug exposure which may also account for individual differences observed in humans.
Figure 5. Heroin inhibits ELK1 phosphorylation in a dose-dependent manner.
A) Levels of phosphorylated ELK1 (pELK1; normalized to total protein or total ELK1) measured in the dorsal striatum of rats 1 hour or 24 hours after final heroin self-administration session. A representative blot is shown in Panel A. Levels pELK1 negatively correlated with each animal’s total heroin intake at both the 1 hour (r=.86) (B) and 24 hour (r=.79) (C) time points. Abbreviations: S, saline; H, heroin. N=5–6 animals/group. Mean ± SEM. * p < 0.05.
Transcriptional repression of ELK1 target genes in the striatum of heroin abusers
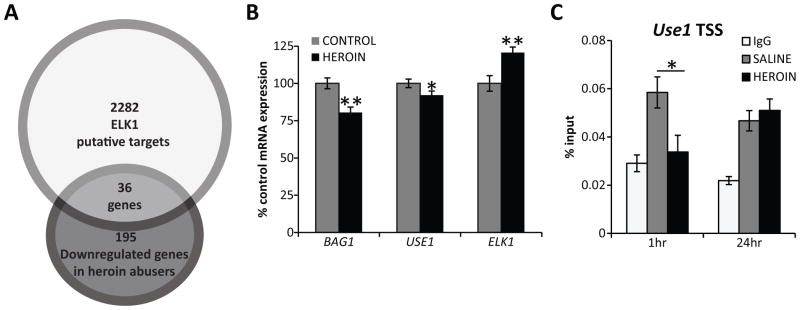
We next addressed whether ELK1 could play a role in transcriptional regulation relevant to heroin abuse. We chose an unbiased strategy in which downstream gene targets could be identified to guide future studies. As such, we utilized a NAc microarray dataset from a cohort of our heroin postmortem population and examined transcription factor binding motifs common to the observed transcriptome alterations. Interestingly, evaluation of common transcription factors for the significantly regulated genes using the Lists2Networks program (55) revealed that ELK1 (p<0.05) and ETS1 (p<0.01) had the most putative target genes overlapping with down-regulated genes (36/195) (Fig. 6A and Supplementary Table 5), confirming the relevance of ELK1 as a transcriptional regulator of gene expression in heroin abuse. Hierarchical clustering on a subset (N=32) of subjects in the two groups that were most different from one another revealed differential expression of 1518 genes, of which 115 were identified as downregulated ELK1 target genes (p<.000001) (Supplementary Fig 1 and Supplementary Table 6). These findings were validated by measuring mRNA expression levels of the ELK1 target genes BAG1 (80.46 ± 3.61%; p<0.01) and USE1 (92.09 ± 2.76%; p<0.05; Fig 6B) in the NAc using Nanostring technology. In addition, Nanostring analyses revealed a significant increase in ELK1 mRNA in heroin subjects compared to controls (120.67 ± 3.72%; p<0.01; Fig 6B), which is in agreement with the increased protein expression of ELK1 previously observed.
Figure 6. Downregulation of ELK1 target genes after repeated heroin exposure.
A) Of the 195 down-regulated genes identified in heroin abusers, approximately 20% were classified as ELK1 putative targets using the List2Networks software analyses. Control N=27; heroin N=22. B) Downregulation of the ELK1 target genes BAG1 and USE1 was confirmed with Nanostring technologies in a subset of subjects. Additionally, ELK1 mRNA levels were increased in heroin subjects. Control N=16; heroin N=20. C) In a rat heroin self-administration model, decreased occupancy of ELK1 was detected at the promoter region of the target gene Use1 at 1 hour after the last heroin session but was normalized 24 hours after the last drug session. N=6–7 animals/group. Mean ± SEM. * p < 0.05, ** p < 0.01.
ELK1 binding sites are located slightly upstream of the transcriptional start site of target genes.(56) To determine if the gene expression changes could be attributed to decreased transcriptional activity of ELK1 directly at the target loci, we used ChIP to assess the occupancy of ELK1 at the promoter regions of downstream targets in the NAc of rats that underwent the self-administration paradigm. We examined published ChIP sequencing data obtained using ELK1, CREB, and SRF antibodies to guide our selection of ELK1 target genes.(56, 57) Such studies suggested Bag1 and Use1 are regulated by ELK1 in a CREB- and SRF-independent manner.(56, 57) We observed no enrichment of ELK1 binding over nonspecific IgG controls at any region of Bag1 studied (data not shown). In animals that were sacrificed 1 hour after the last heroin self-administration session, ELK1 occupancy was significantly decreased at the promoter of Use1 versus saline animals (0.034± 0.007% versus 0.058±0.006; p<.05; Fig 6C), indicating a direct functional regulation of ELK1 on downstream gene targets as a consequence of heroin use. At the 24-hour time-point, ELK1 binding at the Use1 promoter was not significantly different from controls, indicating that heroin did not induce a prolonged decrease in ELK1 occupancy.
Discussion
The current study provides convergent lines of evidence identifying ELK1 disturbances in association with heroin use history and variants of the OPRM1 gene. The findings suggest that repeated activation of MOR, and subsequently the ERK pathway, efficiently controls the availability of ELK1 to regulate striatal transcriptional machinery in a dose-dependent manner. The ELK1 transcription factor has been generally overlooked in the context of addiction, but the current results implicate it as a potential substrate in the pathophysiology of heroin abuse.
Limited insights are available regarding the role of ELK1 in neuropsychiatric disorders, but its molecular regulation has been described. Phosphorylation of ELK1 by ERK1/2 is critical for its translocation into the nucleus where it can exert effects on chromatin and gene expression.(58) The fact that total ELK1 was increased in the putamen of human heroin abusers concomitant with a decrease in pELK1 levels suggested that less ELK1 is trafficked to the nucleus and transcription of ELK1 target genes may be decreased. Indeed, total ELK1 was positively correlated to heroin at 1 hour after heroin intake in animals that directly self-administered heroin, similar to human abusers, and pELK1 levels were negatively correlated with heroin use, irrespective of the time of death following the last drug intake. These findings emphasize an important contribution of the history of repeated heroin use on ELK1 phosphorylation. Interestingly, the absolute levels of pELK1 were elevated 24 hours after heroin use, indicative of an acute withdrawal effect or a potential compensation to counter reduction of pELK1 as a consequence of repeated heroin use. Acute opiate withdrawal 24 hours following heroin self-administration is generally associated with enhanced stress response that involves the activation of the hypothalamic-pituitary axis and glucocorticoid receptor system.(59, 60) Interestingly, glucocorticoid receptor stimulation increases pELK1 (61), suggesting that increased pELK1 24 hours after heroin may be a result of withdrawal-induced stress. Such time course changes could also have distinct functional relevance depending on the cellular localization of pELK1 at these periods since ELK1 localization in dendritic, cytoplasmic and nuclear compartments has significant implications regarding its effects on dendritic sprouting and transcriptional regulation. (62–64)
Although ELK1 has not been widely investigated in addiction, other animal studies have shown that acute administration of cocaine (65, 66) or delta-9-tetrahydrocannabinol (psychoactive component of cannabis)(67) increases pELK1 immediately after drug exposure. Whether these noted differences in pELK1 relate to acute versus chronic drug exposure, type of drugs, cellular localization or other factors in experimental designs of these studies needs to be further explored. However, the fact that ELK1 is altered by several drugs of abuse is intriguing considering the role of ELK1 phosphorylation in neurite outgrowth (62–64) and that dysregulation of structural plasticity is a feature of drugs of abuse.
ELK1 is a member of the ternary complex factor subfamily of ETS transcription factors and intriguingly, our unbiased computational analysis of the NAc microarray data also identified ETS1, the prototype of the ETS1 family, as enriched in the promoter region of down-regulated genes of heroin abusers. In addition to cellular remodeling linked to synaptic plasticity, ETS1 family members are known to play important roles in various cellular functions such as proliferation,(68) differentiation,(69) migration,(70) and apoptosis.(71) Other transcriptional regulators implicated in addiction such as CREB also target some genes common to ELK1. By focusing on gene targets regulated by ELK1 in a CREB- and SRF-independent manner, we confirmed reduction of the genes BAG1 and USE1 identified in the microarray. BAG1 has antiapoptotic effects that confer prosurvival signals to enhance neurite outgrowth activity and is activated downstream of RAF1 and MEK activity.(72–74) Unfortunately, Bag1 shares an ELK1 promoter binding site with another gene in close proximity, Chmp5, which makes the assessment of transcription factor binding difficult to distinguish with ChIP.(56) We were, nevertheless, able to detect ELK1 at the promoter of Use1, a gene known to be antiapoptotic (75), and to act as an E2 ubiquitin-conjugating enzyme to aide in the proteasomal degradation of proteins.(76) ELK1 occupancy at Use1 was decreased 1 hour after heroin self-administration, validating direct gene transcription regulation in the NAc after repeated heroin exposure. ELK1 occupancy at the Use1 promoter was normalized 24 hours later, in line with the dynamic nature of transcription factors. Clearly, the time course of ELK1 regulation is an important question. Moreover, while the current study focused on SRF-independent downstream targets, SRF-dependent ELK1 transcriptional activity is also relevant to addiction and warrants future studies in heroin abuse.
The importance of individual factors to striatal ELK1 expression was also evident in our study, both in relation to drug use history as supported by our animal model and by mutation of the OPRM1 gene. The rs2075572 OPRM1 variant has not been studied in opioid abuse and the functional consequences of this variant on MOR activity are not known. However, a similar genotype-dose effect pattern was evident for the well-known A118G SNP in heroin abusers, suggesting that these mutations relate to general functioning of MOR and its downstream signaling. Although mutations of OPRM1 contributed to variability in the individual ELK1 expression levels, it is important to emphasize that heroin use still enhanced ELK1 at each respective genotype and thus, history of drug use plays a significant role in ELK1 regulation.
While we concentrated on the novel ELK1 findings, there is also clear dysregulation of other components of striatal MOR signaling, including MOR and β-arrestin2. Such impairments may relate to altered drug sensitivity or tolerance to heroin, as β-arrestin2 modulates MOR internalization and desensitization in rodent models.(77) These events contribute significantly to drug tolerance as evidenced by the fact that β-arrestin2 knockout mice do not develop tolerance.(43) A lack of MOR desensitization or proper recycling due to deficits in β-arrestin2 may lead to heightened reward response with repeated heroin use.(78) Human opiate abusers also have reduced expression in the frontal cortex of MOR, β-arrestin2, and GRK2,(79) a kinase that also contributes to MOR desensitization and internalization, which together with the current findings would suggest similar MOR-related pathology in brain areas critically involved in addictive behaviors.
Another key finding indicating compromised integrity of MOR signaling in heroin abuse was the dysregulation of the MAPK/ERK pathway, including differential alterations of ERK1 and ERK2. Most studies have normally assumed that measures of ERK1 are indicative of ERK2 alterations since MEK phosphorylates both these kinases. However, ERK1 inhibits ERK2.(80) Thus increased ERK2 levels observed in the heroin abusers would be in line with compensation for the decreased ERK1 expression. Altogether such aberrant signaling activity of MEK1 and ERK1/2 would be expected to contribute to subsequent downstream changes in ELK1 and gene expression that mediate cellular adaptations to repeated heroin exposure. In summary, the current results demonstrate that the striatum of human heroin abusers is characterized by dysregulation of MOR signaling and the MAPK pathway linked to disturbances of the downstream transcription factor, ELK1. The importance of ELK1 impairment is highlighted by the fact that it targeted the promoter of a large proportion of the repressed genes in the NAc of heroin abusers. The significant contribution of heroin intake history and OPRM1 polymorphisms to individual differences in ELK1 expression also emphasizes the strong link between MOR and ELK1. Overall, these findings suggest that ELK1 may be a key transcriptional substrate in pathophysiological disturbances relevant to drug addiction.
Supplementary Material
Figure S1: Heatmap of gene expression changes in the NAc of human heroin abusers. The hierarchical clustering of differentially expressed genes detected between the control (N=19) and heroin (N=13) groups in the NAc using mRNA microarrays. Dark colors represent normalized low expression and light colors high expression. The x-axis lists subjects represented by a three digit ID number; C-control, H-heroin user; m-male, f-female; age.
Table SI: Demographic data
Table S2: Probes used with Nanostring technologies.
Table S3- Primer pair sequences used in qPCR.
Table S4: Frequency of the rs2075572 OPRM1 polymorphism in human subjects.
Table S5: ELK1 target genes identified in 49 subjects.
Table S6: ELK1 target genes identified in 32 subjects.
Acknowledgments
We thank James Sperry and Nayana Patel for technical assistance as well as Dr. Gang Jin previously at Purdue Pharma L.P. for microarray analysis. This work was funded by grants from the National Institute on Drug Abuse DA015446 (YLH), DA013997 (YXP), and T32DA007135 (MMJ), as well as grant ETT/098/2009 (EK).
Footnotes
Disclosure of conflicts of interest
The authors declare that we have no competing interests.
References
- 1.SAMHSA. Results from the 2009 National Survey on Drug Use and Health: National Findings. Rockville, MD: Office of Applied Studies; 2010. NSDUH Series H-32, DHHS Publication No. SMA 07-4293. [Google Scholar]
- 2.Hulse GK, English DR, Milne E, Holman CD. The quantification of mortality resulting from the regular use of illicit opiates. Addiction. 1999;94:221–229. doi: 10.1046/j.1360-0443.1999.9422216.x. [DOI] [PubMed] [Google Scholar]
- 3.Matthes HW, Maldonado R, Simonin F, Valverde O, Slowe S, Kitchen I, et al. Loss of morphine-induced analgesia, reward effect and withdrawal symptoms in mice lacking the mu-opioid-receptor gene. Nature. 1996;383:819–823. doi: 10.1038/383819a0. [DOI] [PubMed] [Google Scholar]
- 4.Sora I, Elmer G, Funada M, Pieper J, Li XF, Hall FS, et al. Mu opiate receptor gene dose effects on different morphine actions: evidence for differential in vivo mu receptor reserve. Neuropsychopharmacology. 2001;25:41–54. doi: 10.1016/S0893-133X(00)00252-9. [DOI] [PubMed] [Google Scholar]
- 5.Sharma SK, Nirenberg M, Klee WA. Morphine receptors as regulators of adenylate cyclase activity. Proc Natl Acad Sci U S A. 1975;72:590–594. doi: 10.1073/pnas.72.2.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gross RA, Macdonald RL. Dynorphin A selectively reduces a large transient (N-type) calcium current of mouse dorsal root ganglion neurons in cell culture. Proc Natl Acad Sci U S A. 1987;84:5469–5473. doi: 10.1073/pnas.84.15.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tallent M, Dichter MA, Bell GI, Reisine T. The cloned kappa opioid receptor couples to an N-type calcium current in undifferentiated PC-12 cells. Neuroscience. 1994;63:1033–1040. doi: 10.1016/0306-4522(94)90570-3. [DOI] [PubMed] [Google Scholar]
- 8.Fukuda K, Kato S, Morikawa H, Shoda T, Mori K. Functional coupling of the delta-, mu-, and kappa-opioid receptors to mitogen-activated protein kinase and arachidonate release in Chinese hamster ovary cells. J Neurochem. 1996;67:1309–1316. doi: 10.1046/j.1471-4159.1996.67031309.x. [DOI] [PubMed] [Google Scholar]
- 9.Nestler EJ. Molecular mechanisms of drug addiction. Neuropharmacology. 2004;47(Suppl 1):24–32. doi: 10.1016/j.neuropharm.2004.06.031. [DOI] [PubMed] [Google Scholar]
- 10.Luscher C, Malenka RC. Drug-evoked synaptic plasticity in addiction: from molecular changes to circuit remodeling. Neuron. 2011;69:650–663. doi: 10.1016/j.neuron.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie MJ. Cellular neuroadaptations to chronic opioids: tolerance, withdrawal and addiction. Br J Pharmacol. 2008;154:384–396. doi: 10.1038/bjp.2008.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haberstock-Debic H, Kim KA, Yu YJ, von Zastrow M. Morphine promotes rapid, arrestin-dependent endocytosis of mu-opioid receptors in striatal neurons. J Neurosci. 2005;25:7847–7857. doi: 10.1523/JNEUROSCI.5045-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsuang MT, Lyons MJ, Meyer JM, Doyle T, Eisen SA, Goldberg J, et al. Co-occurrence of abuse of different drugs in men: the role of drug-specific and shared vulnerabilities. Arch Gen Psychiatry. 1998;55:967–972. doi: 10.1001/archpsyc.55.11.967. [DOI] [PubMed] [Google Scholar]
- 14.Bevilacqua L, Goldman D. Genes and addictions. Clinical pharmacology and therapeutics. 2009;85:359–361. doi: 10.1038/clpt.2009.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, et al. Association of OPRD1 polymorphisms with heroin dependence in a large case-control series. Addict Biol. 2012 doi: 10.1111/j.1369-1600.2012.00445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi J, Hui L, Xu Y, Wang F, Huang W, Hu G. Sequence variations in the mu-opioid receptor gene (OPRM1) associated with human addiction to heroin. Hum Mutat. 2002;19:459–460. doi: 10.1002/humu.9026. [DOI] [PubMed] [Google Scholar]
- 17.Yuferov V, Fussell D, LaForge KS, Nielsen DA, Gordon D, Ho A, et al. Redefinition of the human kappa opioid receptor gene (OPRK1) structure and association of haplotypes with opiate addiction. Pharmacogenetics. 2004;14:793–804. doi: 10.1097/00008571-200412000-00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drakenberg K, Nikoshkov A, Horvath MC, Fagergren P, Gharibyan A, Saarelainen K, et al. Mu opioid receptor A118G polymorphism in association with striatal opioid neuropeptide gene expression in heroin abusers. Proc Natl Acad Sci U S A. 2006;103:7883–7888. doi: 10.1073/pnas.0600871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Befort K, Filliol D, Decaillot FM, Gaveriaux-Ruff C, Hoehe MR, Kieffer BL. A single nucleotide polymorphic mutation in the human mu-opioid receptor severely impairs receptor signaling. J Biol Chem. 2001;276:3130–3137. doi: 10.1074/jbc.M006352200. [DOI] [PubMed] [Google Scholar]
- 20.Bond C, LaForge KS, Tian M, Melia D, Zhang S, Borg L, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proc Natl Acad Sci U S A. 1998;95:9608–9613. doi: 10.1073/pnas.95.16.9608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang D, Quillan JM, Winans K, Lucas JL, Sadee W. Single nucleotide polymorphisms in the human mu opioid receptor gene alter basal G protein coupling and calmodulin binding. J Biol Chem. 2001;276:34624–34630. doi: 10.1074/jbc.M104083200. [DOI] [PubMed] [Google Scholar]
- 22.Zhang Y, Wang D, Johnson AD, Papp AC, Sadee W. Allelic expression imbalance of human mu opioid receptor (OPRM1) caused by variant A118G. J Biol Chem. 2005;280:32618–32624. doi: 10.1074/jbc.M504942200. [DOI] [PubMed] [Google Scholar]
- 23.Delgado MR. Reward-related responses in the human striatum. Ann of NY Acad of Sci. 2007;1104:70–88. doi: 10.1196/annals.1390.002. [DOI] [PubMed] [Google Scholar]
- 24.Haber SN, Kim KS, Mailly P, Calzavara R. Reward-related cortical inputs define a large striatal region in primates that interface with associative cortical connections, providing a substrate for incentive-based learning. J Neurosci. 2006;26:8368–8376. doi: 10.1523/JNEUROSCI.0271-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jog MS, Kubota Y, Connolly CI, Hillegaart V, Graybiel AM. Building neural representations of habits. Science. 1999;286:1745–1749. doi: 10.1126/science.286.5445.1745. [DOI] [PubMed] [Google Scholar]
- 26.Ellgren M, Spano SM, Hurd YL. Adolescent cannabis exposure alters opiate intake and opioid limbic neuronal populations in adult rats. Neuropsychopharmacology. 2007;32:607–615. doi: 10.1038/sj.npp.1301127. [DOI] [PubMed] [Google Scholar]
- 27.Ren Y, Whittard J, Higuera-Matas A, Morris CV, Hurd YL. Cannabidiol, a nonpsychotropic component of cannabis, inhibits cue-induced heroin seeking and normalizes discrete mesolimbic neuronal disturbances. J Neurosci. 2009;29:14764–14769. doi: 10.1523/JNEUROSCI.4291-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spano MS, Ellgren M, Wang X, Hurd YL. Prenatal cannabis exposure increases heroin seeking with allostatic changes in limbic enkephalin systems in adulthood. Biological Psych. 2007;61:554–563. doi: 10.1016/j.biopsych.2006.03.073. [DOI] [PubMed] [Google Scholar]
- 29.Dai S, Corrigall WA, Coen KM, Kalant H. Heroin self-administration by rats: influence of dose and physical dependence. Pharmacology, biochemistry, and behavior. 1989;32:1009–1015. doi: 10.1016/0091-3057(89)90074-9. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs MM, Okvist A, Horvath M, Keller E, Bannon MJ, Morgello S, et al. Dopamine receptor D1 and postsynaptic density gene variants associate with opiate abuse and striatal expression levels. Mol Psych. 2012 doi: 10.1038/mp.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okvist A, Fagergren P, Whittard J, Garcia-Osta A, Drakenberg K, Horvath MC, et al. Dysregulated postsynaptic density and endocytic zone in the amygdala of human heroin and cocaine abusers. Biol Psych. 2010;69:245–252. doi: 10.1016/j.biopsych.2010.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 34.Rasband WS. ImageJ. Bethesda, Maryland, USA: U. S. National Institutes of Health; 1997–2011. [Google Scholar]
- 35.Aldridge GM, Podrebarac DM, Greenough WT, Weiler IJ. The use of total protein stains as loading controls: an alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J Neurosci Methods. 2008;172:250–254. doi: 10.1016/j.jneumeth.2008.05.00. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin ME, Jin JG, Ji RR, Tong J, Pomonis JD, Lavery DJ, et al. Complement activation in the peripheral nervous system following the spinal nerve ligation model of neuropathic pain. Pain. 2008;137:182–201. doi: 10.1016/j.pain.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 37.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic acids research. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc. 1995;57:289–300. [Google Scholar]
- 39.Lachmann A, Xu H, Krishnan J, Berger SI, Mazloom AR, Ma’ayan A. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotech. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- 42.DiNieri JA, Wang X, Szutorisz H, Spano SM, Kaur J, Casaccia P, et al. Maternal cannabis use alters ventral striatal dopamine D2 gene regulation in the offspring. Biol Psych. 2011;70:763–769. doi: 10.1016/j.biopsych.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bohn LM, Lefkowitz RJ, Gainetdinov RR, Peppel K, Caron MG, Lin FT. Enhanced morphine analgesia in mice lacking beta-arrestin 2. Science. 1999;286:2495–2498. doi: 10.1126/science.286.5449.2495. [DOI] [PubMed] [Google Scholar]
- 44.Gille H, Kortenjann M, Thomae O, Moomaw C, Slaughter C, Cobb MH, et al. ERK phosphorylation potentiates Elk-1-mediated ternary complex formation and transactivation. EMBO J. 1995;14:951–962. doi: 10.1002/j.1460-2075.1995.tb07076.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, et al. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 46.Boglari G, Szeberenyi J. Nuclear translocation of p90Rsk and phosphorylation of CREB is induced by ionomycin in a Ras-independent manner in PC12 cells. Acta Biol Hung. 2002;53:325–334. doi: 10.1556/ABiol.53.2002.3.9. [DOI] [PubMed] [Google Scholar]
- 47.Brami-Cherrier K, Lavaur J, Pages C, Arthur JS, Caboche J. Glutamate induces histone H3 phosphorylation but not acetylation in striatal neurons: role of mitogen- and stress-activated kinase-1. J Neurochem. 2007;101:697–708. doi: 10.1111/j.1471-4159.2006.04352.x. [DOI] [PubMed] [Google Scholar]
- 48.Brami-Cherrier K, Valjent E, Herve D, Darragh J, Corvol JC, Pages C, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- 50.Tomasiewicz HC, Jacobs MM, Wilkinson MB, Wilson SP, Nestler EJ, Hurd YL. Proenkephalin Mediates the Enduring Effects of Adolescent Cannabis Exposure Associated with Adult Opiate Vulnerability. Biol Psych. 2012 doi: 10.1016/j.biopsych.2012.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vaccarino FJ, Bloom FE, Koob GF. Blockade of nucleus accumbens opiate receptors attenuates intravenous heroin reward in the rat. Psychopharmacology. 1985;86:37–42. doi: 10.1007/BF00431681. [DOI] [PubMed] [Google Scholar]
- 52.Mague SD, Blendy JA. OPRM1 SNP (A118G) involvement in disease development, treatment response, and animal models. Drug Alcohol Depend. 2010;108:172–182. doi: 10.1016/j.drugalcdep.2009.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shabalina SA, Zaykin DV, Gris P, Ogurtsov AY, Gauthier J, Shibata K, et al. Expansion of the human mu-opioid receptor gene architecture: novel functional variants. Hum Mut Gen. 2009;18:1037–1051. doi: 10.1093/hmg/ddn439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang L, Kendler KS, Chen X. The mu-opioid receptor gene and smoking initiation and nicotine dependence. Behav Brain Funct. 2006;2:28. doi: 10.1186/1744-9081-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lachmann A, Ma’ayan A. Lists2Networks: integrated analysis of gene/protein lists. BMC Bioinformatics. 2010;11:87. doi: 10.1186/1471-2105-11-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boros J, Donaldson IJ, O’Donnell A, Odrowaz ZA, Zeef L, Lupien M, et al. Elucidation of the ELK1 target gene network reveals a role in the coordinate regulation of core components of the gene regulation machinery. Genome Res. 2009;19:1963–1973. doi: 10.1101/gr.093047.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang X, Odom DT, Koo SH, Conkright MD, Canettieri G, Best J, et al. Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc Natl Acad Sci U S A. 2005;102:4459–4464. doi: 10.1073/pnas.0501076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Besnard A, Galan-Rodriguez B, Vanhoutte P, Caboche J. Elk-1 a transcription factor with multiple facets in the brain. Front Neurosci. 2011;5:35. doi: 10.3389/fnins.2011.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Koob GF. A role for brain stress systems in addiction. Neuron. 2008;59:11–34. doi: 10.1016/j.neuron.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weiss F, Ciccocioppo R, Parsons LH, Katner S, Liu X, Zorrilla EP, et al. Compulsive drug-seeking behavior and relapse. Neuroadaptation, stress, and conditioning factors. Ann of NY Acad Sci. 2001;937:1–26. doi: 10.1111/j.1749-6632.2001.tb03556.x. [DOI] [PubMed] [Google Scholar]
- 61.Gutierrez-Mecinas M, Trollope AF, Collins A, Morfett H, Hesketh SA, Kersante F, et al. Long-lasting behavioral responses to stress involve a direct interaction of glucocorticoid receptors with ERK1/2-MSK1-Elk-1 signaling. Proc Natl Acad Sci U S A. 2011;108:13806–13811. doi: 10.1073/pnas.1104383108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lavaur J, Bernard F, Trifilieff P, Pascoli V, Kappes V, Pages C, et al. A TAT-DEF-Elk-1 peptide regulates the cytonuclear trafficking of Elk-1 and controls cytoskeleton dynamics. J Neurosci. 2007;27:14448–14458. doi: 10.1523/JNEUROSCI.2279-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Salinas S, Briancon-Marjollet A, Bossis G, Lopez MA, Piechaczyk M, Jariel-Encontre I, et al. SUMOylation regulates nucleo-cytoplasmic shuttling of Elk-1. J Cell Bio. 2004;165:767–773. doi: 10.1083/jcb.200310136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Besnard A, Bouveyron N, Kappes V, Pascoli V, Pages C, Heck N, et al. Alterations of molecular and behavioral responses to cocaine by selective inhibition of Elk-1 phosphorylation. J Neurosci. 2011;31:14296–14307. doi: 10.1523/JNEUROSCI.2890-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jenab S, Festa ED, Nazarian A, Wu HB, Sun WL, Hazim R, et al. Cocaine induction of ERK proteins in dorsal striatum of Fischer rats. Brain Res Mol Brain Res. 2005;142:134–138. doi: 10.1016/j.molbrainres.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 66.Valjent E, Corvol JC, Pages C, Besson MJ, Maldonado R, Caboche J. Involvement of the extracellular signal-regulated kinase cascade for cocaine-rewarding properties. J Neurosci. 2000;20:8701–8709. doi: 10.1523/JNEUROSCI.20-23-08701.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valjent E, Pages C, Rogard M, Besson MJ, Maldonado R, Caboche J. Delta 9-tetrahydrocannabinol-induced MAPK/ERK and Elk-1 activation in vivo depends on dopaminergic transmission. Eur J Neurosci. 2001;14:342–352. doi: 10.1046/j.0953-816x.2001.01652.x. [DOI] [PubMed] [Google Scholar]
- 68.Paratore C, Brugnoli G, Lee HY, Suter U, Sommer L. The role of the Ets domain transcription factor Erm in modulating differentiation of neural crest stem cells. Dev Bio. 2002;250:168–180. doi: 10.1006/dbio.2002.0795. [DOI] [PubMed] [Google Scholar]
- 69.Yuasa Y, Okabe M, Yoshikawa S, Tabuchi K, Xiong WC, Hiromi Y, et al. Drosophila homeodomain protein REPO controls glial differentiation by cooperating with ETS and BTB transcription factors. Development. 2003;130:2419–2428. doi: 10.1242/dev.00468. [DOI] [PubMed] [Google Scholar]
- 70.Hasegawa H, Ashigaki S, Takamatsu M, Suzuki-Migishima R, Ohbayashi N, Itoh N, et al. Laminar patterning in the developing neocortex by temporally coordinated fibroblast growth factor signaling. J Neurosci. 2004;24:8711–8719. doi: 10.1523/JNEUROSCI.3070-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Demir O, Aysit N, Onder Z, Turkel N, Ozturk G, Sharrocks AD, et al. ETS-domain transcription factor Elk-1 mediates neuronal survival: SMN as a potential target. Biochimica et biophysica acta. 2011;1812:652–662. doi: 10.1016/j.bbadis.2011.02.012. [DOI] [PubMed] [Google Scholar]
- 72.Kermer P, Digicaylioglu MH, Kaul M, Zapata JM, Krajewska M, Stenner-Liewen F, et al. BAG1 over-expression in brain protects against stroke. Brain Pathol. 2003;13:495–506. doi: 10.1111/j.1750-3639.2003.tb00480.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kermer P, Krajewska M, Zapata JM, Takayama S, Mai J, Krajewski S, et al. Bag1 is a regulator and marker of neuronal differentiation. Cell death and differentiation. 2002;9:405–413. doi: 10.1038/sj.cdd.4400972. [DOI] [PubMed] [Google Scholar]
- 74.Planchamp V, Bermel C, Tonges L, Ostendorf T, Kugler S, Reed JC, et al. BAG1 promotes axonal outgrowth and regeneration in vivo via Raf-1 and reduction of ROCK activity. Brain. 2008;131:2606–2619. doi: 10.1093/brain/awn196. [DOI] [PubMed] [Google Scholar]
- 75.Uemura T, Sato T, Aoki T, Yamamoto A, Okada T, Hirai R, et al. p31 deficiency influences endoplasmic reticulum tubular morphology and cell survival. Mol and Cel Bio. 2009;29:1869–1881. doi: 10.1128/MCB.01089-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aichem A, Pelzer C, Lukasiak S, Kalveram B, Sheppard PW, Rani N, et al. USE1 is a bispecific conjugating enzyme for ubiquitin and FAT10, which FAT10ylates itself in cis. Nat Comm. 2010;1:13. doi: 10.1038/ncomms1012. [DOI] [PubMed] [Google Scholar]
- 77.Marie N, Aguila B, Allouche S. Tracking the opioid receptors on the way of desensitization. Cell Signal. 2006;18:1815–1833. doi: 10.1016/j.cellsig.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 78.Bohn LM, Gainetdinov RR, Sotnikova TD, Medvedev IO, Lefkowitz RJ, Dykstra LA, et al. Enhanced rewarding properties of morphine, but not cocaine, in beta(arrestin)-2 knockout mice. J Neurosci. 2003;23:10265–10273. doi: 10.1523/JNEUROSCI.23-32-10265.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ferrer-Alcon M, La Harpe R, Garcia-Sevilla JA. Decreased immunodensities of micro-opioid receptors, receptor kinases GRK 2/6 and beta-arrestin-2 in postmortem brains of opiate addicts. Brain Res Mol Brain Res. 2004;121:114–122. doi: 10.1016/j.molbrainres.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 80.Mazzucchelli C, Vantaggiato C, Ciamei A, Fasano S, Pakhotin P, Krezel W, et al. Knockout of ERK1 MAP kinase enhances synaptic plasticity in the striatum and facilitates striatal-mediated learning and memory. Neuron. 2002;34:807–820. doi: 10.1016/s0896-6273(02)00716-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Heatmap of gene expression changes in the NAc of human heroin abusers. The hierarchical clustering of differentially expressed genes detected between the control (N=19) and heroin (N=13) groups in the NAc using mRNA microarrays. Dark colors represent normalized low expression and light colors high expression. The x-axis lists subjects represented by a three digit ID number; C-control, H-heroin user; m-male, f-female; age.
Table SI: Demographic data
Table S2: Probes used with Nanostring technologies.
Table S3- Primer pair sequences used in qPCR.
Table S4: Frequency of the rs2075572 OPRM1 polymorphism in human subjects.
Table S5: ELK1 target genes identified in 49 subjects.
Table S6: ELK1 target genes identified in 32 subjects.