Abstract
Background
Epigallocatechin-3-gallate (EGCG) has been documented for its beneficial effects protecting oxidative stress to cardiac cells. Previously, we have shown the EGCG-mediated cardiac protection by attenuating reactive oxygen species and cytosolic Ca2+ in cardiac cells during oxidative stress and myocardial ischemia. Here, we aimed to seek a deeper elucidation of the molecular anti-oxidative capabilities of EGCG in an H2O2-induced oxidative stress model of myocardial ischemia injury using H9c2 rat cardiomyoblasts.
Results
Proteomics analysis was used to determine the differential expression of proteins in H9c2 cells cultured in the conditions of control, 400 μM H2O2 exposure for 30 min with and/or without 10 to 20 μM EGCG pre-treatment. In this model, eight proteins associated with energy metabolism, mitochondrial electron transfer, redox regulation, signal transduction, and RNA binding were identified to take part in EGCG-ameliorating H2O2-induced injury in H9c2 cells. H2O2 exposure increased oxidative stress evidenced by increases in reactive oxygen species and cytosolic Ca2+ overload, increases in glycolytic protein, α-enolase, decreases in antioxidant protein, peroxiredoxin-4, as well as decreases in mitochondrial proteins, including aldehyde dehydrogenase-2, ornithine aminotransferase, and succinate dehydrogenase ubiquinone flavoprotein subunit. All of these effects were reversed by EGCG pre-treatment. In addition, EGCG attenuated the H2O2-induced increases of Type II inositol 3, 4-bisphosphate 4-phosphatase and relieved its subsequent inhibition of the downstream signalling for Akt and glycogen synthase kinase-3β (GSK-3β)/cyclin D1 in H9c2 cells. Pre-treatment with EGCG or GSK-3β inhibitor (SB 216763) significantly improved the H2O2-induced suppression on cell viability, phosphorylation of pAkt (S473) and pGSK-3β (S9), and level of cyclin D1 in cells.
Conclusions
Collectively, these findings suggest that EGCG blunts the H2O2-induced oxidative effect on the Akt activity through the modulation of PIP3 synthesis leading to the subsequent inactivation of GSK-3β mediated cardiac cell injury.
Keywords: EGCG, H9c2, Oxidative stress, Proteomics analysis, Survival pathway
Background
Oxidative stress has been associated with hypoxia or myocardial ischemia, and likely contributes to the progression of cardiovascular diseases [1]. Accumulating evidence also indicates that redox-sensitive signalling pathways via the effects of generation of reactive oxygen species (ROS) or reactive nitrogen species (RNS) or reactive lipid derived aldehydes (LDAs) are essentially involved in the pathological stress of heart cells [2]. Accordingly, molecular targeting for anti-oxidative interventions on redox signalling pathways may provide a therapeutic approach to ameliorate the risk and progression for heart diseases.
Myocardial ischemia injury involving brief regional ischemia followed by prolonged reperfusion (IR) is the result of an imbalance between myocardial oxygen supply and demand [3]. Such myocardial ischemia stress can cause oxidative stress in myocardium, in which the diminished cellular antioxidant defence system accompanied by the increased ROS production triggers the irreversible cell death [4-6]. However, the detailed mechanism of ROS-induced cardiac cell death during myocardial IR injury remains to be determined. A cell line of H9c2 rat cardiomyoblasts treated with H2O2 has been used as an in vitro cellular model for cardiac tissues in response to oxidative stress associated with heart IR injury [7-11]. Using this H2O2-induced oxidative stress model, several studies using proteomics analyses have been reported to identify target proteins associated with oxidative stress with or without antioxidant intervention [7-9].
Green tea polyphenols (GTPs), including epicatechin (EC), epigallocatechin (EGC), epicatechin-3-gallate (ECG), and epigallocatechin-3-gallate (EGCG), have potent properties of antioxidant and radical-scavenger, which may partially account for their anti-atherogenic effects [12-14]. EGCG is the most physiologically potent compound, and predominantly accounts for the biological effects of green tea [15]. Although studies have provided convincing evidence to support the cardioprotective effects of GTPs, the end effectors that mediate cardiac protection are only beginning to be addressed.
The present study aimed to seek a deeper elucidation of the potential proteins for the EGCG-mediated cardioprotection against the H2O2-induced oxidative stress in H9c2 rat cardiomyoblasts by using a proteomics study. Differential protein expression in control cells with or without treatment were distinguished by two-dimensional electrophoresis (2-DE). After image analysis, the proteins were co-detected, normalized, and quantified. Protein spots cutting off with 1.5 fold difference were selected for protein identification with matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) by peptide mass fingerprinting. The proteins identified were then used to generate an interaction map and to establish interaction networks. Based on the hypothetical model with interaction networks, the present study proposed a putative mechanism for EGCG-induced antioxidant intervention on the H2O2-induced oxidative stress in H9c2 cells.
Methods
Chemicals and reagents
H9c2 cell lines were purchased from American Type Culture Collection (ATCC, CRL-1446) (Rockville, MD). All reagents used were ACS or MB grade. EGCG, purchased from Sigma, was prepared as a stock solution of 10 mM by dissolving the compound in deionized water.
Cell culture, EGCG and/or H2O2 treatments, MTT assay
H9c2 cells were cultured in Dulbecco’s modified essential medium (DMEM, Gibco, Invitrogen Taiwan Ltd., Taipei, Taiwan) containing 10% fetal bovine serum (FBS) (Gibco), 25 mM D-glucose, 2 mM L-glutamine, 1 mM sodium pyruvate, 1% streptomycin (100 μg/ml) (Gibco), and 1% penicillin (100 U/ml) (Gibco) at pH 7.4 in a 5% CO2 incubator at 37°C. Cell viability was measured using the MTT (3-(4, 5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) cell proliferation assay (ATCC, Manassas, VA, USA). Cells (105) were seeded onto 6-cm plates in DMEM-10% FBS. After adhering overnight, the cells were changed to serum-free medium with or without EGCG for 30 min in a 5% CO2 incubator at 37°C and then washed with phosphate buffer solution (PBS). The washed cells were treated with different concentrations of H2O2 in serum-free DMEM for 30 min in a 5% CO2 incubator at 37°C. After washing with PBS, the cells were incubated in serum-free DMEM for 24 h in a 5% CO2 incubator at 37°C. After 24 h incubation, MTT was then added to the cells at a final concentration of 0.5 mg/ml and the mixture was incubated at 37°C for 4 hours. The optical density of the purple MTT formazan product was measured at 570 nm using a microplate reader.
Determination of cellular Ca2+ levels
Fura 2-AM (fura 2-tetra-acetoxymethyl ester; Molecular Probes, Eugene, OR) was used as the fluorescent indicator. H9c2 cells were dissolved in PBS containing 2 mM fura 2-AM and incubated for 45 min at room temperature and then for 30 min at 37°C, during which time the fura 2-AM was trapped inside by esterase cleavage. The cells were then washed twice with PBS and diluted to a density of 2×106 cells/ml in PBS. Recordings were made in a Perkin-Elmer LS 50B spectrofluorimeter equipped with an accessory to measure Ca2+ (Beaconsfield, Buckinghamshire, England). The dye trapped inside the cells was excited every second by exposure to alternating 340 and 380 nm light beams and the intensity of light emission at 510 nm was measured, allowing the monitoring of both the light intensity and the 340 nm fluorescence/380 nm ratio (F340/F380). EGCG was added to the cuvette using a minimum 100-fold concentrated stock solution to avoid large volume variations [16]. The 340/380 ratio (R) was calculated and converted to the corresponding levels of [Ca2+]i as described by Grynkiewicz et al. [17], using a Kd of 0.14 μM:
where Rmin and Rmax are the ratios measured by the release of intracellular dye with 2 mM EGTA in 0.1% Triton X-100 (Rmin) followed by the addition of 2.1 mM Ca2+ (Rmax), whereas Sf2/Sb2 is the ratio of the 380 nm signals in Ca2+-free and Ca2+-replete solutions, respectively.
Measurement of intracellular ROS generation by fluorescence spectrophotometry
Intracellular ROS levels were assessed using 2’, 7’-dichlorofluorescein diacetate (DCF-DA) [18]. Cells (1.2 × 106) loaded with DCF-DA in 3 ml PBS at a final concentration of 10 μM were incubated at 37°C for 1 h. After incubation, the cells were then washed three times with PBS by centrifugation at 300 × g at 4°C for 5 min. The cells re-suspended with PBS and brought to a density of 105 cells/ml were measured for DCF-DA fluorescence changes every 10 min after the addition of H2O2 or EGCG by fluorescence spectrophotometry. The fluorescence excitation maximum for DCF-DA was 495 nm, and the corresponding emission maximum was 527 nm.
Sample preparation and two-dimensional electrophoresis
After treatment, the cells were washed twice with cold PBS and lysed in 2-DE lysis buffer containing 7 M urea, 2 M thiourea, 4% (w/v) 3-[(3-cholamidopropyl) dimethylammonio]-1-propane sulfonate, 2% immobilized pH gradient (IPG) buffer (GE Healthcare UK Ltd., England) and 40 mM DTT. Protein concentration was determined by Bradford protein assay kit (Bio-Rad) according to manufacturer’s procedure. Immobilized nonlinear pH gradient strips (pH 4–7, 24 cm) were rehydrated with 450 μg protein at room temperature overnight (at least 12 h). Isoelectric focusing was then performed using an Ettan IPGphor 3 instrument (GE Healthcare) for a total of 60 kVh at 20°C. After isoelectric focusing, strips were equilibrated in 6 M urea, 75 mM Tris–HCl (pH 8.8), 29.3% (v/v) glycerol, 2% (w/v) SDS and 0.002% (w/v) bromophenol blue with 1% (w/v) DTT for 15 min and then in the same buffer containing 2.5% (w/v) iodoacetamide for 15 min. The equilibrated IPG strips were transferred onto 10% polyacrylamide gels and sealed with 0.5% (w/v) low-melting-point agarose in SDS running buffer containing 0.02% (w/v) bromophenol blue. The gels were run in an Ettan DALTsix electrophoresis system (GE Healthcare) at 40 mA per gel at 20°C until the dye reached the bottom of the gels.
Gels staining, image analysis and MALDI-TOF MS analysis
After the electrophoresis, gels were stained with Bio-safe™ Coomassie G-250 Stain (Bio-Rad) according to the manufacturer’s protocol. Then, stained gels were scanned with Scanmaker 9800XL (Microtek) using a resolution of 300 dpi. Spot detection, gel matching, and spot quantification were performed by using ImageMaster™ 2D Platinum 7.0 (GE healthcare). The criteria used for selection of candidate protein spots were: (1) the protein spots with >1.5-fold increased or decreased intensity between H2O2 group and control group (Ctrl.), and (2) the protein spots with >1.5-fold recovery between EGCG + H2O2 group and H2O2 group. The proteins of interest were then excised, destained, dehydrated and in-gel digested with trypsin. The resulting peptides were concentrated using Zip-Tip C18 (Millipore). 1 μ1 of peptide was mixed with 1 μ1 of matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid), spotted onto a target plate and subjected to mass spectrometer. The mass fingerprint search was using the MASCOT search engine (Matrix Science, London, UK; http://www.matrixscience.com) against SwissPort/TrEMBL database. The parameters used for searching were: Rattus; allow one missed cleavage by trypsin; carbamidomethylation of cysteine, partial methionine oxidation and mass tolerance of 100 ppm. Proteins identification was based on MASCOT Mowse scores (p < 0.05) and the calculated MW and pI had to be in common with the observed MW and pI on 2-DE.
Gels staining, image analysis and MALDI-TOF MS analysis
After the electrophoresis, gels were stained with Bio-safe™ Coomassie G-250 Stain (Bio-Rad) according to the manufacturer’s protocol. Then, stained gels were scanned with Scanmaker 9800XL (Microtek) using a resolution of 300 dpi. Spot detection, gel matching, and spot quantification were performed by using ImageMaster™ 2D Platinum 7.0 (GE healthcare). The criteria used for selection of candidate protein spots were: (1) the protein spots with >1.5-fold increased or decreased intensity between H2O2 group and control group (Ctrl.), and (2) the protein spots with >1.5-fold recovery between EGCG + H2O2 group and H2O2 group. The proteins of interest were then excised, destained, dehydrated and in-gel digested with trypsin. The resulting peptides were concentrated using Zip-Tip C18 (Millipore). 1 μ1 of peptide was mixed with 1 μ1 of matrix solution (α-cyano-4-hydroxycinnamic acid, 5 mg/mL in 50% (v/v) acetonitrile/0.1% (v/v) trifluoroacetic acid), spotted onto a target plate and subjected to mass spectrometer. The mass fingerprint search was using the MASCOT search engine (Matrix Science, London, UK; http://www.matrixscience.com) against SwissPort/TrEMBL database. The parameters used for searching were: Rattus; allow one missed cleavage by trypsin; carbamidomethylation of cysteine, partial methionine oxidation and mass tolerance of 100 ppm. Proteins identification was based on MASCOT Mowse scores (p < 0.05) and the calculated MW and pI had to be in common with the observed MW and pI on 2-DE.
Real-time polymerase chain reaction
Total RNA was isolated using TRIzol® Reagent (Invitrogen) according to the manufacturer’s protocol. For reverse transcription, 2 μg of total RNA was used for reverse transcription with Moloney Murine Leukemia Virus Reverse Transcriptase (M-MLV-RT) (Genemark) using oligo-dT. Samples were run in triplicate using the SYBR qPCR Kit (Genemark) and the ABI Prism 7300 Sequence Detection System and software (Applied Biosystems). The primers used for qPCR were listed in Table 1.
Table 1.
Primers used for real time quantitative PCR to detect gene expression in H9c2 cells
| Accession no. | mRNA name | Primer sequence (5’ → 3’) | Size of products (bp) |
|---|---|---|---|
|
NM_032416.1 |
Aldehyde dehydrogenase 2 family (mitochondrial) (Aldh2) |
Forward: TGGCTGATCTCATCGAACGG (360–379) |
134 |
| Reverse: CCAGCCAGCATAATAGCGGA (493–474) | |||
|
NM_012554.3 |
Enolase 1, (alpha) (Eno1) |
Forward: CCTACTGCCAGAACTTCACCA (102–122) |
208 |
| Reverse: GAGACACCCTTCCCCATGAA (309–290) | |||
|
NM_057141.1 |
Heterogeneous nuclear ribonucleoprotein K (Hnrnpk) |
Forward: CACCTTGCTTTGTGGTCACTG (1700–1720) |
232 |
| Reverse: TTAGTTTAGGGGTGGGCTGG (1931–1912) | |||
|
NM_001007149.1 |
Staufen, RNA binding protein, homolog 2 (Drosophila) (Stau2) |
Forward: CAGAGCGGGGTCATTTCTCG (25–44) |
220 |
| Reverse: GGATGCTATGGAAACGGGCT (244–225) | |||
|
NM_022521.3 |
Ornithine aminotransferase (Oat) |
Forward: CAGGGTGAAGCGGGTGTTAT (803–822) |
262 |
| Reverse: CGTGCTCGCCTGGTTTAATG (1064–1045) | |||
|
NM_053917.1 |
Inositol polyphosphate-4-phosphatase, type II (Inpp4b) |
Forward: ATGGAAAAGATGCCGCCTGA (2739–2758) |
239 |
| Reverse: TCGTCTCTCAGGATGGAGCA (2977–2958) | |||
|
NM_053512.2 |
Peroxiredoxin 4 (Prdx4) |
Forward: GCCAAGATTTCCAAGCCAGC (268–287) |
284 |
| Reverse: CTTATTGGCCCCAGTCCTCC (551–532) | |||
|
NM_130428.1 |
Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) (Sdha) |
Forward: ATGGGCGAACCTACTTCAGC (793–812) |
84 |
| Reverse: AAGGTAAACCAGCCCGAGTG (876–857) |
Western blot analysis
After treatment, the cells were washed twice with cold PBS and lysed in cell lysis buffer containing 20 mM Tris–HCl (pH 7.5), 150 mM NaCl, 1% Triton X-100, 1 mM phenylmethanesulfonyl fluoride, 2 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, 0.3 μM aprotinin, 130 μM bestatin, 14 μM proteinase inhibitor E-64, 1 mM EDTA, 1 μM leupetin and 1% phosphatase inhibitor cocktail 2/3 (Sigma). Protein concentration was determined by Bradford protein assay kit (Bio-Rad). One hundred microgram of samples were resolved on 12% SDS-PAGE gels and then transferred onto a PVDF membrane. The immuo-blotting procedure was as described previously [11]. The membranes were blocked with 5% bovine serum albumin (BSA) and incubated with anti-Inpp4b (Santa Cruz; 1: 1000 dilution), anti-phospho-AKT (Ser473) (Sigma, 1: 500 dilution), anti-phospho-AKT (Thr308) (Santa Cruz, 1: 500 dilution), anti-phospho-GSK-3β (Ser9) (Santa Cruz; 1: 1000 dilution), anti-cyclin D1 (Santa Cruz; 1: 1000 dilution) and anti-GAPDH antibody (Santa Cruz; 1: 1000 dilution), followed by incubation with AP-conjugated anti-rabbit or anti-mouse IgG secondary antibodies (Santa Cruz; 1:5000). Proteins specifically recognized by the antibody were visualized using the 5-bromo-4-chloro-3-indolyl phosphate-nitro blue tetrazolium substrate kit (invitrogen). Band intensities were quantified using Quantity One software (Bio-Rad).
Measurements of Aldh activity
Aldh activity was measured at 25°C in 33 mM sodium pyrophosphate containing 0.8 mM NAD+, 15 μM propionaldehyde and 0.1 ml of cellular extract (50 μg soluble protein). Propionaldehyde, the substrate of Aldh, was oxidized into propionic acid by Aldh, while NAD+ was reduced to NADH to quantitatively indicate the Aldh activity. Production of NADH was determined by spectrophotometric absorbance at 340 nm. Aldh activity was expressed as nmol NADH/min per mg protein. An extinction coefficient of 6.22/mM per cm for NADH was used for the calculation of reaction rates [19].
Statistical analysis
Excel 2013 (Microsoft office) was used to perform statistical analyses. Quantitative values are presented as mean ± standard error (mean ± SEM). Statistical significance between more than two groups was tested using one-way ANOVA, while comparisons between two groups were performed using Student’s t test. Differences were considered to be statistically significant when p < 0.05 or less.
Results
The proteomic strategy used to evaluate EGCG-mediated cardioprotection against H2O2-induced oxidative stress in H9c2 rat cardiomyoblasts
In this study, H2O2 treatment of H9c2 rat cardiomyocytes was used as a model for oxidative stress associated with heart IR injury. Upon H2O2 treatment from 0 to 1000 μM for 30 min, a dose-dependent decrease in cell viability occurred in H9c2 cells with a 50% decrease of cell viability occurred at 400 μM H2O2 (Figure 1a). On the other hand, the toxicity of EGCG yielding 50% cell death for H9c2 cells was found to appear at 50 μM (Figure 1b). EGCG pre-treatment with 10 or 20 μM for 30 min effectively improved viability of cells in prior to their exposure to 400 μM H2O2 (Figure 1c). To understand further the molecular events for EGCG-mediated anti-oxidative intervention on the H2O2-induced oxidative stress, H9c2 cells cultured in the medium of control, 400 μM H2O2 with or without 20 μM EGCG pre-treatment (Figure 1d) were used to differentiate their protein expression profile by 2-DE analyses (Figure 1e).
Figure 1.
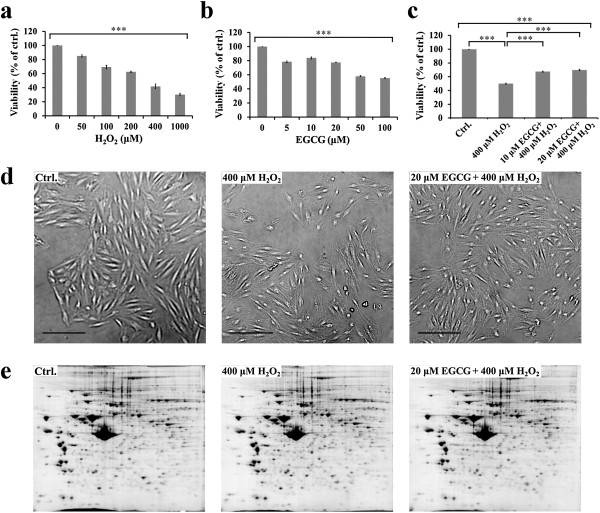
A proteomic study of the effects of EGCG on H2O2-induced oxidative stress in H9c2 cells. (a) MTT assay of cell viability after incubation with 0, 50, 100, 200, 400 and 1000 μM H2O2 for 30 min (b) MTT assay of cell viability after incubation with 0, 5, 10, 20, 50 and 100 μM EGCG for 30 min (c) H9c2 cells were pre-treated with 10 and 20 μM EGCG for 30 min followed by 400 μM H2O2 for 30 min, cell viability was measured by MTT assay (d) Phase contrast microscopy showing cell morphology of H9c2 cells in the condition of control (left), 400 μM H2O2 exposure for 30 min (middle), and 20 μM EGCG pre-treatment for 30 min followed by 400 μM H2O2 exposure for 30 min (right). (Scale Bar = 200 μm). (e) 2-DE gel images showing protein distribution in the condition of control (left), 400 μM H2O2 exposure for 30 min (middle), and 20 μM EGCG pre-treatment for 30 min followed by 400 μM H2O2 exposure for 30 min (right). In a, the values are the mean ± SEM, with ***p < 0.001.
2-DE analysis on differential protein expression in control, and H2O2-treated H9c2 cells with and without EGCG pretreatment
2-DE global protein expression analysis resolved more than 330 protein spots showing differential expression among three conditions (Figure 2a). In order to understand H9c2 cells in response to H2O2-induced oxidative stress and EGCG-mediated antioxidant interventions, the criteria setting with >1.5-fold increased or decreased intensity between H2O2 group and control group, and >1.5-fold recovery between EGCG pretreatment group and H2O2 group were used to select candidate protein spots on the 2-DE gels. According to the thresholded settings, 8 protein spots confirmed by three-dimensional image analysis (Figure 2b) were selected for protein identification with MALDI-TOF mass spectrometry by peptide mass fingerprinting (Figure 3a). Identified proteins were listed in Table 2. To establish a hypothetical model for interaction networks, the proteins identified were imported into the EMBL Search Tool for the Retrieval of Interacting Proteins (STRING) database (http://string-db.org/) to generate an interaction map (Figure 3b). According to functionally annotations derived from the reported database, these differentially expressed proteins are implicated in cellular energetic metabolism, including: α-enolase (Eno1), aldehyde dehydrogenase-2 (Aldh2), and ornithine aminotransferase (Oat), mitochondrial electron transfer, i.e. succinate dehydrogenase ubiquinone flavoprotein subunit (Sdha), redox regulation, i.e. peroxiredoxin-4 (Prdx4), Akt signal transduction, i.e. Type II inositol 3,4-bisphosphate 4-phosphatase (Inpp4b), RNA binding, i.e. heterogeneous nuclear ribonucleoprotein K (HnRNP K) and Staufen homolog 2 (Stau2) (Table 2).
Figure 2.
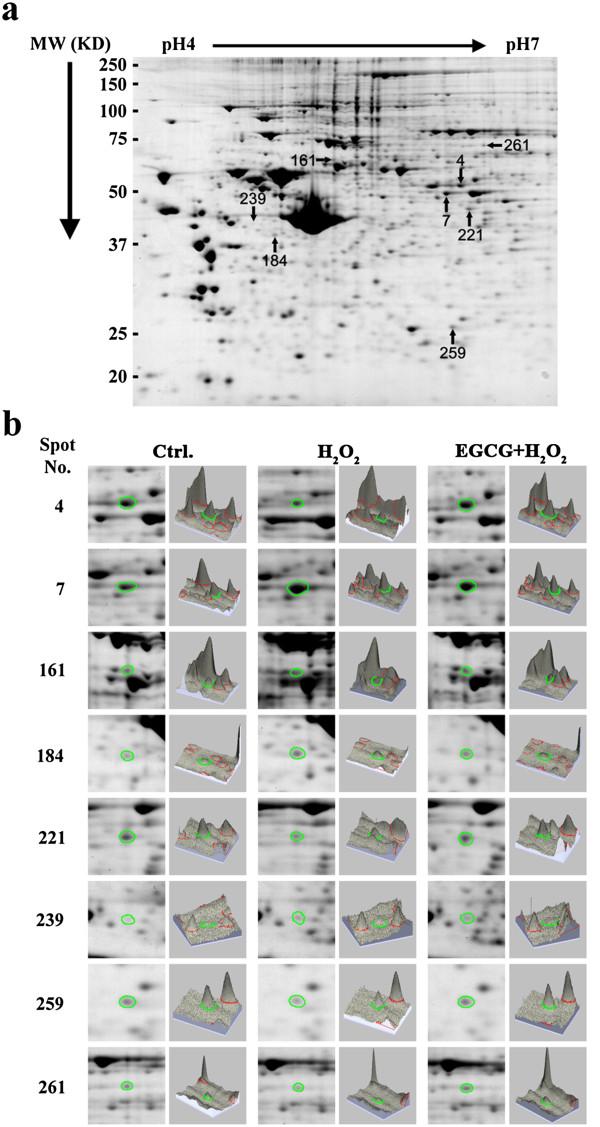
2D analysis. (a) A coomassie blue stained of 2-DE map showing the differentially protein spot profiles in H9c2 cells. The differentially expressed proteins are annotated with spot numbers. (b) Representative images of each spot in 2-DE map (left) and three-dimensional spot images (right).
Figure 3.
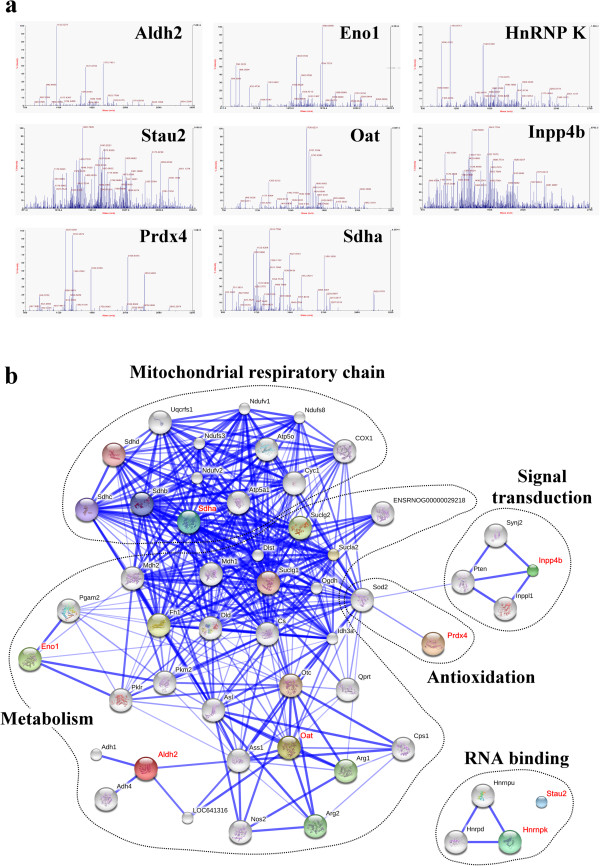
Molecular identification and hypothetical protein-protein interactions. (a) Peptide mass fingerprinting of Aldh2, Eno1, HnRNP K, Stau2, Oat, Inpp4b, Prdx4 and Sdha were identified by MALDI-TOF. (b) The identified proteins in Table 2 were input into STRING database (http://string-db.org/) and an interaction map was generated.
Table 2.
List of identified differentially expressed proteins obtained with 2D-DIGE coupled with MALDI-TOF MS analysis
| Spot no. | Swiss-Prot no. | Protein name | MW | p I | No. match peptides | Cov.(%) | Score | Function | H 2 O 2 /Ctrl | EGCG/H 2 O 2 |
|---|---|---|---|---|---|---|---|---|---|---|
|
Constitutively upregulated
|
|
|
|
|
|
|
|
|
||
| 7 |
P04764 |
Alpha-enolase (Eno1) |
47,440 |
6.16 |
18/48 |
43 |
153/51 |
Metabolism |
+1.88 |
−1.77 |
| 184 |
Q68SB1 |
Double-stranded RNA-binding protein Staufen homolog 2 (Stau2) |
62,870 |
9.54 |
14/61 |
23 |
59/51 |
RNA binding |
+1.75 |
−2.46 |
| 239 |
Q9QWG5 |
Type II inositol 3,4-bisphosphate 4-phosphatase (Inpp4b) |
106,205 |
5.86 |
12/44 |
14 |
57/51 |
Signal transduction |
+2.24 |
−1.80 |
|
Constitutively downregulated
|
|
|
|
|
|
|
|
|
||
| 4 |
P11884 |
Aldehyde dehydrogenase, mitochondrial (Aldh2) |
56,966 |
6.63 |
15/40 |
33 |
141/51 |
Metabolism |
−1.99 |
+2.02 |
| 161 |
P61980 |
Heterogeneous nuclear ribonucleoprotein K (Hnrnpk) |
51,230 |
5.39 |
10/39 |
28 |
76/51 |
RNA binding |
−2.43 |
+2.01 |
| 221 |
P04182 |
Ornithine aminotransferase, mitochondrial (Oat) |
48,701 |
6.53 |
13/38 |
30 |
123/51 |
Metabolism |
−1.94 |
+1.87 |
| 259 |
Q9Z0V5 |
Peroxiredoxin-4 (Prdx4) |
31,216 |
6.18 |
17/34 |
57 |
221/51 |
Antioxidation |
−3.71 |
+2.89 |
| 261 | Q920L2 | Succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial (Sdha) | 72,596 | 6.75 | 31/52 | 51 | 305/51 | Electron transport | −3.21 | +2.34 |
Average ratios of differential expression (p < 0.05) across H9c2 cells, H9c2 cells treated with and/or without 10 to 20 μM EGCG pre-treatment followed by 400 μM H2O2 exposure were calculated from triplicate gels.
Effects of H2O2 and EGCG on oxidative stress associated with cellular metabolism
H2O2 exposure increased oxidative stress in H9c2 cells, evidenced by increases in ROS and cytosolic Ca2+ overload (Figure 4a). In addition, H2O2 exposure decreased the protein level but not mRNA expression for antioxidant protein, i.e. Prdx4, in H9c2 cells; whereas, EGCG pretreatment prevented the decrease of the protein level without effect on the mRNA expression for Prdx4 in H2O2-treated cells (Figure 4b). When H9c2 cells were treated with H2O2, the protein level of glycolytic protein, Eno1, was increased with its decreased mRNA expression in cells (Figure 4b). EGCG pretreatment reversed the H2O2-decreased protein level with no effect on the mRNA expression for Eno1 in the H2O2-treated cells (Figure 4c). In contrast, mitochondrial proteins involved in aerobic energy production, including Aldh2, Oat, and Sdha were decreased in H9c2 cells with the H2O2-induced oxidative stress, but not changed in the H2O2-treated cells with EGCG pretreatment, as compared to cells in the control condition (Figure 4d). For these mitochondrial proteins, only Aldh2 mRNA expression was decreased by H2O2-induced oxidative stress but recovered by EGCG pretreatment (Figure 4d). In addition, total cellular Aldh activity (nmole/min/mg protein) was measured as 377.6 ± 26.0, 217.0 ± 21.1, and 301.5 ± 18.7 in H9c2 cells under the conditions of control, H2O2 treatment with and without EGCG, respectively (Figure 4e). These results suggested that under the H2O2-induced oxidative stress, H9c2 cells undergo the inflicted cellular changes of energy production by switching aerobic metabolism to anaerobic metabolism. Moreover, EGCG pretreatment could induce antioxidant intervention and protect cardiac cells from the H2O2-induced oxidative stress.
Figure 4.
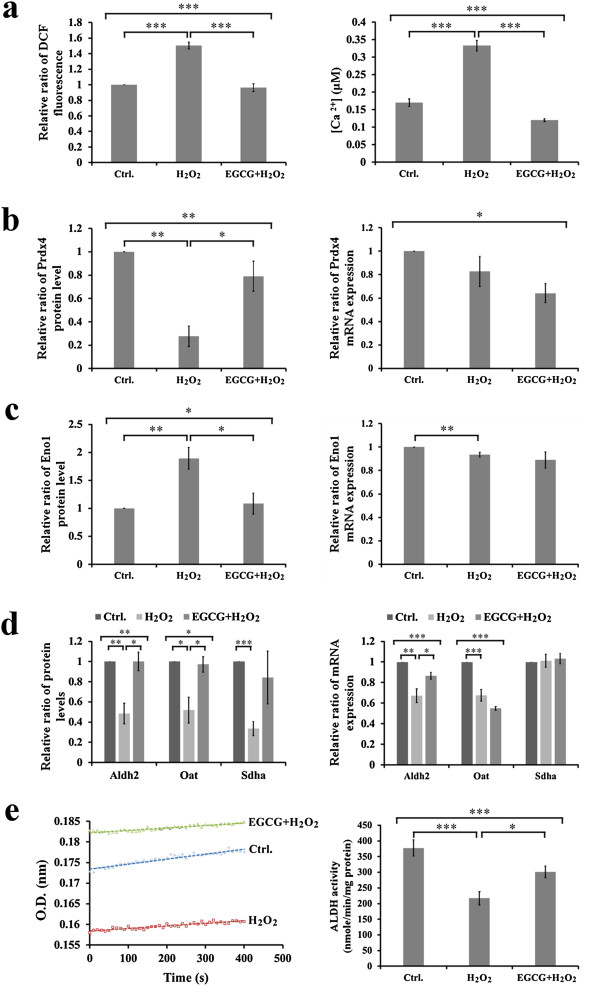
Effects of H2O2 and EGCG on oxidative stress associated with cellular metabolism. (a) Measurements of intracellular ROS formation by DCF-DA in H9c2 cells (left). Cellular Ca2+ levels were measured using the Fura-2 fluorescence dye (right). (b) Representative result of Prdx4 protein levels in 2-DE gel and mRNA expression (c) Representative result of Eno1 protein levels in 2-DE gel and mRNA expression (d) Representative results of Aldh2, Oat and Sdha protein levels in 2-DE gel and mRNA expression. (e) ALDH activity was measured by spectrophotometer at 340 nm (left). Quantitative results showed protective effect of EGCG in H2O2-induced H9c2 cells (right). All values are the mean ± SEM, with *p < 0.05; **p < 0.01; ***p < 0.001.
Effects of H2O2 and EGCG on PI3K/Akt/GSK3β signaling pathway
Inpp4b, one of the enzymes involved in membrane phosphatidylinositol (PI) metabolism, has been shown to act as one of phosphoinositide 3-kinase (PI3K) inhibitors for the subsequent activation of Akt prosurvival signalling pathway [20-22]. In the present study, Inpp4b protein level and mRNA expression were increased when cells were exposed to H2O2 (Figure 5a). Concomitantly, the levels of phosphorylated Akt (S473), phosphorylated GSK-3b (S9), and cyclin D1 were decreased in H2O2-treated cells (Figure 5b). This result is consistent with the finding that oxidative stress regulates the activity of the cell survival factor Akt through the regulation of PI(3,4,5)P3 and PI(3,4)P2 synthesis [23]. In addition, EGCG pretreatment counteracted the H2O2-increased Inpp4b expression in H9c2 cells (Figure 5a).
Figure 5.
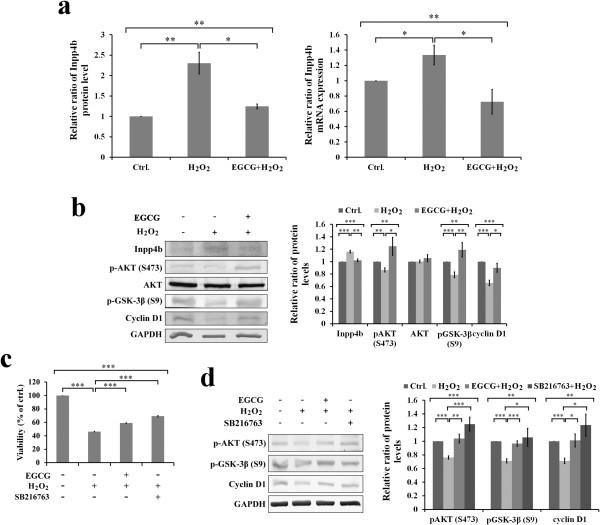
Effects of H2O2 and EGCG on PI3K/AKT/GSK3β signaling pathway. (a) Representative result of Inpp4b protein level in 2-DE gel and mRNA expression (b) Western blotting (right) and quantitative analysis (left) showing effects of EGCG on Inpp4b, p-AKT (S473), AKT, p-GSK-3β (S9), cyclin D1 in H2O2-induced H9c2 cells. (c) MTT assay showing the improvement of H2O2-suppressed cell viability by EGCG and/or GSK-3β inhibitor, SB216763 pre-treatment in H2O2-induced H9c2 cells. (d) Western blotting (right) and quantitative analysis (left) showing effects of EGCG and/or GSK-3β inhibition by GSK-3β inhibitor, SB216763, on the phosphorylation of Akt (S473), GSK-3β (S9) and cyclin D1 in H2O2-induced H9c2 cells. All values are the mean ± SEM, with *p < 0.05; **p < 0.01; ***p < 0.001.
Recently, we have shown that the Akt prosurvival pathway associated with glycogen synthase kinase-3β (GSK-3β) signalling takes part in EGCG-mediated cardoioprotection in an H2O2-induced H9c2 cell injury [11]. Consistently, immunoblot analyses showed that EGCG attenuated the H2O2-induced increases Inpp4b and relieved its subsequent inhibition of the downstream signalling for Akt and GSK-3β/cyclin D1 in H9c2 cells (Figure 5b). Pre-treatment with EGCG or GSK-3β inhibitor (SB 216763) significantly improved the H2O2-induced suppression on cell viability (Figure 5c), phosphorylation of pAkt (S473) and pGSK-3β (S9), and level of cyclin D1 (Figure 5d) in cells.
Figure 6 shows effects of H2O2 and EGCG on the Thr308 (T308) phosphorylation of Akt in H9c2 cells. In contrast to Akt phosphorylation at S473, H2O2 exposure significantly increased Akt phosphorylation at T308 in H9c2 cells. With EGCG pretreatment, T308 phosphorylation was suppressed by 20% in H2O2-treated H9c2 cells.
Figure 6.
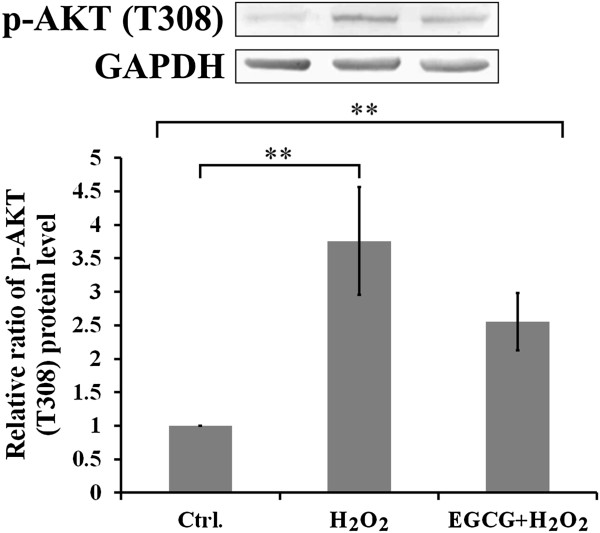
Effects of H2O2 and EGCG on p-AKT (T308). Western blotting (upper) and quantitative analysis (lower) showing effects of EGCG on the p-AKT (T308) in H2O2-induced H9c2 cells. All values are the mean ± SEM, with **p < 0.01.
Discussion
Previously, we have demonstrated the cardio-protection of green tea polyphenols (GTPs) against oxidative stress associated with myocardial ischemic injury by reducing cytosolic Ca2+ overload and generation of ROS via the Akt/GSK-3β/β-catenine and caveolae signaling in a rat surgical model of myocardial ischemia and in an H2O2-induced oxidative stress model of H9c2 rat cardiomyoblasts [11,24-26]. In the present study, using the H9c2 cell model of H2O2-induced oxidative stress for a proteomics study (Figures 1, 2), we identified proteins involved in energy metabolism, mitochondrial electron transfer, redox regulation, signal transduction, and RNA binding that might take part in EGCG-ameliorating H2O2-induced injury in H9c2 cells (Figure 3, Table 2).
During hypoxia or ischemia, ATP depletion impairs the Ca2+ uptake capacity of the sarcoplasmic reticulum (SR), leading to intracellular Ca2+ accumulation [27]. The rise in Ca2+ leads to mitochondrial Ca2+ accumulation, particularly during reperfusion when oxygen is reintroduced. Reintroduction of oxygen causing damage to the electron transport chain results in increased mitochondrial generation of ROS [27]. Both mitochondrial Ca2+ overload and increased ROS can result in opening of the mitochondrial permeability transition pore, which further compromises cellular energetics [28]. In this study, not only increasing ROS formation and cytosolic Ca2+ overload (Figure 4a), H2O2 exposure also resulted in decreasing the level of antioxidant, Prox4 in H9c2 cells (Figure 4b) as well as altering the expression for cellular energy production by decreasing the expression for mitochondrial metabolism (i.e. Aldh2, Oat, and Sdha) (Figure 4c) but increasing the expression for glycolytic metabolism (i.e. Eno1) (Figure 4d). Apparently, oxidative stress strongly correlates with mitochondrial dysfunction and likely contributes to the decline in mitochondrial bioenergetics [29]. Moreover, the activity of ALDH was found to decrease significantly in the H2O2-treated H9c2 cells (Figure 4e). This might suggest that excessive ROS leading to the formation of potentially toxic aldehydes induces inactivation of Aldh2 such as to impair the cardiac functions [30]. The present study also showed that EGCG pretreatment prevented the decrease of antioxidant, Prdx4 in H2O2-treated H9c2 cells (Figure 4b), and avoided the H2O2-decreased mitochondrial proteins (Figure 4c) with -increased glycolytic protein, Eno1 (Figure 4d), as well as ameliorated the Aldh2 activity during the H2O2-induced oxidative stress (Figure 4e). Consistently, a study with cultured rat cardiomyocytes exposed to different periods of hypoxia (H), followed by reoxygenation (R), demonstrated that GTPs acts to counteract the H/R damage-induced switch to the biosynthesis of highly unsaturated fatty acids [31], pointing out the importance of GTPs in providing good antioxidant defence not only after, but mainly prior to, the onset of H [31].
It has been shown that oxidative stress induced the PI3K/Akt dependent apoptosis in cardiac cells [32,33]. PI3K produces two lipid products that PI(3,4,5)P3 contributes predominantly to Thr308 (T308) phosphorylation and membrane-associated activation of Akt, but PI(3,4)P2 contributes mostly to Ser473 (S473) phosphorylation and cytoplasmic activation of Akt [34]. In this study, the H2O2-increased Inpp4b in concomitant with the decrease in protein levels of phosphorylated Akt (S473), phosphorylated GSK-3β (S9), and cyclin D1 appeared in H9c2 cells (Figure 5). This result might also suggest that the H2O2-induced oxidative stress caused to decrease the level of PI(3,4)P2 for turning on activation of cytosolic Akt phosphorylation at S473 such as to relieve the subsequent inhibition on the downstream target of GSK-3β/cyclin D1 in H9c2 cells (Figure 5). It has been shown that GSK-3 inhibition limits myocardial IR injury and stimulates glycogen synthesis, repartitions glucose away from glycolysis, reduces proton production from glucose metabolism, and attenuates intracellular Ca2+ overload [35]. Moreover, several studies have shown that H2O2-induced oxidative stress can trigger T308 phosphorylation for membrane-associated activation of Akt, by the PI3K dependent pathway in lymphocytes [36-38]. The present study also indicated that H2O2 exposure modulates the PI3K signalling events for Akt phosphorylation at T308 in H9c2 cells, and this Akt phosphorylation at T308 is partly suppressed by EGCG pretreatment (Figure 6).
The present study using cardiac proteomic analysis has identified EGCG-induced cardio-protection against H2O2-induced oxidative stress through the Akt/GSK-3β pathway in cultured H9c2 cells. However, the limitation of this study was to identify the modified target proteins associated with anti-oxidative effect of EGCG. The future work using redox proteomics might further help identify and quantify EGCG-mediated changes within the proteome both in redox signaling and under oxidative stress conditions.
Conclusions
In summary, the results obtained with proteomic analyses that EGCG blunts the H2O2-induced oxidative effect on the Akt activity through the modulation of PI(3,4)P2 and PI (3,4,5)P3 synthesis leading to the subsequent inhibition of GSK-3β mediated cardiac cell injury.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CWC carried out all experiments, and drafted the manuscript. HSR participated in the design of the study, and drafted the manuscript. CCH participated in the measurements of Aldh activity. HBD participated in the design of the study. LYM conceived of the study, and participated in the design and coordination and helped to draft the manuscript and final MS submission. All authors read and approved the final manuscript.
Contributor Information
Wei-Cheng Chen, Email: weicheng1112@yahoo.com.tw.
Shih-Rong Hsieh, Email: Season@vghtc.gov.tw.
Chun-Hwei Chiu, Email: chchiu@dragon.nchu.edu.tw.
Ban-Dar Hsu, Email: bdhsu@life.nthu.edu.tw.
Ying-Ming Liou, Email: ymlion@dragon.nchu.edu.tw.
Acknowledgements
This work was supported by the National Science Council of Taiwan government (to Y-M L., Grants: NSC 100-2320-B-005-001, NSC 101-2320-B-005-001).
References
- Santos CX, Anilkumar N, Zhang M, Brewer AC, Shah AM. Redox signaling in cardiac myocytes. Free Radic Biol Med. 2011;50:777–793. doi: 10.1016/j.freeradbiomed.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall SB, Oh JY, Diers AR, Landar A. Oxidative modification of proteins: an emerging mechanism of cell signaling. Front Physiol. 2012;3:369. doi: 10.3389/fphys.2012.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdouw PD, van den Doel MA, de Zeeuw S, Duncker DJ. Animal models in the study of myocardial ischaemia and ischaemic syndromes. Cardiovasc Res. 1998;39:121–135. doi: 10.1016/S0008-6363(98)00069-8. [DOI] [PubMed] [Google Scholar]
- Burgoyne JR, Mongue-Din H, Eaton P, Shah AM. Redox signaling in cardiac physiology and pathology. Circ Res. 2012;111:1091–1106. doi: 10.1161/CIRCRESAHA.111.255216. [DOI] [PubMed] [Google Scholar]
- Ferdinandy P, Schulz R, Baxter GF. Interaction of cardiovascular risk factors with myocardial ischemia/reperfusion injury, preconditioning, and postconditioning. Pharmacol Rev. 2007;59:418–458. doi: 10.1124/pr.107.06002. [DOI] [PubMed] [Google Scholar]
- Anversa P, Kajstura J. Myocyte cell death in the diseased heart. Circ Res. 1998;82:1231–1233. doi: 10.1161/01.RES.82.11.1231. [DOI] [PubMed] [Google Scholar]
- Chou HC, Chen YW, Lee TR, Wu FS, Chan HT, Lyu PC, Timms JF, Chan HL. Proteomics study of oxidative stress and Src kinase inhibition in H9C2 cardiomyocytes: a cell model of heart ischemia-reperfusion injury and treatment. Free Radic Biol Med. 2010;49:96–108. doi: 10.1016/j.freeradbiomed.2010.04.001. [DOI] [PubMed] [Google Scholar]
- Law CH, Li JM, Chou HC, Chen YH, Chan HL. Hyaluronic acid-dependent protection in H9C2 cardiomyocytes: a cell model of heart ischemia-reperfusion injury and treatment. Toxicology. 2013;303:54–71. doi: 10.1016/j.tox.2012.11.006. [DOI] [PubMed] [Google Scholar]
- Chen YW, Chou HC, Lin ST, Chen YH, Chang YJ, Chen L, Chan HL. Cardioprotective effects of quercetin in cardiomyocyte under ischemia/reperfusion injury. Evid Based Complement Alternat Med. 2013;2013:364519. doi: 10.1155/2013/364519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou HC, Chan HL. 5-Methoxytryptophan-dependent protection of cardiomyocytes from heart ischemia reperfusion injury. Arch Biochem Biophys. 2014;543:15–22. doi: 10.1016/j.abb.2013.12.014. [DOI] [PubMed] [Google Scholar]
- Hsieh SR, Hsu CS, Lu CH, Chen WC, Chiu CH, Liou YM. Epigallocatechin-3-gallate-mediated cardioprotection by Akt/GSK-3beta/caveolin signalling in H9c2 rat cardiomyoblasts. J Biomed Sci. 2013;20:86. doi: 10.1186/1423-0127-20-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stangl V, Dreger H, Stangl K, Lorenz M. Molecular targets of tea polyphenols in the cardiovascular system. Cardiovasc Res. 2007;73:348–358. doi: 10.1016/j.cardiores.2006.08.022. [DOI] [PubMed] [Google Scholar]
- Mak JC. Potential role of green tea catechins in various disease therapies: progress and promise. Clin Exp Pharmacol Physiol. 2012;39:265–273. doi: 10.1111/j.1440-1681.2012.05673.x. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Yokozawa T. Direct scavenging of nitric oxide and superoxide by green tea. Food Chem Toxicol. 2002;40:1745–1750. doi: 10.1016/S0278-6915(02)00169-2. [DOI] [PubMed] [Google Scholar]
- L’Allemain G. Multiple actions of EGCG, the main component of green tea. Bull Cancer. 1999;86:721–724. [PubMed] [Google Scholar]
- Hsu YC, Liou YM. The anti-cancer effects of (−)-epigallocatechin-3-gallate on the signaling pathways associated with membrane receptors in MCF-7 cells. J Cell Physiol. 2011;226:2721–2730. doi: 10.1002/jcp.22623. [DOI] [PubMed] [Google Scholar]
- Grynkiewicz G, Poenie M, Tsien RY. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- Maciel EN, Vercesi AE, Castilho RF. Oxidative stress in Ca(2+)-induced membrane permeability transition in brain mitochondria. J Neurochem. 2001;79:1237–1245. doi: 10.1046/j.1471-4159.2001.00670.x. [DOI] [PubMed] [Google Scholar]
- Li SY, Li Q, Shen JJ, Dong F, Sigmon VK, Liu Y, Ren J. Attenuation of acetaldehyde-induced cell injury by overexpression of aldehyde dehydrogenase-2 (ALDH2) transgene in human cardiac myocytes: role of MAP kinase signaling. J Mol Cell Cardiol. 2006;40:283–294. doi: 10.1016/j.yjmcc.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Agoulnik IU, Hodgson MC, Bowden WA, Ittmann MM. INPP4B: the new kid on the PI3K block. Oncotarget. 2011;2:321–328. doi: 10.18632/oncotarget.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozulic L, Hemmings BA. PIKKing on PKB: regulation of PKB activity by phosphorylation. Curr Opin Cell Biol. 2009;21:256–261. doi: 10.1016/j.ceb.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Bozulic L, Surucu B, Hynx D, Hemmings BA. PKBalpha/Akt1 acts downstream of DNA-PK in the DNA double-strand break response and promotes survival. Mol Cell. 2008;30:203–213. doi: 10.1016/j.molcel.2008.02.024. [DOI] [PubMed] [Google Scholar]
- Ma K, Cheung SM, Marshall AJ, Duronio V. PI(3,4,5)P3 and PI(3,4)P2 levels correlate with PKB/akt phosphorylation at Thr308 and Ser473, respectively; PI(3,4)P2 levels determine PKB activity. Cell Signal. 2008;20:684–694. doi: 10.1016/j.cellsig.2007.12.004. [DOI] [PubMed] [Google Scholar]
- Hsieh SR, Tsai DC, Chen JY, Tsai SW, Liou YM. Green tea extract protects rats against myocardial infarction associated with left anterior descending coronary artery ligation. Pflugers Arch. 2009;458:631–642. doi: 10.1007/s00424-009-0655-1. [DOI] [PubMed] [Google Scholar]
- Liou YM, Hsieh SR, Wu TJ, Chen JY. Green tea extract given before regional myocardial ischemia-reperfusion in rats improves myocardial contractility by attenuating calcium overload. Pflugers Arch. 2010;460:1003–1014. doi: 10.1007/s00424-010-0881-6. [DOI] [PubMed] [Google Scholar]
- Liou YM, Kuo SC, Hsieh SR. Differential effects of a green tea-derived polyphenol (−)-epigallocatechin-3-gallate on the acidosis-induced decrease in the Ca(2+) sensitivity of cardiac and skeletal muscle. Pflugers Arch. 2008;456:787–800. doi: 10.1007/s00424-008-0456-y. [DOI] [PubMed] [Google Scholar]
- Murphy E, Steenbergen C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol Rev. 2008;88:581–609. doi: 10.1152/physrev.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng TI, Jou MJ. Oxidative stress caused by mitochondrial calcium overload. Ann N Y Acad Sci. 2010;1201:183–188. doi: 10.1111/j.1749-6632.2010.05634.x. [DOI] [PubMed] [Google Scholar]
- Judge S, Leeuwenburgh C. Cardiac mitochondrial bioenergetics, oxidative stress, and aging. Am J Physiol Cell Physiol. 2007;292:C1983–C1992. doi: 10.1152/ajpcell.00285.2006. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang H, Hao P, Xue L, Wei S, Zhang Y, Chen Y. Inhibition of aldehyde dehydrogenase 2 by oxidative stress is associated with cardiac dysfunction in diabetic rats. Mol Med. 2011;17:172–179. doi: 10.2119/molmed.2010.00114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordoni A, Angeloni C, Leoncini E, Danesi F, Maranesi M, Biagi PL, Hrelia S. Hypoxia/reoxygenation alters essential fatty acids metabolism in cultured rat cardiomyocytes: protection by antioxidants. Nutr Metab Cardiovasc Dis. 2005;15:166–173. doi: 10.1016/j.numecd.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Shi YP, Wu D, Ji YJ, Wang X, Chen HL, Wu SS, Huang DJ, Jiang W. Salidroside protects against hydrogen peroxide-induced injury in cardiac H9c2 cells via PI3K-Akt dependent pathway. DNA Cell Biol. 2011;30:809–819. doi: 10.1089/dna.2010.1183. [DOI] [PubMed] [Google Scholar]
- Angeloni C, Spencer JP, Leoncini E, Biagi PL, Hrelia S. Role of quercetin and its in vivo metabolites in protecting H9c2 cells against oxidative stress. Biochimie. 2007;89:73–82. doi: 10.1016/j.biochi.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Scheid MP, Marignani PA, Woodgett JR. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol. 2002;22:6247–6260. doi: 10.1128/MCB.22.17.6247-6260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar MA, Wang L, Clanachan AS. Cardioprotection by GSK-3 inhibition: role of enhanced glycogen synthesis and attenuation of calcium overload. Cardiovasc Res. 2010;86:478–486. doi: 10.1093/cvr/cvp421. [DOI] [PubMed] [Google Scholar]
- Lahair MM, Howe CJ, Rodriguez-Mora O, McCubrey JA, Franklin RA. Molecular pathways leading to oxidative stress-induced phosphorylation of Akt. Antioxid Redox Signal. 2006;8:1749–1756. doi: 10.1089/ars.2006.8.1749. [DOI] [PubMed] [Google Scholar]
- Lindvall J, Islam TC. Interaction of Btk and Akt in B cell signaling. Biochem Biophys Res Commun. 2002;293:1319–1326. doi: 10.1016/S0006-291X(02)00382-0. [DOI] [PubMed] [Google Scholar]
- Cheung SMS, Kornelson JC, Al-Alwan M, Marshall AJ. Regulation of phosphoinositide 3-kinase signaling by oxidants: Hydrogen peroxide selectively enhances immunoreceptor-induced recruitment of phosphatidylinositol (3,4) bisphosphate-binding PH domain proteins. Cell Signal. 2007;19:902–912. doi: 10.1016/j.cellsig.2006.10.013. [DOI] [PubMed] [Google Scholar]


