Abstract
A novel marine thermophilic and heterotrophic Anaerolineae bacterium in the phylum Chloroflexi, strain SW7T, was isolated from an in situ colonization system deployed in the main hydrothermal vent of the Taketomi submarine hot spring field located on the southern part of Yaeyama Archipelago, Japan. The microbial community associated with the hydrothermal vent was predominated by thermophilic heterotrophs such as Thermococcaceae and Anaerolineae, and the next dominant population was thermophilic sulfur oxidizers. Both aerobic and anaerobic hydrogenotrophs including methanogens were detected as minor populations. During the culture-dependent viable count analysis in this study, an Anaerolineae strain SW7T was isolated from an enrichment culture at a high dilution rate. Strain SW7T was an obligately anaerobic heterotroph that grew with fermentation and had non-motile thin rods 3.5–16.5 μm in length and 0.2 μm in width constituting multicellular filaments. Growth was observed between 37–65°C (optimum 60°C), pH 5.5–7.3 (optimum pH 6.0), and 0.5–3.5% (w/v) NaCl concentration (optimum 1.0%). Based on the physiological and phylogenetic features of a new isolate, we propose a new species representing a novel genus Thermomarinilinea: the type strain of Thermomarinilinea lacunofontalis sp. nov., is SW7T (=JCM15506T=KCTC5908T).
Keywords: Chloroflexi, Anaerolineae, thermophile, hydrothermal, anaerobe
Distribution of the putative non-phototrophic Chloroflexi class Anaerolineae has been revealed by culture-independent analyses in diverse marine, terrestrial and artificial environments including hydrothermal environments, and it is known to be one of the uncultivated bacterial lineages (28, 32). In this decade, 9 species, Anaerolinea thermophila, Anaerolinea thermolimosa, Levilinea saccharolytica, Leptolinea tardivitalis, Bellilinea caldifistulae, Longilinea arvoryzae, Thermanaerothrix daxensis and Ornatilinea apprima, have been isolated (6, 21, 32). The members of A. thermophila, A. thermolimosa, L. saccharolytica, L. tardivitalis and B. caldifistulae were isolated from methanogenic sludges under mesophilic or thermophilic conditions. In contrast to these species, L. arvoryzae was obtained from a rice field soil, and two species of T. daxensis and O. apprima were derived from terrestrial subsurface hot aquifers (6, 21, 31). Furthermore, isolation of two marine strains from an enrichment reactor of subseafloor sediments was reported recently (10). These species have similar physiological characteristics, such as being obligately anaerobic, mesophilic to moderately thermophilic, and multicellular filamentous heterotrophs utilizing carbohydrates and amino acids (or peptides). All the Anaerolineae species except for O. apprima are slowly growing microorganisms with generation times of 10–100 hours under the optimum growth conditions (6, 21, 32).
On the other hand, no Anaerolineae isolates have been reported from hydrothermal environments although the class has been recognized as one of the significant bacterial populations associated with these environments based on SSU rRNA gene clone analyses (28). The relatively slow growth of this group, and co-occurrence of diverse heterotrophic bacteria in hydrothermal vent environments might have prevented enrichment and isolation of the Anaerolineae species. In hydrothermal environments, the predominance of thermophilic and heterotrophic Thermococcaceae archaea and other thermophilic heterotrophs growing faster than Anaerolineae species has likely inhibited the enrichment and isolation of Anaerolineae species. We fortunately isolated the marine thermophilic strain of the Anaerolineae from a shallow submarine hydrothermal field in the southern part of the Yaeyama Archipelago, Japan during culture-dependent viable counting analysis of the microbial ecosystem associated with hydrothermal activity. We report here the microbial community structure attached to an in situ colonization system (ISCS) deployed in the main hydrothermal vent of the hydrothermal field, which was the isolation source of the Anaerolineae strain. Furthermore, we describe the physiological and partial chemotaxonomic characterization of a novel strain belonging to the class Anaerolineae.
Materials and Methods
Sample collection
Taketomi submarine hot spring shallow hydrothermal field (24° 20′ 9″N, 124° 06′ 10″E) is located in a coral lagoon in the southern part of the Yaeyama Archipelago, Japan (4, 15). The geochemistry of venting fluids, microbial mat formation in the hydrothermal environments, and diverse novel bacterial strains have already been reported (8, 9, 16, 27). An in situ colonization system with a self-recording temperature probe (STR-ISCS) (25) was set in the main vent emission for 5 days. Pumiceous materials were placed in the ISCS to support microbial attachment (Fig. S1). Placement and retrieval of the STR-ISCS were conducted by scuba divers (June 2006). The temperature of the main vent emission in this hydrothermal field determined by the STR-ISCS was a constant 52°C at a water depth of 17 m. The retrieved pumice in the STR-ISCS was sub-sampled for cultivation and stored anaerobically (with 0.05% neutralized Na2S) in a Schott glass bottle under 100% N2 (100 kPa), sealed with butyl rubber stoppers. Samples were stored at 5°C. The sample for molecular analyses was stored at −80°C.
Enrichment and purification
The abundance of viable archaea and bacteria represented by a variety of physiological and metabolic characteristics was determined by a series of serial dilution cultures under various cultivation conditions. In this study, cultures were grown in a 15 mL test tube containing 3 mL medium. MJYPS medium (29) was used at 85 and 55°C for thermophilic to hyperthermophilic fermentative sulfur-reducing heterotrophs, MMJ medium (26) was used at 70°C for thermophilic methanogens, MMJSO medium (20) was used at 85, 70 and 55°C for thermophilic to hyperthermophilic sulfate reducers, MMJS medium (18) was used at 85 and 70°C for thermophilic to hyperthermophilic sulfur reducers, MMJHS medium (25) with three types of head space gases of 80% H2 and 20% CO2 (200 kPa), and 79% H2, 20% CO2 and 1% O2 (200 kPa) was used at 85, 70 and 55°C for thermophilic to hyperthermophilic, anaerobic to microaerophilic autotrophs (nitrate-reducing and microaerophilic hydrogen oxidizers and sulfur oxidizers), and MMJYPS medium (19) was used at 70 and 55°C for strictly anaerobic thermophilic mixotrophs. Compositions of these media are summarized in the supplementary material. The microorganisms present in the most diluted series of the medium at each temperature were isolated by the subsequent extinction–dilution method (29).
In order to obtain an isolate of thermophilic Anaerolineae strain, MJYPS medium was used for serial dilution cultivation at 55°C. Since the growth of a potential strain was unstable, modified MJY (MJY medium supplemented with 0.1% NaHCO3) medium under head space gas of N2 and CO2 mixture (16) was used for further isolation and characterization. MJY medium consists of MJ synthetic seawater with 0.1% yeast extract. The medium was prepared as follows: 1) Modified MJY medium with resazurin was autoclaved under N2 gas; 2) The medium was pressurized with N2/CO2 gas mixture (80:20) at 150 kPa; 3) Neutralized Na2S solution (final 0.05%) was added to the medium. Pure culture was obtained by the dilution to extinction technique. The first dilution to extinction was carried out at 45°C, and the following dilution to extinction was conducted at 65°C. Purity of the isolate was tested by microscopic observation for cultures obtained at different growth temperatures (30–70°C) with MJY, repeating SSU rRNA gene direct sequencing and SSU rRNA gene clone analysis described below. In addition, the absence of Thermococcaceae in the culture was also confirmed by a cultivation test using MJYPS medium at 70°C.
Microscopic observations
Cells were routinely observed using an Olympus BX51 microscope (Tokyo, Japan). Scanning electron microscope observation was carried out using JSM-6700F (JEOL, Tokyo, Japan) as described previously (3). Transmission electron micrographs of negatively stained cells and thin cell sections were obtained as described by Zillig et al. (34). Cells grown in modified MJY medium at 60°C in the late exponential phase were used for transmission electron microscope observations using JEOL JEM-1210 at an accelerating voltage of 80 kV.
Nucleic acid analyses
Environmental DNA was extracted using the Ultra Clean Soil DNA Purification Mega Kit (Mo Bio Laboratories, Solana Beach, CA, USA). The archaeal and bacterial SSU rRNA genes were amplified from the DNA assemblage using LA Taq polymerase with GC buffer (Takara Bio, Otsu, Japan) with primer sets of Arch21F-U907R and B27F-U907R, respectively (2, 11) (Table S1). Gene fragments of mcrA were also obtained with a primer set of ME3MF and ME2r’ (7, 17) using SYBR Premix Ex Taq II (Takara Bio). The DNA amplification conditions are summarized in Table S1. PCR amplification was performed using a thermal cycler GeneAmp 9700 (Perkin-Elmer, Waltham, MA, USA). The amplified gene fragments were cloned into pCR2.1 vector (Invitrogen, Carlsbad, CA, USA) and were sequenced with M13 primer by the deoxynucleotide chain-termination method with a DNA sequencer model 3130XL (Applied Biosystems, Carlsbad, CA, USA).
The archaeal and prokaryotic SSU rRNA genes were quantified according to a previously published method (24) with minor modifications using the 7500 Real Time PCR System (Applied Biosystems) (17). Sets of primers and probes used for archaeal and total prokaryotic SSU rRNA genes were Arch349F-516F-806R and Uni340F-516F-806R, respectively, and PCR conditions are summarized in Table S1. Quantification of mcrA with a primer set of ME3MF and ME2r’ was also conducted using SYBR Premix Ex Taq II as described previously (17). Abundance of each gene was determined as an average of duplicate or triplicate analyses.
Genomic DNA for PCR amplification from isolates was purified using the Illustra bacteria genomicPrep Mini Spin Kit (GE Healthcare, Buckinghamshire, England). The SSU rRNA gene was amplified by PCR using LA Taq polymerase with GC buffer. Primers Bac27F and 1492R (2, 11) were used for PCR amplification (Table S1). Amplified SSU rRNA gene fragment served as a direct template for sequencing analysis.
In order to know the phylogenetic positions of the SSU rRNA genes from the environmental DNA and isolates, these sequences were analyzed by the Blast search program in DDBJ, and ARB software (14). Alignments of SSU rRNA gene sequences from the Chloroflexi species, strain SW7T and environmental DNA were generated using ARB software (14), and the SSU rRNA gene trees using unambiguous residues were constructed by the PhyML method using SeaView4 software (5). Alignment of mcrA sequences and the neighbor-joining phylogenetic tree using unambiguous nucleic acid residues were constructed by Clustal X ver 2.0 (12).
The G+C content of genomic DNA from a Chloroflexi strain SW7T prepared as in Lauer et al. (1986) (13) was determined by direct analysis of deoxyribonucleotides by HPLC (30).
Growth characteristics of Chloroflexi strain
Growth of the isolate was determined by microscopic observation in most cases. Utilization of possible electron donors (H2 and yeast extract) and acceptors (sulfur (0.3 w/v%), thiosulfate (0.1%), sulfate (0.3%), sulfite (0.03%), l-cystine (0.03%), glutathione (oxidized form) (0.03%), nitrate (0.1%), nitrite (0.03%)) was examined with modified MJY medium. Cultures were grown in a 15 mL test tube containing 3 mL medium during the tests for growth characteristics, except for the experiments that determined the growth rates described below.
To determine the growth range of temperature, NaCl concentration and pH, cultures were grown in a 100 mL serum bottle containing 33 mL medium with the head-space (N2/CO2) gas described above in temperature control drying ovens. Modified MJY medium was used for cultivation tests for temperature (30–70°C) and NaCl concentration (0–5%). The effect of initial pH on growth was examined at 60°C using modified MJY medium. Various pHs (pH 5–8) were adjusted by changing NaHCO3 concentrations in the MJY medium (0 or 0.1%), CO2 concentrations in head-space gas (0–20%) and the pressure of head-space gas (100–170 kPa). Aggregate formation and precipitation of medium inhibited cell counting and the protein concentration measurement, respectively. Thus, growth rate was determined from the cellular ATP concentration with respective intervals as follows. Cells in 1 mL culture were filtrated through a 0.2 μm polycarbonate filter (12 mm in diameter), and cellular ATP concentration was measured using ATP analyzer AF-100 (TOADKK, Tokyo, Japan) following the manufacturer’s instructions. ATP concentration was measured in duplicate.
The utilization of carbon sources was tested using MJY medium without yeast extract supplemented with the vitamin mixture (1) and the following carbon sources at 60°C: yeast extract, tryptone, peptone, Casamino acids (Difco), gelatin, chitin, starch, glucose, fructose, maltose, galactose, lactose, cellobiose, xylose, sucrose, rhamnose, mannose, ethanol, methanol, glycerol, acetate, pro-pionate, pyruvate, formate, fumarate, citrate, malate, succinate, tartarate, glutamate, glycine, alanine and xylan (each substrate was tested at 2 concentrations; 0.02% and 0.1%). The substrate utilization test with 0.01% yeast extract was also performed for substrates that did not support growth as a sole carbon source in the absence of yeast extract. Head-space gas (N2/CO2) was prepared as described above.
The sensitivity of antibiotics such as ampicillin, chloramphenicol, erythromycin, kanamycin, penicillin G, novobiocin, spectinomycin, tetracycline, streptomycin, vancomycin and rifampicin at 50 μg mL−1 was tested at 60°C using modified MJY medium.
Fatty acid analysis of Chloroflexi strain
The cellular fatty acid composition of strain SW7T was analyzed with cells grown in modified MJY medium at 60°C in the late exponential phase. Lyophilized cells were suspended in 1 mL anhydrous methanolic HCl and heated at 100°C for 3 h. The fatty acid methyl esters (FAMEs) were extracted three times with n-hexane. Concentrated FAMEs were analyzed using a gas chromatography-mass spectrometer (Xcalibur for Trace DSQ; Thermo Fisher Scientific, Waltham, MA, USA).
Nucleotide sequence accession numbers
The GenBank/EMBL/DDBJ accession number for the SSU rRNA gene sequence of Chloroflexi strain SW7T is AB669272. Environmental SSU rRNA and mcrA gene sequences, and SSU rRNA genes from representative strains of viable populations are also deposited in the public database with accession numbers AB752310 to AB752343.
Results
Environmental SSU rRNA gene and mcrA community structures
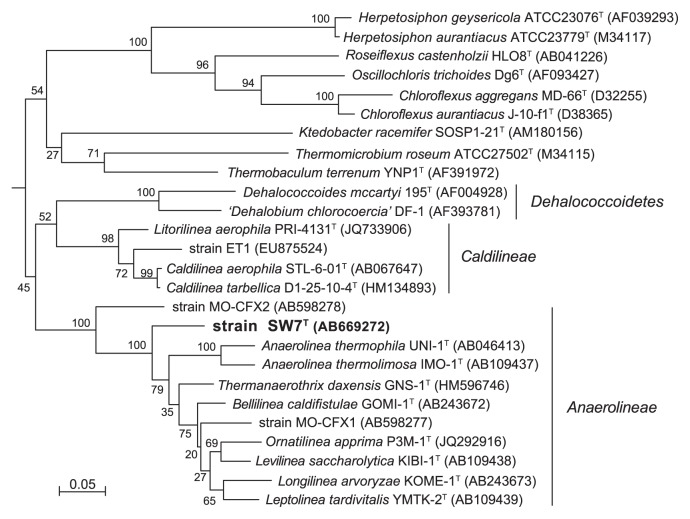
Quantitative PCR analyses indicated that gene abundances of total prokaryotic SSU rRNA gene, archaeal SSU rRNA gene and mcrA were 2.4×109, 4.8×107 and 3.1×103 copies g pumice in the ISCS−1, respectively. Compositions of bacterial and archaeal SSU rRNA gene and mcrA communities associated with the ISCS settled in the main vent emission are summarized in Fig. 1 and Table S2 and S3. Bacterial SSU rRNA gene community was predominated by Chloroflexi phylotypes belonged to Anaerolineae (Fig. S2), and deltaproteobacterial phylotypes were also detected as dominant populations. Except for one phylotype closely related to the gammaproteobacterial sulfur-oxidizing genus Sulfurivirga, other phylotypes detected in the bacterial SSU rRNA gene clone analysis were likely derived from heterotrophic bacterial populations. The archaeal SSU rRNA gene clone library was predominated by hyperthermophilic Thermococcaceae and the thermophilic Deep-sea Hydrothermal Vent Euryarchaeotic Group (DHVEG) represented by “Candidatus Acidulliprofundum” (22). Unusually, most of the Themococcaceae sequences represented by the phylotype TKM_WI_A50 were closely related to Palaeococcus ferriphilus (99.8% similarity) while Thermococcus or Pyrococcus species have been known to predominate in most of the hydrothermal vent ecosystems (28). One sequence grouped into methanogenic or methanotrophic Lost City Methanosarcinales was identified in the analysis. On the other hand, the mcrA community consisted of three phylogroups, ANME I, ANME I-like and unclassified sediment clusters, and this composition suggested that more than 70% of the mcrA sequences detected in this study derived from anaerobic methanotrophs (ANMEs) but not methanogens (Fig. 1, Fig S3, Table S3).
Fig. 1.
Bacterial and archaeal SSU rRNA genes and mcrA gene community structures obtained from the pumiceous materials in the ISCS deployed in the main hydrothermal vent of the Taketomi hydrothermal field. Numbers in parentheses indicate the number of clones sequenced.
Viable populations
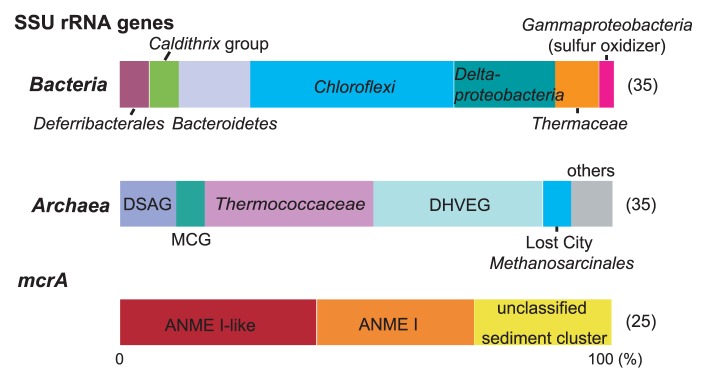
Heterotrophic Thermococcaceae and Anaerolineae members and chemolithoautotrophic sulfur-oxidizing gamma-proteobacterium Sulfurivirga sp. were found to be the most predominantly cultivated populations in the hydrothermal vent (Fig. 2, Table S4). Hydrogenotrophic viable populations of chemolithoautotrophic Persephonella sp. and mixotrophic Deferribacter sp. were obtained, although these viable numbers were more than 3 orders of magnitude smaller than those of fermentative heterotrophs and sulfur-oxidizing chemolithoautotrophs. Thermophilic methanogens and sulfate reducers were not detected.
Fig. 2.
Viable microbial populations associated with the ISCS deployed in the main hydrothermal vent of the Taketomi hydrothermal field.
The Anaerolineae member obtained in the viable count was phylogenetically distant from the previously isolated and/or characterized strains in this family. Thus, we tried to purify a strain and characterize it in detail, as described below.
Purification and morphology of the Anaerolineae strain
From a diluted series of enrichment cultures at 55°C with MMJYS medium, into which pumiceous materials recovered from the STR-ISCS had been inoculated, an aggregate of non-motile thin rods was observed. Positive growth of the mono-morphotype was observed at the dilution rate of <10−9 cells mL−1 pumice without sulfide production, and no growth was observed at dilution rates of <10−8 cells mL−1 pumice (Fig. 2, Table S4). After several successive cultures, we determined a partial SSU rRNA gene sequence of the enrichment by direct sequencing of the PCR product without ambiguous residues. The result suggested that a single Anaerolineae species grew predominantly in the enrichment. Since the growth of the thin rods was unstable in the MMJYS medium and elemental sulfur was not reduced during culture, we examined several media and found that modified MJY medium could support stable growth of the thin rods. We then used the modified MJY medium for further purification and culture analyses. Since colony formation of the thin rods was not observed using a role-tube method, we applied the dilution to extinction technique serially for purification. Before purification using the serial dilution to extinction technique, we confirmed the contamination of hyperthermophilic Thermococcaceae-like and moderately thermophilic fermenters in the enrichment using MMJYPS and MJY medium incubated at 70 and 40°C, respectively, by microscopic observation; therefore, serial dilution to extinction was conducted at 40°C and then at 65°C to eliminate Thermococcaceae-like and moderately thermophilic fermenters, respectively. After purification, the absence of Thermococcaceae-like and moderately thermophilic fermenters was confirmed by the culture test as described above with no PCR amplification of the archaeal SSU rRNA gene. Purity of the isolate was also examined by sequencing the 5′ or 3′ end of 94 SSU rRNA gene fragments. Then, we determined the almost full length (1,435 bp) of the SSU rRNA gene sequence without ambiguous residues, and thus obtained strain SW7T (=JCM 15506T=KCTC5908T).
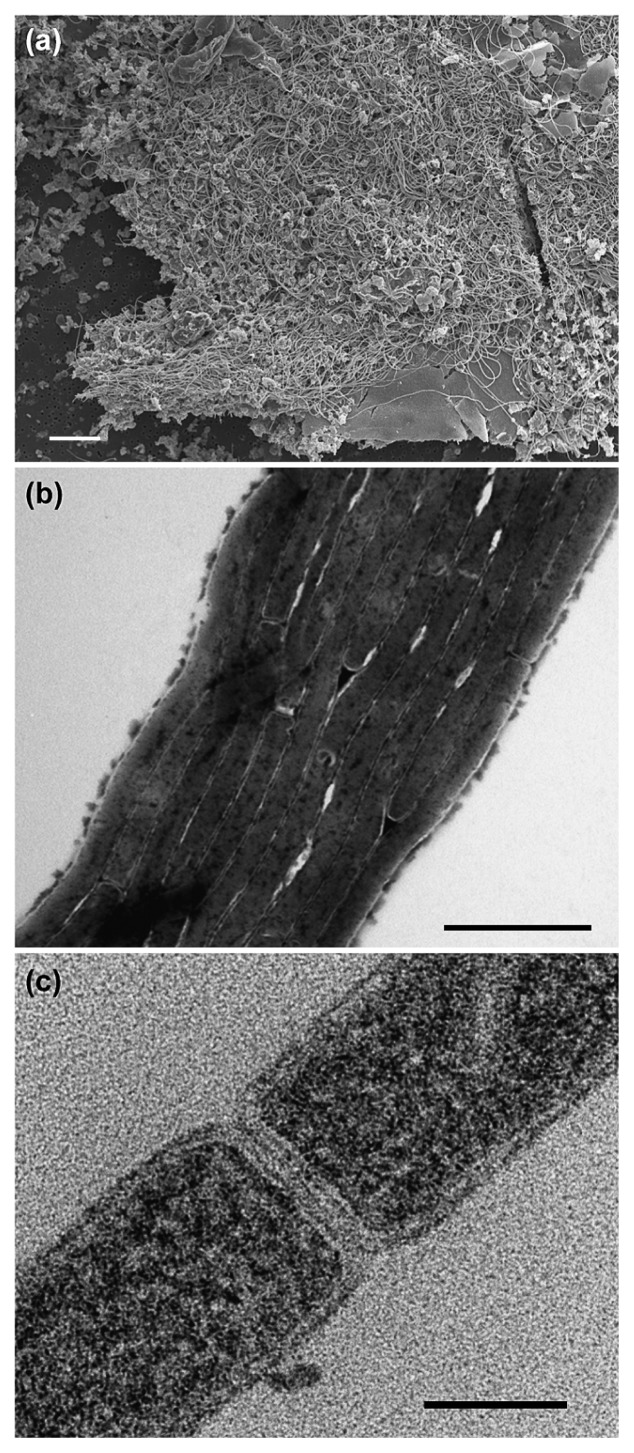
Thin rod cells of strain SW7T constituted multicellular filaments and formed large aggregates up to 1 cm in longest diameter (Fig. 3) at the bottom of the 15 mL test tube. Cells were about 3.5–16.5 μm in length, 0.2 μm in width without a flagellum and did not show gliding motility (Fig. 3). Spores were not found under any growth conditions or with any observation techniques.
Fig. 3.
Scanning electron micrograph of SW7T cells (a). Transmission electron micrographs of negatively stained (b) and thin section (c) cells of strain SW7T. Scale bars indicate 10 μm (a), 1 μm (b) and 0.1 μm (c).
Growth characteristics
Strain SW7T grew only by fermentation and did not utilize molecular oxygen (O2) or other potential electron acceptors. The isolate was sensitive to O2 and could not grow in the medium without reducing regents. Strain SW7T grew in modified MJY medium over a temperature range of 37–65°C (optimum; 50–60°C) (Fig. 4). Growth was observed at 37–65°C, but growth at 65°C was unstable and sometimes stopped. No growth was observed at 30 and 70°C. The growth pH range was 5.5–7.3 (optimum pH was 6.0), and no growth occurred at pH 5.0 and 7.6. Effect of NaCl concentration on growth in modified MJY medium was tested, and growth was observed 0.5–3.5% (w/v) NaCl concentration (optimum 1.0%) (Fig. 4). The fastest doubling time was 4.6 h, observed at 60°C in modified MJY medium with 1.0% NaCl concentration. In the substrate utilization test, tryptone, peptone, casein, gelatin, chitin, glutamate, alanine, mannitol and citrate supported growth in the absence of yeast extract. Substrate utilization in the presence of 0.01% yeast extract was also tested only for substrates that were not utilized as a sole carbon source in the absence of yeast extract, but no substrates supported the growth of the strain.
Fig. 4.
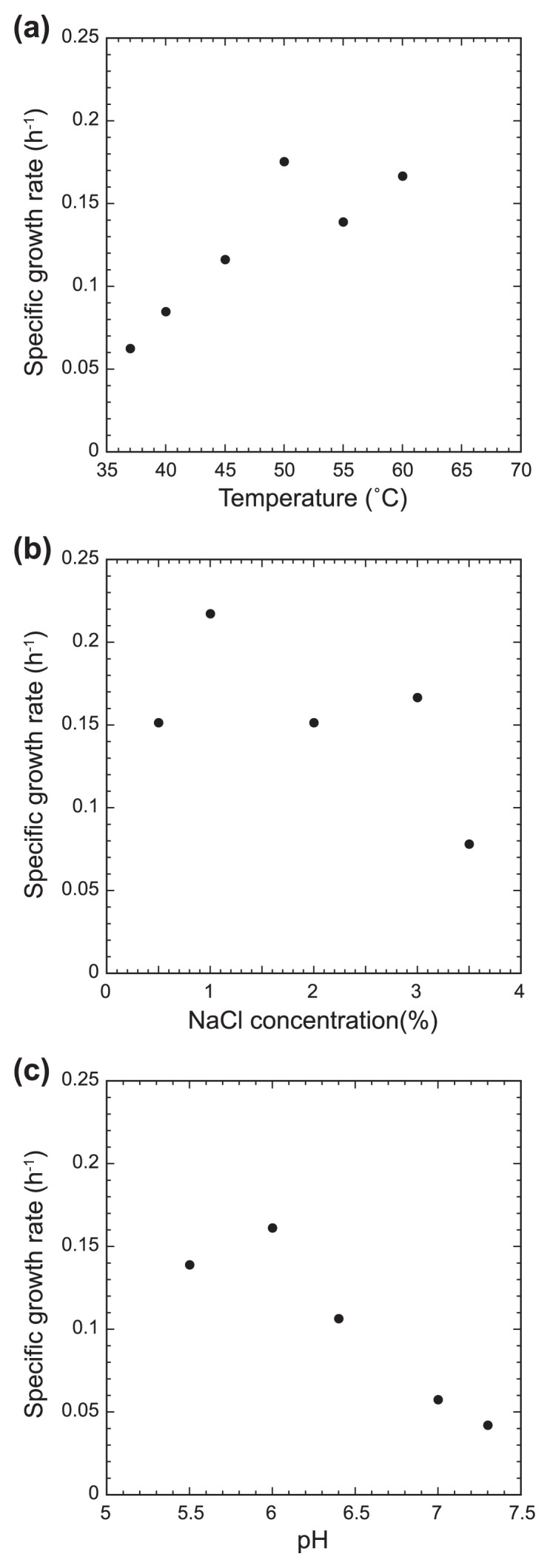
The effect of temperature (a), NaCl concentration (b) and (c) pH on growth of strain SW7T. Growth at 65°C was unstable and a reliable growth rate was not obtained. No growth occurred at 35 and 70°C, pH 5.0 and 7.6, and 0 and 4.0% NaCl concentration.
The sensitivity of antibiotics was tested at 60°C using modified MJY medium. Growth was inhibited by chloramphenicol, erythromycin, novobiocin, spectinomycin and vancomycin at 50 μg mL−1. Strain SW7T was insensitive to ampicillin, penicillin G, kanamycin, streptomycin, tetracycline and rifampicin at 50 μg mL−1.
SSU rRNA gene phylogenetic analysis
An almost complete SSU rRNA gene sequence (1,435 bp) was obtained after repeating isolation and was analyzed by FASTA algorithm in DDBJ or EMBL. Similar sequences from characterized species to strain SW7T belonged to the class Anaerolineae in the Chloroflexi subphylum I such as Thermanaerothrix dexensis (88.7%), Levilinea saccharolytica (88.2%), Ornatilinea apprima (88.2%), Bellilinea caldifistulae (87.3%), Anaerolinea thermophila (87.4%), Leptolinea tardivitalis (87.1%), Longilinea arvoryzae (87.3%) and Anaerolinea thermolimosa (86.9%). The SSU rRNA gene similarity value between the SW7T and Caldilinea aerophila representing the class Caldilineae was 82.5%. Similarity values between SW7T and environmental SSU rRNA gene sequences were also lower than 90% (data not shown).
Phylogenetic analysis based on SSU rRNA gene sequences from cultivated Chloroflexi species indicate that SW7T belongs to the class Anaerolineae (Fig. 5). In the phylogenetic tree including environmental SSU rRNA gene sequences, strain SW7T belonged to a cluster that was distant from branches including environmental Anaerolineae sequences obtained in this study (Fig. S2).
Fig. 5.
Phylogenetic tree of the phylum Chloroflexi based on SSU rRNA gene sequence by PhyML using 1,173 homologous sequence positions for each organism. Numbers indicate bootstrap values from 100 trials. Numbers in parentheses are GenBank/EMBL/DDBJ accession number. Bar indicates 5 substitutions per 100 nucleotides. Desulfovibrio vulgaris DSM644 (M34399) was used as an out-group.
Fatty acid composition and DNA base composition
The fatty acid composition of strain SW7T was C12:0 (1.4%), C16:0 (47.0%), C16:1 (5.2%), C18:0 (37.3%) and C18:1 (9.1%). The abundance of C18:0 likely reflected the relatively higher growth temperature also observed in thermophilic T. daxensis but not detected in the mesophilic Anaerolineae species (21, 32). Unsaturated fatty acids only found in strain SW7T were not observed in other Aanerolineae species (Table 1).
Table 1.
Characteristics of cultivated species belong to the class Anaerolineae in the phylum Chloroflexi
| Character | Thermomarinilinea lacunofontalis SW7T | Anaerolinea thermophila UNI-1T | Anaerolinea thermolimosa IMO-1T | Levilinea saccharolytica KIBI-1T | Leptolinea tardivitalis YMTK-2T | Bellilinea caldifistulae GOMI-1T | Longilinea arvoryzae KOME-1T | Thermanaerothrix daxensis GNS-1T | Ornatilinea apprima P3M-1T |
|---|---|---|---|---|---|---|---|---|---|
| Cell diameter# | 0.2 | 0.2–0.3 | 0.3–0.4 | 0.4–0.5 | 0.15–0.2 | 0.2–0.4 | 0.4–0.6 | 0.2–0.3 | 0.3–0.7 |
| Temperature range (°C) | 37–65 | 50–60 | 42–55 | 25–50 | 25–50 | 45–65 | 30–40 | 50–73 | –400 |
| Optimum temperature (°C) | 50–60 | 55 | 50 | 37–40 | 37 | 55 | 37 | 65 | 42–45 |
| NaCl range (%) | 0.5–3.5 | <1.0 | <1.5 | <3.0 | <1.5 | ≤3.0 | <1.5 | 0–10 | 0–2.0 |
| Optimum NaCl (%) | 1.0 | 0–<0.5 | 0–<0.25 | 0–<0.25 | 0–<0.25 | 0– | 0– | 2.0 | 0.1 |
| pH range | 5.5–7.3 | 6.0–8.0 | 6.5–7.5 | 6.0–7.2 | 6.0–7.2 | 6.0–8.5 | 5.0–7.5 | 5.8–8.5 | 6.5–9.0 |
| Optimum pH | 6.0 | around 7.0 | around 7.0 | around 7.0 | around 7.0 | around 7.0 | around 7.0 | 7.0 | 7.5–8.0 |
| Doubling time (h) | 4.6 | 72 (48)* | 48 (10)* | 56 (56)* | 50 (50)* | 45 (29)* | 92 (38)* | 100 | 6 |
| Major cellular fatty acids | C16:0, C18:0, C18:1 | C16:0, C15:0, C14:0 | ai-C17:0, i-C15:0, C16:0 | C14:0, i-C15:0, C16:0 | branched C17:0, C16:0, C14:0 | C16:0, C14:0, i-C15:0 | i-C15:0, ai-C15:0, C14:0 | C16:0, C18:0, i-C17:0, C20:0, i-C15:0, ai-C17:0, C14:0 | branched i-C15:0, ai-C15:0 |
| DNA G+C content (mol %) | 59.9 | 54.5 | 53.3 | 59.5 | 48.2 | 54.7 | 54.5 | 57.6 | 55 |
| Utilization of: | |||||||||
| yeast extract | + | + | + | + | + | + | + | + | + |
| tryptone | + | + | − | + | + | − | − | + | N.D. |
| peptone | + | N.D. | + | N.D. | + | N.D. | + | − | − |
| starch | − | + | + | − | + | − | − | − | − |
| glucose | − | + | + | + | + | + | − | + | + |
| mannose | − | + | + | + | + | + | − | + | N.D. |
| galactose | − | + | + | + | + | + | − | + | − |
| fructose | − | + | + | + | + | + | + | + | − |
| arabinose | − | + | + | − | + | + | − | + | − |
| xylose | − | + | + | + | + | + | − | + | + |
| ribose | − | + | + | + | + | + | − | + | N.D. |
| pyruvate | − | + | + | + | + | + | − | + | − |
| Origin | Shallow sea hydrothermal vent | Thermophilic UASB sludge | Thermophilic UASB sludge | Mesophilic UASB sludge | Mesophilic UASB sludge | Thermophilic anaerobic sludge | Rice paddy soil | Deep terrestrial hot aquifer (60°C) | Deep terrestrial hot aquifer (47°C) |
| References | This study | 22, 31 | 31, 32 | 31, 32 | 31, 32 | 30, 31 | 30, 31 | 6 | 20 |
All strains show multicellular filamentous morphology
Doubling time shown in parentheses obtained under syntrophic growth with hydrogenotrophic methanogens. Substrate utilization tests were examined in the presence of yeast extract.
+, positive; −, negative; N.D., not determined
The DNA G+C content of strain SW7T was 59.9 mol%, which is similar to L. saccharolytica, but 2–6 mol% higher than other Anaerolineae species. No significant relationship between the DNA G+C content and growth temperature was observed among the Anaerolineae species (Table 1).
Discussion
Microbial ecosystem of the hydrothermal environment
Both culture-dependent and -independent analyses showed that the microbial community associated with the hydrothermal vent was predominated by heterotrophic organisms such as Anaerolineae, Bacteroidetes and Thermaceae in Bacteria, and Thermococcaceae and the DHVEG in Archaea. The next dominant population was likely sulfur-oxidizing Gammaproteobacteria, represented by the Sulfurivirga sp. The hydrogenotrophic population including methanogens and hydrogen-oxidizing bacteria such as Persephonella sp. was apparently smaller than heterotrophic and sulfur-oxidizing populations (Figs. 1, 2). The lower abundance of hydrogenotrophic organisms in this environment is consistent with the relatively low hydrogen concentration in the venting fluids in this hydrothermal field (9).
We also noted the relatively high abundance of the Anaerolineae species in the microbial ecosystem that hydrogenotrophic organisms shared scare population. Previously, the relatively high abundance of Anaerolineae phylotypes/strains in man-made environments has been observed under methanogenic conditions, and some of the Anaerolineae strains from such environments have been obtained from syntrophic enrichment with methanogens (32). The observation in this microbial ecosystem suggests that such metabolic association with hydrogenotrophic organisms was not necessary for the high abundance of Anaerolineae species in this hydrothermal environment.
Characterization of Chloroflexi strain SW7T
Similarity and phylogenetic analyses of the SSU rRNA gene sequence with previously isolated strains indicated that strain SW7T belongs to the Chloroflexi class Anaerolineae. Strain SW7T shows typical features commonly observed among the Anaerolineae species, such as anaerobic and fermentative metabolism and cellular morphology. Relatively low SSU rRNA gene sequence similarities (below 90%) between SW7T and the Anaerolineae species and its distinct genomic G+C content among the Anaerolineae species (Table 1) indicate that strain SW7T is genetically different from previously characterized species in the class Anaerolineae. Furthermore, the phylogenetic position of strain SW7T in the SSU rRNA gene phylogenetic tree suggests that the strain represents a novel lineage in the class Anaerolineae (Fig. 5).
Significant physiological dissimilarities between strain SW7T and Anaerolineae species were also noted. Optimum NaCl concentration (1.0%) for growth and inability of growth in the absence of NaCl in strain SW7T showed that it is a typical marine microorganism, while NaCl inhibits the growth of other terrestrial Anaerolineae strains except for halophilic T. daxensis (Table 1). The relatively fast generation time (4.6 h) is also distinct from the long generation time (up to 100 hours) of slowly growing members within the Anaerolineae except for O. apprima (6 h). The inability to ferment using sugars as the sole carbon and energy source in strain SW7T has not been reported for other Anaerolineae species. The fatty acid composition of the new strain, which is characterized by the relatively high abundance of unsaturated fatty acids (C16:1 and C18:1) and the existence of C18:0, is different from those of the previously described thermophilic strains in the Anaerolineae (Table 1). Considering these phylogenetic and physiological differences between strain SW7T and other characterized strains in the Anaerolineae, we conclude here that strain SW7T represents a novel species in a novel genus, for which the name Thermomarinilinea lacunofontalis is proposed; the type strain is SW7T (=JCM 15506T=KCTC5908T).
Description of Thermomarinilinea gen. nov
Thermomarinilinea (Ther.mo.ma.ri.ni.li’ne.a. Gr. fem. n. therme heat; L. adj. marinus of the sea; L. fem. n. linea line; N.L. fem. n. the “thermophilic marine line”).
Gram negative. Non-motile filamentous cells. Multicellular. Spores are not observed. Grow under obligately anaerobic conditions with fermentation. Thermophilic and neutrophilic. Growth occurs up to 65°C. NaCl is required for growth. Major fatty acids are C16:0 and C18:0. SSU rRNA gene phylogenetic analysis indicates that the genus belongs to the class Anaerolineae in the phylum Chloroflexi. Isolated from shallow sea hydrothermal environment. The type species is Thermomarinilinea lacunofontalis. The G + C content of genomic DNA of the type species was 59.9 mol% (HPLC).
Description of Thermomarinilinea lacunofontalis sp. nov
(la.cu’no.fon’talis.. L. adj. lacutosus lagoonal; L. adj. fontalis of a spring; N.L. adj. lacunofontalis the “lagoonal spring”, as the strain was isolated from a hot spring in a coral lagoon).
Cells observed are 3.5–16.5 μm in length and 0.2 μm in width. Temperature for growth ranges between 37–65°C with optimum at 50–60°C, but unstable growth at 65°C. Growth occurs at 0.5–3.5% of NaCl concentration, and optimum is 1.0%. The growth pH range is pH 5.5–7.3, and optimum growth is observed at pH 6.0. Obligately anaerobic. Growth occurs with yeast extract, tryptone, peptone, casein, gelatin, chitin, glutamate, alanine, mannitol and citrate. The fatty acid composition was C12:0 (1.4%), C16:0 (47.0%), C16:1 (5.2%), C18:0 (37.3%) and C18:1 (9.1%). The G + C content of genomic DNA was 59.9 mol% (HPLC). The type strain SW7T (= JCM15506T = KCTC5908T) was isolated from Taketomi submarine hot spring shallow hydrothermal field in the southern part of Yaeyama Archipelago, Japan.
Supplementary Material
Acknowledgements
We would like to thank Dr Katsuyuki Uematsu for assistance in taking electron micrographs. Part of this study was supported by a Grant-in-Aid from the Ministry of Education, Culture, Sports, Science & Technology of Japan (No. 18658135).
References
- 1.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeLong EF. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fujiwara Y, Kawato M, Noda C, Kinoshita G, Yamanaka T, Fujita Y, Uematsu K, Miyazaki J. Extracellular and mixotrophic symbiosis in the whale-fall mussel Adipicola pacifica: a trend in evolution from extra- to intracellular symbiosis. PLoS One. 2010;5:e11808. doi: 10.1371/journal.pone.0011808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Furushima Y, Nagao M, Suzuki A, Yamamoto H, Maruyama T. Periodic behavior of the bubble jet (geyser) in the Taketomi submarine hot springs of the southern part of Yaeyama Archipelago, Japan. Marine Technol Soc J. 2009;43:13–22. [Google Scholar]
- 5.Gouy M, Guindon S, Gascuel O. SeaView version 4: a multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 6.Grégoire P, Fardeau ML, Joseph M, Guasco S, Hamaide F, Biasutti S, Michotey V, Bonin P, Ollivier B. Isolation and characterization of Thermanaerothrix daxensis gen. nov., sp. nov., a thermophilic anaerobic bacterium pertaining to the phylum “Chloroflexi”, isolated from a deep hot aquifer in the Aquitaine Basin. Syst Appl Microbiol. 2011;34:494–497. doi: 10.1016/j.syapm.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Hales BA, Edwards C, Ritchie DA, Hall G, Pickup RW, Saunders JR. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hirayama H, Fuse H, Abe M, Miyazaki M, Nakamura T, Furusima Y, Yamamoto H, Nunoura T, Takai K. Methylomarinum vadi gen. nov., sp. nov., a marine methanotroph isolated from two distinct marine environments in Japan. Int J Syst Evol Microbiol. 2013;63:1073–1082. doi: 10.1099/ijs.0.040568-0. [DOI] [PubMed] [Google Scholar]
- 9.Hirayama H, Sunamura M, Takai K, et al. Culture-dependent and -independent characterization of microbial communities associated with a shallow submarine hydrothermal system occurring within a coral reef off Taketomi Island, Japan. Appl Environ Microbiol. 2007;73:7642–7656. doi: 10.1128/AEM.01258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imachi H, Aoi K, Tasumi E, et al. Cultivation of methanogenic community from subseafloor sediments using a continuous-flow bioreactor. ISME J. 2011;5:1913–1925. doi: 10.1038/ismej.2011.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lane DJ. 16S–23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Techniques in Bacterial Systematics. Wiley; Chichester: 1985. pp. 115–175. [Google Scholar]
- 12.Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 13.Lauerer G, Kristjansson JK, Langworthy TA, König H, Stetter KO. Methanothermus sociabilis sp. nov., a second species within the Methanothermaceae growing at 97°C. Syst Appl Microbiol. 1986;8:100–105. [Google Scholar]
- 14.Ludwig W, Strunk O, Westram R, et al. ARB: a software environment for sequence data. Nucl Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamura T, Yamazaki SS, Sakai K, Yamasaki H, Furushima Y, Yamamoto H. Acroporid corals growing over a methane-bubbling hydrothermal vent. Southern Ryukyu Archipelago Coral Reefs. 2006;25:382. [Google Scholar]
- 16.Nunoura T, Hirai M, Imachi H, Miyazaki M, Makita H, Hirayama H, Furushima Y, Yamamoto H, Takai K. Kosmotoga arenicorallina sp. nov. a thermophilic and obligately anaerobic heterotroph isolated from a shallow hydrothermal system occurring within a coral reef, southern part of the Yaeyama Archipelago, Japan, reclassification of Thermococcoides shengliensis as Kosmotoga shengliensis comb. nov., and emended description of the genus Kosmotoga. Arch Microbiol. 2010;192:811–819. doi: 10.1007/s00203-010-0611-7. [DOI] [PubMed] [Google Scholar]
- 17.Nunoura T, Oida H, Miyazaki J, Miyashita A, Imachi H, Takai K. Quantification of mcrA by fluorescent PCR in methanogenic and anaerobic methanotrophic microbial communities. FEMS Microbiol Ecol. 2008;64:240–247. doi: 10.1111/j.1574-6941.2008.00451.x. [DOI] [PubMed] [Google Scholar]
- 18.Nunoura T, Oida H, Miyazaki M, Suzuki Y. Thermosulfidibacter takaii gen. nov. sp. nov. a thermophilic hydrogen oxidizing, sulfur reducing bacterium isolated from a deep-sea hydrothermal field, Southern Okinawa Trough within the phylum Aquificae. Int J Syst Evol Microbiol. 2008;58:659–665. doi: 10.1099/ijs.0.65349-0. [DOI] [PubMed] [Google Scholar]
- 19.Nunoura T, Oida H, Miyazaki M, Suzuki Y, Takai K, Horikoshi K. Marinitoga okinawensis sp. nov. a novel thermophilic and anaerobic heterotroph isolated from a deep-sea hydrothermal field, Southern Okinawa Trough. Int J Syst Evol Microbiol. 2007;57:467–471. doi: 10.1099/ijs.0.64640-0. [DOI] [PubMed] [Google Scholar]
- 20.Nunoura T, Oida H, Miyazaki M, Suzuki Y, Takai K, Horikoshi K. Desulfothermus okinawensis sp. nov. a thermophilic and heterotrophic sulfate-reducing bacterium isolated from a deep-sea hydrothermal field. Int J Syst Evol Microbiol. 2007;57:2360–2364. doi: 10.1099/ijs.0.64781-0. [DOI] [PubMed] [Google Scholar]
- 21.Podosokorskaya OA, Bonch-Osmolovskaya EA, Novikov AA, Kolganova TV, Kublanov IV. Ornatilinea apprima gen. nov., sp. nov., a novel cellulolytic representative of class Anaerolineae. Int J Syst Evol Microbiol. 2013;63:86–92. doi: 10.1099/ijs.0.041012-0. [DOI] [PubMed] [Google Scholar]
- 22.Reysenbach AL, Liu Y, Banta AB, Beveridge TJ, Kirshtein JD, Schouten S, Tivey MK, Von Damm KL, Voytek MA. A ubiquitous thermoacidophilic archaeon from deep-sea hydrothermal vents. Nature. 2006;442:444–447. doi: 10.1038/nature04921. [DOI] [PubMed] [Google Scholar]
- 23.Sekiguchi Y, Yamada T, Hanada S, Ohashi A, Harada H, Kamagata Y. Anaerolinea thermophila gen. nov., sp. nov. and Caldilinea aerophila gen. nov., sp. nov., novel filamentous thermophiles that represent a previously uncultured lineage of the domain Bacteria at the subphylum level. Int J Syst Evol Microbiol. 2003;53:1843–1851. doi: 10.1099/ijs.0.02699-0. [DOI] [PubMed] [Google Scholar]
- 24.Takai K, Horikoshi K. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl Environ Microbiol. 2000;66:5066–5072. doi: 10.1128/aem.66.11.5066-5072.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takai K, Inagaki F, Nakagawa S, Hirayama H, Nunoura T, Sako Y, Nealson KH, Horikoshi K. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol Lett. 2003;218:167–174. doi: 10.1111/j.1574-6968.2003.tb11514.x. [DOI] [PubMed] [Google Scholar]
- 26.Takai K, Inoue A, Horikoshi K. Methanothermococcus okinawensis sp. nov., a thermophilic, methane-producing archaeon isolated from a Western Pacific deep-sea hydrothermal vent system. Int J Syst Evol Microbiol. 2002;52:1089–1095. doi: 10.1099/00207713-52-4-1089. [DOI] [PubMed] [Google Scholar]
- 27.Takai K, Miyazaki M, Nunoura T, Hirayama H, Oida H, Furushima Y, Yamamoto H, Horikoshi K. Sulfurivirga caldicuralii gen. nov., sp. nov., a novel microaerobic, thermophilic, thiosulfate-oxidizing chemolithoautotroph, isolated from a shallow marine hydrothermal system occurring in a coral reef, Japan. Int J Syst Evol Microbiol. 2006;56:1921–1929. doi: 10.1099/ijs.0.64297-0. [DOI] [PubMed] [Google Scholar]
- 28.Takai K, Nakagawa S, Reysenbach A-L, Hoek J. Microbial ecology of mid-ocean ridges and back-arc basins. In: Christie DM, Fisher CR, Lee S-M, Givens S, editors. Back-arc Spreading Systems: Geological, Biological, Chemical and Physical Interactions. American Geophysical Union; Washington DC: 2006. pp. 185–213. (Geophysical Monograph Series 166). [Google Scholar]
- 29.Takai K, Sugai A, Itoh T, Horikoshi K. Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int J Syst Evol Microbiol. 2000;50:489–500. doi: 10.1099/00207713-50-2-489. [DOI] [PubMed] [Google Scholar]
- 30.Tamaoka J, Komagata K. Determination of DNA base composition by reverse-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 31.Yamada T, Imachi H, Ohashi A, Harada H, Hanada S, Kamagata Y, Sekiguchi Y. Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol. 2007;57:2299–2306. doi: 10.1099/ijs.0.65098-0. [DOI] [PubMed] [Google Scholar]
- 32.Yamada T, Sekiguchi Y. Cultivation of uncultured Chloroflexi subphyla: significance and ecophysiology of formerly uncultured Chloroflexi‘subphylum I’ with natural and bio-technological relevance. Microbes Environ. 2009;24:205–216. doi: 10.1264/jsme2.me09151s. [DOI] [PubMed] [Google Scholar]
- 33.Yamada T, Sekiguchi Y, Hanada S, Imachi H, Ohashi A, Harada H, Kamagata Y. Anaerolinea thermolimosa sp. nov., Levilinea saccharolytica gen. nov., sp. nov. and Leptolinea tardivitalis gen. nov., sp. nov., novel filamentous anaerobes, and description of the new classes Anaerolineae classis nov. and Caldilineae classis nov. in the bacterial phylum Chloroflexi. Int J Syst Evol Microbiol. 2006;56:1331–1340. doi: 10.1099/ijs.0.64169-0. [DOI] [PubMed] [Google Scholar]
- 34.Zillig W, Holz I, Janekovic D, et al. Hyperthermus butylicus, a hyperthermophilic sulfur-reducing archaebacterium that ferments peptides. J Bacteriol. 1990;172:3959–3965. doi: 10.1128/jb.172.7.3959-3965.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.