Abstract
Fluorescence in situ hybridization (FISH) has become a standard technique in environmental microbiology. More than 20 years have passed since this technique was first described, and it is currently used for the detection of ribosomal RNA, messenger RNA, and functional genes encoded on chromosomes. This review focuses on the advancement and applications of FISH combined with catalyzed reporter deposition (CARD, also known as tyramide signal amplification or TSA), in the detection of environmental microorganisms. Significant methodological improvements have been made in CARD-FISH technology, including its combination with other techniques and instruments.
Keywords: catalyzed reporter deposition (CARD), fluorescence in situ hybridization (FISH), single cell detection, tyramide signal amplification (TSA)
Introduction
Since it was first developed (20), fluorescence in situ hybridization (FISH) has become one of the most routinely used molecular techniques in environmental microbiology. FISH can be used to detect, identify, and enumerate environmental microorganisms without requiring culture, and therefore it has been used to help elucidate the microbial ecology of many habitats, including soil, sediments, aquatic environments, and engineered sludge (reviewed in refs. 7, 8, 53). Nevertheless, there are several problems in the application of FISH, primarily insufficient sensitivity due to the low number of target molecules in cells, low probe permeability of cells, and poor probe hybridization efficiency (7). Many methods have been devised to overcome these problems (reviewed in refs. 9, 86, 88). This review will focus on the technical advancement and applications of a sensitive FISH technique, catalyzed reporter deposition (CARD)-FISH, also known as tyramide signal amplification (TSA)-FISH (Table 1). The applications of CARD-FISH will be discussed, not only in rRNA-targeted phylogenetic identification but also in linking microbial phylogeny to physiology and metabolic activity.
Table 1.
Important technical developments in the history of CARD-FISH for environmental microorganisms
| Year | Description | Reference |
|---|---|---|
| 1989 | • First CARD publication (for immunoassay) | 15 |
| • Development of FISH for microorganisms | 20 | |
| 1991 | • FISH applied to uncultured microorganisms | 4 |
| 1997 | • CARD-FISH for environmental microorganisms | 39, 68 |
| 1998 | • CARD-FISH with polyribonucleotide probes for mRNA detection | 85 |
| 2002 | • Low-melting point agarose-embedding protocol | 56 |
| • Detection of DNA synthesizing (BrdU incorporated) bacteria by CARD-FISH | 57 | |
| • CARD-FISH with oligonucleotide probes for mRNA detection | 10 | |
| • Development of clone-FISH | 71 | |
| 2003 | • CARD-FISH combined with FACS | 14 |
| 2004 | • Simultaneous detection of rRNA and mRNA by CARD-FISH with polyribonucleotide probes | 58 |
| • CARD-FISH combined with microautoradiography | 75 | |
| 2006 | • Two-pass TSA-FISH with oligonucleotide probes for mRNA detection | 37 |
| 2008 | • EL-FISH and HISH for nanoSIMS analysis | 13, 51 |
| • Magneto-FISH for cell capturing | 60 | |
| 2010 | • Two-pass TSA-FISH with oligonucleotide probes for gene detection | 30 |
| • Simultaneous detection of rRNA and gene by CARD-FISH with polydeoxyribonucleotide probes | 48 | |
| 2012 | • Two-pass TSA-FISH with polydeoxyribonucleotide probes for gene detection | 31 |
| • mRNA-FISH combined with FACS | 50 |
Catalyzed Reporter Deposition: CARD
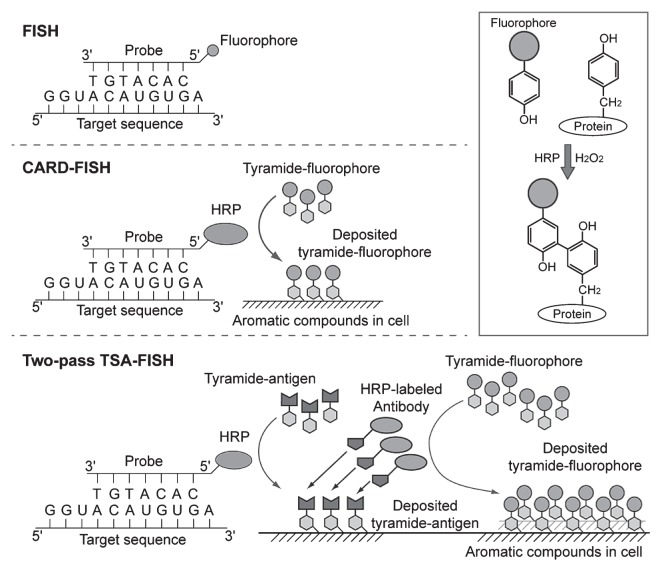
CARD was first reported more than twenty years ago as a novel method of signal amplification for immunoassays and membrane immunoassays (15, 16), and was later applied to FISH (33, 66). The principle of CARD is as follows: in the presence of hydrogen peroxide, horseradish peroxidase (HRP) converts tyramide into a radical intermediate. This radical tyramide nonspecifically reacts with aromatic compounds, such as tyrosine and tryptophan, in cells or blocking reagents (Fig. 1). This radical reaction occurs only near the HRP molecule and on a very short timescale. As a result, a great number of tyramides are deposited around the HRP molecule. Tyramides with conjugates (e.g., fluorescein, cyanine dyes, Alexa fluor dyes, biotin, etc.) for the CARD amplification system are commercially available from several companies. Alternatively, tyramide conjugates can be prepared by simple and rapid chemical reactions (27).
Fig. 1.
Schematic depiction of FISH, CARD-FISH, and two-pass TSA-FISH. The scheme in the box depicts tyramide immobilization on tyrosine.
Fundamental developments in CARD-FISH
The first application of CARD-FISH in environmental microbiology was reported by two different groups in 1997. Schönhuber et al. (68) reported a direct method using HRP-labeled probes (Fig. 1), while Lebaron et al. (39) described an indirect method using biotinylated probes and HRP-labeled streptavidin. Both studies showed significant signal amplification after the CARD reaction, with more than 10-fold stronger signals than mono-fluorescently labeled probes. The direct method is simpler than the indirect method as it omits the immunological reaction step and is therefore more popular in environmental microbiology.
Improving sensitivity and reducing background
Many strategies have been adopted to further improve CARD-FISH signals, mainly by amending the CARD working solution. The addition of 10–30% dextran sulfate has positive effects on signal localization (82) and signal intensity (37). This is attributed to the effect of volume exclusion, a result of the trapping of solvent water molecules by long polymer rods (83); however, dextran sulfate sometimes introduces spotty background signals dispersed over the entire slide (82). This problem is overcome by washing at elevated temperatures (45–60°C) (30, 82). The addition of an inorganic salt and/or an organic reagent enhances CARD-FISH signals (17). Inorganic salts include NaCl, MgCl2, KCl, CaCl2, sodium phosphate, sodium acetate, ammonium acetate, and ammonium sulfate. Most preferably, the concentration of the inorganic reagent ranges from at least 2 M to saturation. Preferred organic reagents are described in the paper (17): the preferred enhancer for non-fluorescent reagents is N-(5-hydroxypentyl)-3-(p-hydroxyphenyl) propionamide and that for fluorescent reagents is p-iodophenyl boronic acid. The concentration of the organic reagent ranges from approximately 1×10−6 to 1×10−4 M. This discovery was unexpected because organic reagents act as inhibitors rather than as enhancers (17), and the detailed mechanisms are not fully understood. For application to environmental micro-organisms, the addition of NaCl (2 M) and p-iodophenyl boronic acid (at 20 times the tyramide concentration) to the CARD working solution with fluorescently labeled tyramides has been reported (29, 59). The concentration of tyramide in the working solution is also an important factor. Detection rates for environmental bacteria were increased by elevating tyramide concentration in the working solution; however, too much tyramide causes high background fluorescence (56).
To minimize background fluorescence, the concentration of HRP-labeled probes is an important factor. Many FISH protocols use fluorescently labeled probes at concentrations of 0.5–1 μM or 2–5 ng μL−1 (3, 6, 36, 46, 61); however, these probe concentrations resulted in nonspecific fluorescent signals following CARD-FISH (56). To reduce the background, lower probe concentrations (0.1 μM or 0.5 ng μL−1) were used for CARD-FISH. Furthermore, a blocking reagent is often involved in the CARD working solution to minimize nonspecific fluorescent deposition (72).
Two-pass TSA-FISH is another approach for enhancing signal intensity (Fig. 1). Two-pass TSA-FISH involves two sequential TSA reactions. In the first reaction, dinitrophenyl (DNP)-labeled tyramide is deposited instead of a fluorophore-labeled tyramide. After incubation with an HRP-conjugated anti-DNP-antibody, the second TSA reaction is carried out with a fluorophore-labeled tyramide. This technique was first applied to eukaryotic cells (81) and later to prokaryotic cells (37). In two-pass TSA-FISH, the addition of both dextran sulfate and blocking reagent is especially important to maximize signal intensity and minimize nonspecific staining (37).
Permeabilization
Because HRP is approximately 5–6 nm with a molecular weight of approximately 40 kDa, probes labeled with this large molecule penetrate fixed cells very poorly compared with fluorescently labeled probes (molecular weights of fluorochromes are generally 500–1,000 Da); therefore, before applying CARD-FISH, permeabilization protocols must be optimized for the group of interest. This is often laborious because the range for “optimum” permeabilization is very narrow. Even though some prokaryotic species can be detected without any treatments, e.g., Planctomycetes in marine sediments (29) and methanogens with an s-layer (38), most prokaryotic cells need to be pretreated for probe penetration.
Optimization of the fixation procedure is the first step in optimizing the permeabilization process. Fixation with protein denaturing reagents (i.e., ethanol fixation) typically produces better permeability than that with cross-linking reagents (i.e., paraformaldehyde fixation) (68); however, some prokaryotes exhibit a loss of fluorescent signals following ethanol fixation (38). The concentration, fixation time, and temperature are also important. Pizzetti et al. (64) reported that a higher detection rate for Planctomycetes was obtained by FISH than by CARD-FISH when samples were fixed with 2% formaldehyde, but the opposite results were obtained when samples were fixed with 1% paraformaldehyde. Furthermore, storage conditions and term also affect the permeability. Long-term storage of samples resulted in higher detection rates because permeability inexplicably increased during storage (38, 93).
Prior to permeabilization, cells are immobilized on slides or filters using low-melting point agarose to prevent major cell loss during permeabilization and CARD-FISH (9, 56). In the first report on agarose embedding, no bacterial cell loss was observed even after stringent lysozyme treatment (10 mg mL−1 for 90 min at 37°C) when samples were embedded in agarose (56). Agarose embedding is currently included in the majority of CARD-FISH protocols.
Enzymatic treatments using lysozyme, achromopeptidase, proteinase K, and pseudomurein endopeptidase are often employed for permeabilization. Lysozyme is the most commonly used enzyme for treatment as it catalyzes the hydrolysis of 1,4-beta-linkages between N-acetylmuramic acid and N-acetyl-D-glucosamine residues in peptidoglycans. Many Gram-negative bacteria are permeabilized by lysozyme treatment (e.g., 42, 55, 56); however, because several Gram-positive cultures are resistant to lysozyme (18), and because lysozyme only partly digests the murein multilayers of fixed Gram-positive cells (72), some Gram-positive bacteria cannot be permeabilized by lysozyme treatment alone. Simultaneous treatment with achromopeptidase and lysozyme is effective for lysis of these Gram-positive bacteria (11, 22). Sekar et al. (72) introduced achromopeptidase treatment following lysozyme treatment for permeabilization of Gram-positive Actinobacteria. Interestingly, this achromopeptidase treatment is only effective when conducted after lysozyme treatment, but not before (72). Achromopeptidase hydrolyzes lysyl peptide bonds (79), and lysozyme treatment likely improves the accessibility of achromopeptidase to the peptide bonds. This treatment is used for permeabilization of many microbial cells (e.g., Planctomycetes [64, 90] and Archaea [29]). Proteinase K is also used in many protocols. Although some studies have found that proteinase K treatment is difficult to control and causes unstable results (55, 69), this treatment is effective for many Archaea (38, 42, 47, 65, 75). Pseudomurein endopeptidase is effective for permeabilization of methanogens with pseudomurein. The glycan strands of pseudomurein consist of alternating β(1→3)-linked N-acetyl-D-glucosamine and N-acetyl-L-talosaminuronic acid residues, instead of the N-acetyl-D-muranosamine of murein and thus, the pseudomurein cell wall structure is resistant to the three enzymes described above, as well as to many other enzymes (84). Pseudomurein endopeptidase was originally isolated from (pro)phages in Methanothermobacter wolfei (PeiW) (44) and Methanothermobacter marburgensis (PeiP) (63), and the enzyme-encoding genes have since been cloned (45). Treatment with recombinant PeiW effectively permeabilized fixed methanogens with pseudomurein for FISH (52) and CARD-FISH (31, 38) applications.
Chemical treatments are also used for permeabilization. Sodium dodecyl sulfate (SDS) is a detergent usually used in hybridization buffer to improve probe accessibility to rRNA (12). In addition to this purpose, increased concentration of SDS (e.g., 1%) in hybridization buffer successfully permeabilized Methanotorris igneus (formaly Methanococcus igneus) for penetration of HRP-labeled probes (5). Furthermore, HCl treatment is effective for marine Archaea (47, 90). SDS or SDS/dithiothreitol treatment followed by lysozyme treatment has also been reported (19, 54).
Microwave treatment has been applied for the detection of pmoA (the alpha subunit of the particulate methane monooxygenase) mRNA in tissue sections of Bathymodiolus puteoserpentis (59), and for the detection of ANME-2c-group organisms in marine sediment (60). This type of physical treatment is preferable to enzymatic treatments, especially when the cell structures and effective permeabilizers are unknown. Very recently, microwave treatment in Tris-EDTA buffer (pH 9.0) was reported for the detection of Bacteria and Archaea (77). It is suggested that Tris binds to lipopolysaccharides and replaces stabilizing Ca2+ and Mg2+ (80). EDTA removes these divalent cations by chelation and therefore the interaction between lipopolysaccharide molecules is reduced; however, Tris-EDTA permeabilization cannot stand alone. The authors noted that hybridization should also be conducted in a microwave oven, rather than a normal oven, for successful application (77).
Endogenous peroxidase
The CARD reaction is initiated by the oxidation of hydrogen peroxide to the hydroxyl radical by peroxidase activity; therefore, endogenous peroxidase should be inactivated prior to hybridization. False-positive signals caused by endogenous peroxidase cannot be explained by the mere presence of peroxidase (55). Unfortunately, there is no comprehensive method to inactivate endogenous peroxidase activity in various environments and thus it should be optimized for each sample. Treatment with H2O2 in water or methanol is often used. Ishii et al. (29) reported that 0.15% H2O2 in methanol successfully inactivated endogenous peroxidase activities without decreasing the number of 4′,6-diamidino-2-phenylindole (DAPI)-stained cells, whereas higher concentrations (up to 3%) of H2O2 resulted in a significant decrease in the number of DAPI-stained cells (29). Our research group also found that CARD-FISH signals were significantly weaker following pre-treatment with higher concentrations of H2O2 (unpublished results). Nucleic acids are likely hydrolyzed by the strong oxidant H2O2. In contrast, the strong endogenous peroxidase activity in anaerobic ammonium oxidation (anammox bacterial cells needed to be treated with 3% H2O2 for sea samples (90) or 30% H2O2 for bioreactor samples (55). In addition, diethylpyrocarbonate (56) and HCl (72) treatments are also useful for inactivation.
Thermal stability of HRP
It was traditionally thought that HRP should be handled below 42°C. Thus, hybridization of HRP-labeled probes is carried out at lower temperatures (e.g., at 35°C or 40°C), rather than the standard temperature of 46°C, to prevent inactivation of HRP (37, 61). However, a more recent study investigated the thermal stability of the HRP molecule attached to oligonucleotides and found that HRP is not inactivated, even after hybridization at 55°C for 2 h and washing at 57°C for 30 min (29); therefore, conventional hybridization and washing temperatures (46°C and 48°C) can also be used. Nevertheless, hybridization stringency, i.e., the formamide concentration, should be re-optimized for HRP-labeled probes because the dissociation profile of HRP-labeled probes differs significantly from that of fluorescently labeled probes (28).
Phylogenetic staining
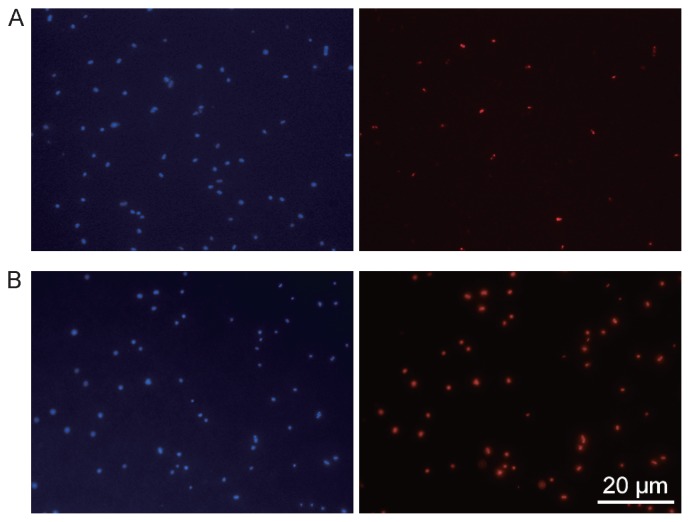
CARD-FISH has been used for phylogenetic staining of microorganisms in many environments, including marine, freshwater, soil, sediment, and engineered sludge (e.g., 19, 23, 25, 26, 29, 34, 38, 42, 47, 54–56, 64, 65, 67, 72, 75, 90, 91), and a basic protocol for CARD-FISH target to rRNA is described in Table 2. Microorganisms living in oligotrophic environments often have a low rRNA content; hence, CARD-FISH is especially superior to FISH under these circumstances because of its high sensitivity. For example, significantly higher detection rates of marine bacteria were achieved by CARD-FISH (85–100%) compared to FISH using Cy3-labeled probes (19–66%) (56) (also see differences in Figure 2). Similar results were seen for bacteria in sediment (29, 56) and freshwater (72). Conversely, CARD-FISH detection of bacteria in activated sludge (68), anammox cells in marine environments (90), methanogenic archaea in anaerobic granular sludge (38), and Planctomycetes in marine water (64) showed lower detection rates than those obtained by FISH. As described above, permeabilization is always an issue for application of CARD-FISH, and detection rates using CARD-FISH may be lower than those using FISH when the target organisms contain sufficient rRNA for conventional FISH detection.
Table 2.
Basic protocol for CARD-FISH for rRNA
Cell fixation
on glass slide
|
Fig. 2.
Detection of marine bacteria by FISH with Cy3-labeled probes (A) and by CARD-FISH with the deposition of tyramide-Cy3 (B). Lysozyme treatment was carried out prior to hybridization. Exposure times were 1 s for FISH and 0.1 s for CARD-FISH. Stronger signals and higher detection rates were obtained after CARD-FISH. Left (DAPI-stained cells) and right (FISH signals) epifluorescent micrographs show identical fields.
Strong CARD-FISH signals also enable reliable detection of microorganisms with autofluorescence. Cyanobacteria have yellowish-red autofluorescence and are difficult to detect using FISH with fluorescently-labeled probes (69). CARD-FISH with fluorescein-labeled tyramide successfully overcame the background (autofluorescence) noise of cyanobacteria (1, 69, 89).
Phylogenetic staining that targeted tmRNA (transfer-messenger RNA) was also developed to overcome the limitations of phylogenetic classification using the rRNA gene (i.e., poor resolution of closely related strains, subspecies) (70). The tmRNA molecule was first discovered in Escherichia coli (40). It is a stable RNA that is present at fewer than 1,000 copies per cell. Because of its low copy number, tmRNA is difficult to detect using mono-fluorescently labeled probes, but it can be detected successfully with CARD-FISH and can be used as a marker for phylogenetic analysis (70).
Functional identification
The number of target molecules required to detect microorganisms by CARD-FISH with HRP-labeled oligonucleotide probes has been quantified, and the detection limit was found to be dozens of molecules (28); therefore, highly-expressed mRNA can be detected by CARD-FISH, but genes or mRNA with low expression cannot. Further signal amplification should be performed for the detection of mRNA and genes by CARD-FISH. Two-pass TSA-FISH, as described above, is one way to increase signal intensity.
Alternatively, the use of multiply-labeled polynucleotide probes instead of oligonucleotide probes is an option, but their characteristics are different (Table 3). Polynucleotide probes are typically generated by PCR (double-stranded DNA [dsDNA] polynucleotides) or reverse transcription (RNA polynucleotides). dsDNA and RNA polynucleotide probes are often used to detect genes and mRNA, respectively. Dozens of haptens or fluorophores are incorporated into the polynucleotides during synthesis, resulting in higher sensitivity than single-molecule-labeled oligonucleotides. Nevertheless, the specificity of polynucleotide probes is lower than that of oligonucleotide probes. Oligonucleotide probes can distinguish single base mismatches, although it depends on the sequence and the position of the mismatch (3). Even if single base mismatches are not distinguishable, the use of competitor probes makes this possible (29, 37, 42, 46). Polynucleotide probes can also distinguish closely related species under very stringent hybridization conditions; however, the signal intensity is significantly decreased under these conditions, diminishing the merits of the polynucleotide probe (78). This difference in specificity is a result of the difference in hybridization behavior between oligonucleotides and polynucleotides. For example, the melting temperature of an oligonucleotide is sequence-dependent, whereas that of a polynucleotide is base content-dependent (76). The threshold for discrimination using polynucleotide probes has been reported to be in the range of 70–89% of sequence identity (24, 31, 43); therefore, polynucleotide probes are frequently used for phylogenetic discrimination at higher taxonomic levels (21). The sequence identities of functional genes within a genus are often low. In the case of the mcrA (the alpha subunit of the methyl-coenzyme M reductase) gene, the average nucleotide sequence identities within a genus and family are only 88.9% and 79%, respectively (74); therefore, polynucleotide probes are useful for functional staining. Recently, Moraru et al. (49) proposed a concept and software for polynucleotide probe design. Another drawback of polynucleotides is lower permeability than oligonucleotides because of differences in size. In addition, design flexibility is lower, and polynucleotide probes are not commercially available.
Table 3.
Comparison of oligonucleotides and polynucleotides
| Oligonucleotides | Polynucleotides | |
|---|---|---|
| Specificity | Higher. Single mismatch can be distinguished with or without competitor probes (depends on sequences and hybridization conditions). | Lower. The threshold for discrimination using polynucleotide probes has been reported to have 70–89% sequence identity. |
| Sensitivity | Lower. The number of molecules to be labeled is low (usually one or two). | Higher. Probes can be labeled with many molecules. |
| Design flexibility | Higher. Probes can be designed for a conserved region of a gene and also for a species-specific region. | Lower. Probe sequences depend on the template DNA. |
| Commercial availability | Yes. Fluorescently-, hapten-, and HRP-labeled probes can be purchased. | No. Probes need to be generated via in vitro transcription or PCR. |
| Permeability (nucleotides only) | Higher. | Lower. |
| Definition of Tm (Dissociation behavior) | Sequence-dependent. The point at which half the oligonucleotides are dissociated. | Base content-dependent. The point at which half the base pairs are dissociated. |
mRNA-FISH
The detection of mRNA is particularly challenging because these molecules are unstable and only present at very low copy numbers. Preparation of control samples is the first highest hurdle. Clone-FISH, which was originally developed for rRNA-FISH (71), is often used for the optimization of hybridization conditions for both oligonucleotide probes (37) and polyribonucleotide probes (58) for mRNA-FISH. The first report of the detection of mRNA by CARD-FISH was in 1998 (85). In that study, digoxigenin (DIG)-labeled RNA polynucleotide probes and HRP-conjugated anti-DIG-antibodies were used to detect iap (gene encoding invasion-associated protein) mRNA in Listeria monocytogenes cells. Four years later, nahAc (naphthalene dioxygenase) mRNAs in bacteria in contaminated groundwater were detected using biotinylated oligonucleotide probes and HRP-labeled streptavidin (10). These authors introduced DNase treatment prior to hybridization to improve the accessibility to mRNA, and extended exposure times (up to 10 s) to acquire digital images of fluorescent signals. The weak signals were overcome by the development of two-pass TSA-FISH (37). Archaeal mcrA mRNA was detected with intensified signals using HRP-labeled oligonucleotide probes, and the discrimination of single-base mismatches was also achieved using competitor probes. Recently, mRNA-FISH combined with microsensor measurements has been reported (35). This method has the potential to connect microenvironments and microbial activities as defined by mRNA expression.
Linking physiology and phylogeny using simultaneous mRNA and rRNA staining is also an intriguing challenge. In 2004, the simultaneous detection of mRNA and rRNA was reported (58). In the protocol of Pernthaler et al. (58), mRNA-FISH targeting pmoA was performed using DIG-labeled RNA polynucleotide probes. After inactivating the HRP used for the first CARD reaction, rRNA-FISH was subsequently carried out using HRP-labeled oligonucleotide probes. It is better to perform mRNA-FISH first for the simultaneous detection of mRNA and rRNA because mRNA is typically present at a low copy number and is very unstable. Using this method, archaeal ammonia oxidizers in soil were quantified by simultaneous detection of archaeal amoA (the alpha subunit of the ammonia monooxygenase) mRNA and rRNA, and their high relative abundance among the overall archaeal community were demonstrated (65).
More recently, sequential mRNA FISH followed by fluorescence-assisted cell sorting (SmRFF) has been developed, and active nitrite reducers in an activated sludge sample were successfully identified (50). This technique is useful to identify phylogenetically unknown mRNA-expressers in environments.
Gene-FISH
The detection of genes encoded on chromosomes has been developed recently (30, 31, 48). DNA is much more stable than mRNA, but the copy number of a given gene is usually very low. The detection of genes can provide insight into microbial functional potential, whereas the detection of mRNA demonstrates the transcriptional activities of functional genes in microbial cells. Gene detection using the TSA reaction was first reported by Kawakami et al. (30), who used two-pass TSA-FISH with oligonucleotide probes targeting the mcrA gene. Locked nucleic acid-incorporated DNA probes were used to improve hybridization efficiency and specificity (36, 92). These authors demonstrated that two-pass TSA-FISH can detect a single-copy gene on a chromosome with specificity sufficient to discriminate a single-base mismatch. The detection efficiency was approximately 15% when the single-copy gene was targeted; however, efficiency increased to more than 50% when a multi-copy gene, the rRNA gene, was targeted. Recently, a high detection efficiency method (more than 98%) has been developed, two-pass TSA-FISH with dsDNA polynucleotide probes, and applied to detect the apsA (the alpha subunit of adenosine-5′-phosphosulfate kinase) and mcrA genes in sludge samples (31).
The simultaneous detection of genes and rRNA was reported by Moraru et al. (48). The authors used CARD-FISH with dsDNA polynucleotide probes to detect the crenarchaeotal amoA gene in a seawater sample. The detection efficiency of this method was approximately 40%. To maximize gene detection efficiency, four dsDNA poly-nucleotide probes, targeting different sites on the target gene, were prepared and applied simultaneously (62).
In combination with other techniques, Kenzaka et al. (32) deposited biotinylated tyramide and subsequently applied nanogold-conjugated streptavidin. For further signal amplification, they employed gold enhancement and successfully detected the green fluorescent gene and ampicillin resistance gene on plasmids using a scanning electron microscope.
Cell isolation
Fluorescence-activated cell sorting (FACS) is a popular method for cell isolation. CARD-FISH was first used for the isolation of marine picoeukaryotes (14). For these experiments, all procedures were performed using cells in suspension. Cells were concentrated by centrifugation at various steps of the experiment, e.g., during washing. Using this method, cell loss can be avoided; however, the wide range of cell sizes makes it impossible to recover cells quantitatively by centrifugation. The g forces required to sediment small cells might damage larger cells (73), and cell concentration efficiency by centrifugation changes based on the properties of the buffer used (e.g., salt concentration and viscosity), temperature, and other variables. As an alternative, a protocol for cell recovery following CARD-FISH using a membrane filter was developed (73). In this method, the agarose-embedding procedure is omitted because it interferes with the detachment of cells from filters, and agarose particles interfere with flow cytometric analysis. For cell detachment, filters were incubated in various solutions, followed by vortexing or sonication. The authors found that incubation in NaCl-Tween solution followed by vortexing was the best strategy for detaching cells from their seawater samples (cell recovery efficiency was approximately 70%). The ideal conditions may differ based on the sample of interest. Recently, cell sorting following mRNA-FISH was reported, as described above (50). In that report, mRNA-FISH was carried out for cells embedded in agarose on a glass slide, and the cells were recovered from the slide into suspension in pre-warmed PBS buffer.
An interesting method using CARD-FISH and paramagnetic beads for cell isolation (magneto-FISH) was developed recently to investigate the syntrophic association of cell aggregates (60). In magneto-FISH, fluorescein-labeled tyramide is deposited after the in situ hybridization of HRP-labeled probes. Subsequently, monoclonal mouse anti-fluorescein antibody-labeled pan-mouse paramagnetic beads are used to label the target cells with magnetic beads. The target cells, and cells associated with the target cells, can be separated from other cells using a magnet. Isolated cells can then be used for subsequent analyses (e.g., sequencing and isotopic studies). Because tyramide is deposited on intercellular proteins as well as on cell surface proteins, the paramagnetic beads (5 μm in the aforementioned study) do not need to penetrate the small microbial cells. Magneto-FISH does not require any special instruments, is inexpensive, and can be used as a substitute for FACS.
Linking metabolic activity and phylogeny
CARD-FISH can also be combined with techniques such as microautoradiography (MAR) and with instruments such as nanoSIMS (secondary ion mass spectrometry), which are used to evaluate microbial metabolic activities using isotopic or radioisotopic substrates. Unlike FISH-MAR, CARD-FISH-MAR can be applied to oligotrophic samples because of the greater sensitivity of CARD-FISH (26, 34, 75). A detailed technical description of this protocol is reviewed elsewhere (2).
More recently, CARD-FISH has been used for nanoSIMS analyses. The principles of nanoSIMS and its applications for environmental microbiology are reviewed elsewhere (87). Because nanoSIMS is not a microscope-based method, it cannot be used to observe fluorescent signals. In simultaneous isotopic measurements and phylogenetic identification of microbial cells by nanoSIMS, halogen elements such as iodine, fluorine, and bromine are often used. SIMSISH (SIMS in situ hybridization) uses 5′-iodo-2′-deoxycytidine instead of 2′-deoxycytidine in oligonucleotide probes (41); however, this method has limited sensitivity because the number of substitutable bases depends on the probe sequence. Enhanced elemental labeling (EL)-FISH (13) and halogen in situ hybridization (HISH) (51) were developed as more sensitive methods. Both of these techniques are based on CARD-FISH, but fluorochromes that contain a halogen element (e.g., 544Br, BODIPY TMR-X, and Oregon Green 488-X) are coupled with tyramine and are used for the CARD reaction. Following CARD-FISH, a large number of halogen elements are deposited in cells, resulting in sensitive detection by nanoSIMS.
CARD has also been used to identify DNA-synthesizing bacteria in marine environments (57). Bromodeoxyuridine (BrdU) is a halogenated nucleotide analogue of tritiated thymidine and is incorporated during DNA synthesis in bacteria. After incubating cells in BrdU-containing medium, BrdU-incorporated cells were detected using HRP-labeled anti-BrdU-antibody and the CARD reaction.
Conclusion
CARD-FISH is one of the most important molecular tools for enhancing our understanding of environmental micro-organisms. Because of its increased sensitivity and the many recent technical developments, CARD-FISH can now be used for the detection of not only rRNA, but also mRNA and genes encoded on chromosomes in microorganisms in various environments. This review describes the fundamental principle, applications, and problems of CARD-FISH; however, the application of CARD-FISH requires some technical advances to overcome several drawbacks, as described in this review (e.g., low probe permeability and endogenous peroxidase activity). Further technical developments in CARD-FISH, including the combination with recent leading-edge technologies, will provide new insights into environmental microbiology with single-cell resolution.
Acknowledgements
The author thanks Tsuyoshi Yamaguchi at Nagaoka University of Technology for providing the FISH and CARD-FISH images of marine microorganisms.
References
- 1.Abed RMM, Schönhuber W, Amann RI, Garcia-Pichel F. Picobenthic cyanobacterial populations revealed by 16S rRNA-targeted in situ hybridization. Environ Microbiol. 2002;4:375–382. doi: 10.1046/j.1462-2920.2002.00307.x. [DOI] [PubMed] [Google Scholar]
- 2.Alonso C. Tips and tricks for high quality MAR-FISH preparations: focus on bacterioplankton analysis. Syst Appl Microbiol. doi: 10.1016/j.syapm.2012.02.005. in press. [DOI] [PubMed] [Google Scholar]
- 3.Amann RI, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol. 1990;172:762–770. doi: 10.1128/jb.172.2.762-770.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R, Springer N, Ludwig W, Görtz HD, Schleifer KH. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- 5.Amann RI, Zarda B, Stahl DA, Schleifer KH. Identification of individual prokaryotic cells by using enzyme-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1992;58:3007–3011. doi: 10.1128/aem.58.9.3007-3011.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amann RI. In situ identification of micro-organisms by whole cell hybridization with rRNA-targeted nucleic acid probes. In: Akkermans ADL, van Elsas JD, de Bruijn FJ, editors. Molecular Microbial Ecology Manual. 3.3.6. Kluwer Academic Publishers; Dodrecht, The Netherlands: 1995. pp. 1–15. [Google Scholar]
- 7.Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Amann RI, Fuchs BM, Behrens S. The identification of microorganisms by fluorescence in situ hybridisation. Curr Opin Biotechnol. 2001;12:231–236. doi: 10.1016/s0958-1669(00)00204-4. [DOI] [PubMed] [Google Scholar]
- 9.Amann RI, Fuchs BM. Single-cell identification in microbial communities by improved fluorescence in situ hybridization techniques. Nat Rev Micro. 2008;6:339–348. doi: 10.1038/nrmicro1888. [DOI] [PubMed] [Google Scholar]
- 10.Bakermans C, Madsen EL. Detection in coal tar waste-contaminated groundwater of mRNA transcripts related to naphthalene dioxygenase by fluorescent in situ hybridization with tyramide signal amplification. J. Microbiol Methods. 2002;50:75–84. doi: 10.1016/s0167-7012(02)00015-5. [DOI] [PubMed] [Google Scholar]
- 11.Barsotti O, Morrier JJ, Freney J, Renaud F, Benay G, Decoret D, Dumont J. Achromopeptidase for rapid lysis of oral anaerobic gram-positive rods. Oral Microbiol Immunol. 1988;3:86–88. doi: 10.1111/j.1399-302x.1988.tb00088.x. [DOI] [PubMed] [Google Scholar]
- 12.Behrens S, Fuchs BM, Mueller F, Amann RI. Is the in situ accessibility of the 16S rRNA of Escherichia coli for Cy3-labeled oligonucleotide probes predicted by a three-dimensional structure model of the 30S ribosomal subunit? Appl Environ Microbiol. 2003;69:4935–4941. doi: 10.1128/AEM.69.8.4935-4941.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Behrens S, Lösekann T, Pett-Ridge J, Weber PK, Ng W-O, Stevenson BS, Hutcheon ID, Relman DA, Spormann AM. Linking microbial phylogeny to metabolic activity at the single-cell level by using enhanced element labeling-catalyzed reporter deposition fluorescence in situ hybridization (EL-FISH) and NanoSIMS. Appl Environ Microbiol. 2008;74:3143–3150. doi: 10.1128/AEM.00191-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biegala IC, Not F, Vaulot D, Simon N. Quantitative assessment of picoeukaryotes in the natural environment by using taxon-specific oligonucleotide probes in association with tyramide signal amplification-fluorescence in situ hybridization and flow cytometry. Appl Environ Microbiol. 2003;69:5519–5529. doi: 10.1128/AEM.69.9.5519-5529.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bobrow MN, Harris TD, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification: application to immunoassays. J. Immunol Methods. 1989;125:279–285. doi: 10.1016/0022-1759(89)90104-x. [DOI] [PubMed] [Google Scholar]
- 16.Bobrow MN, Shaughnessy KJ, Litt GJ. Catalyzed reporter deposition, a novel method of signal amplification. II. Application to membrane immunoassays. J. Immunol Methods. 1991;137:103–112. doi: 10.1016/0022-1759(91)90399-z. [DOI] [PubMed] [Google Scholar]
- 17.Bobrow MN, Adler KE, Roth KA. Enhanced catalyzed reporter deposition. 6,593,100 US patent. 2003
- 18.Chassy BM, Giuffrida A. Method for the lysis of Gram-positive, asporogenous bacteria with lysozyme. Appl Environ Microbiol. 1980;39:153–158. doi: 10.1128/aem.39.1.153-158.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chatzinotas A, Sandaa RA, Schönhuber W, Amann R, Daae FL, Torsvik V, Zeyer J, Hahn D. Analysis of broad-scale differences in microbial community composition of two pristine forest soils. Syst Appl Microbiol. 1998;21:579–587. doi: 10.1016/S0723-2020(98)80070-2. [DOI] [PubMed] [Google Scholar]
- 20.DeLong EF, Wickham GS, Pace NR. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 21.DeLong EF, Taylor LT, Marsh TL, Preston CM. Visualization and enumeration of marine planktonic archaea and bacteria by using polyribonucleotide probes and fluorescent in situ hybridization. Appl Environ Microbiol. 1999;65:5554–5563. doi: 10.1128/aem.65.12.5554-5563.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ezaki T, Suzuki S. Achromopeptidase for lysis of anaerobic gram-positive cocci. J Clin Microbiol. 1982;16:844–846. doi: 10.1128/jcm.16.5.844-846.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferrari BC, Tujula N, Stoner K, Kjelleberg S. Catalyzed reporter deposition-fluorescence in situ hybridization allows for enrichment-independent detection of microcolony-forming soil bacteria. Appl Environ Microbiol. 2006;72:918–922. doi: 10.1128/AEM.72.1.918-922.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fichtl KM. Doctor thesis (Tech. Univ. München) 2005. Polynucleotide probe based enrichment of bacterial cells: development of probes for species of clinical relevance; pp. 1–216. [Google Scholar]
- 25.Hao DM, Tashiro T, Kato M, et al. Population dynamics of Crenarchaeota and Euryarchaeota in the mixing front of river and marine waters. Microbes Environ. 2010;25:126–132. doi: 10.1264/jsme2.me10106. [DOI] [PubMed] [Google Scholar]
- 26.Herndl GJ, Reinthaler T, Teira E, van Aken H, Veth C, Pernthaler A, Pernthaler J. Contribution of Archaea to total prokaryotic production in the deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopman AH, Ramaekers FC, Speel EJ. Rapid synthesis of biotin-, digoxigenin-, trinitrophenyl-, and fluorochrome-labeled tyramides and their application for in situ hybridization using CARD amplification. J Histochem Cytochem. 1998;46:771–777. doi: 10.1177/002215549804600611. [DOI] [PubMed] [Google Scholar]
- 28.Hoshino T, Yilmaz LS, Noguera DR, Daims H, Wagner M. Quantification of target molecules needed to detect micro-organisms by fluorescence in situ hybridization (FISH) and catalyzed reporter deposition-FISH. Appl Environ Microbiol. 2008;74:5068–5077. doi: 10.1128/AEM.00208-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii K, Mußmann M, MacGregor B, Amann RI. An improved fluorescence in situ hybridization protocol for the identification of bacteria and archaea in marine sediments. FEMS Microbiol Ecol. 2004;50:203–212. doi: 10.1016/j.femsec.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 30.Kawakami S, Kubota K, Imachi H, Yamaguchi T, Harada H, Ohashi A. Detection of single copy genes by two-pass tyramide signal amplification fluorescence in situ hybridization (Two-Pass TSA-FISH) with single oligonucleotide probes. Microbes Environ. 2010;25:15–21. doi: 10.1264/jsme2.me09180. [DOI] [PubMed] [Google Scholar]
- 31.Kawakami S, Hasegawa T, Imachi H, Yamaguchi T, Harada H, Ohashi A, Kubota K. Detection of single-copy functional genes in prokaryotic cells by two-pass TSA-FISH with poly-nucleotide probes. J. Microbiol Methods. 2012;88:218–223. doi: 10.1016/j.mimet.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Kenzaka T, Ishidoshiro A, Tani K, Nasu M. Scanning electron microscope imaging of bacteria based on DNA sequence. Lett Appl Microbiol. 2009;49:796–799. doi: 10.1111/j.1472-765X.2009.02680.x. [DOI] [PubMed] [Google Scholar]
- 33.Kerstens HM, Poddighe PJ, Hanselaar AG. A novel in situ hybridization signal amplification method based on the deposition of biotinylated tyramine. J Histochem Cytochem. 1995;43:347–352. doi: 10.1177/43.4.7897179. [DOI] [PubMed] [Google Scholar]
- 34.Klausen C, Nicolaisen MH, Strobel BW, Warnecke F, Nielsen JL, Jørgensen NOG. Abundance of actinobacteria and production of geosmin and 2-methylisoborneol in Danish streams and fish ponds. FEMS Microbiol Ecol. 2005;52:265–278. doi: 10.1016/j.femsec.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 35.Kofoed MVW, Nielsen DÅ, Revsbech NP, Schramm A. Fluorescence in situ hybridization (FISH) detection of nitrite reductase transcripts (nirS mRNA) in Pseudomonas stutzeri biofilms relative to a microscale oxygen gradient. Syst. Appl. Microbiol. 2012 Jan 31; doi: 10.1016/j.syapm.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Kubota K, Ohashi A, Imachi H, Harada H. Improved in situ hybridization efficiency with locked-nucleic-acid-incorporated DNA probes. Appl Environ Microbiol. 2006;72:5311–5317. doi: 10.1128/AEM.03039-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kubota K, Ohashi A, Imachi H, Harada H. Visualization of mcr mRNA in a methanogen by fluorescence in situ hybridization with an oligonucleotide probe and two-pass tyramide signal amplification (two-pass TSA-FISH) J. Microbiol Methods. 2006;66:521–528. doi: 10.1016/j.mimet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 38.Kubota K, Imachi H, Kawakami S, Nakamura K, Harada H, Ohashi A. Evaluation of enzymatic cell treatments for application of CARD-FISH to methanogens. J. Microbiol Methods. 2008;72:54–59. doi: 10.1016/j.mimet.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 39.Lebaron P, Catala P, Fajon C, Joux F, Baudart J, Bernard L. A new sensitive, whole-cell hybridization technique for detection of bacteria involving a biotinylated oligonucleotide probe targeting rRNA and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3274–3278. doi: 10.1128/aem.63.8.3274-3278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee SY, Bailey SC, Apirion D. Small stable RNAs from Escherichia coli: evidence for the existence of new molecules and for a new ribonucleoprotein particle containing 6S RNA. J Bacteriol. 1978;133:1015–1023. doi: 10.1128/jb.133.2.1015-1023.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li T, Wu T-D, Mazéas L, Toffin L, Guerquin-Kern J-L, Leblon G, Bouchez T. Simultaneous analysis of microbial identity and function using NanoSIMS. Environ Microbiol. 2008;10:580–588. doi: 10.1111/j.1462-2920.2007.01478.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lin X, Wakeham SG, Putnam IF, Astor YM, Scranton MI, Chistoserdov AY, Taylor GT. Comparison of vertical distributions of prokaryotic assemblages in the anoxic Cariaco Basin and Black Sea by use of fluorescence in situ hybridization. Appl Environ Microbiol. 2006;72:2679–2690. doi: 10.1128/AEM.72.4.2679-2690.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ludwig W, Dorn S, Springer N, Kirchhof G, Schleifer KH. PCR-based preparation of 23S rRNA-targeted group-specific polynucleotide probes. Appl Environ Microbiol. 1994;60:3236–3244. doi: 10.1128/aem.60.9.3236-3244.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luo Y, Pfister P, Leisinger T, Wasserfallen A. The genome of archaeal prophage ΨM100 encodes the lytic enzyme responsible for autolysis of Methanothermobacter wolfeii. J Bacteriol. 2001;183:5788–5792. doi: 10.1128/JB.183.19.5788-5792.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luo Y, Pfister P, Leisinger T, Wasserfallen A. Pseudomurein endoisopeptidases PeiW and PeiP, two moderately related members of a novel family of proteases produced in Methanothermobacter strains. FEMS Microbiol Lett. 2002;208:47–51. doi: 10.1111/j.1574-6968.2002.tb11059.x. [DOI] [PubMed] [Google Scholar]
- 46.Manz W, Amann R, Ludwig W, Wagner M, Schleifer K. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria—problems and solutions. Syst Appl Microbiol. 1992;15:593–600. [Google Scholar]
- 47.Molari M, Manini E. Reliability of CARD-FISH procedure for enumeration of Archaea in deep-sea surficial sediments. Curr Microbiol. 2012;64:242–250. doi: 10.1007/s00284-011-0056-5. [DOI] [PubMed] [Google Scholar]
- 48.Moraru C, Lam P, Fuchs BM, Kuypers MMM, Amann R. GeneFISH—an in situ technique for linking gene presence and cell identity in environmental microorganisms. Environ Microbiol. 2010;12:3057–3073. doi: 10.1111/j.1462-2920.2010.02281.x. [DOI] [PubMed] [Google Scholar]
- 49.Moraru C, Moraru G, Fuchs BM, Amann R. Concepts and software for a rational design of polynucleotide probes. Environ Microbiol Rep. 2011;3:69–78. doi: 10.1111/j.1758-2229.2010.00189.x. [DOI] [PubMed] [Google Scholar]
- 50.Mota CR, So MJ, de los Reyes FL. Identification of nitrite-reducing bacteria using sequential mRNA fluorescence in situ hybridization and fluorescence-assisted cell sorting. Microb Ecol. 2012;64:256–267. doi: 10.1007/s00248-012-0018-x. [DOI] [PubMed] [Google Scholar]
- 51.Musat N, Halm H, Winterholler B, et al. A single-cell view on the ecophysiology of anaerobic phototrophic bacteria. Proc Natl Acad Sci USA. 2008;105:17861–17866. doi: 10.1073/pnas.0809329105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakamura K, Terada T, Sekiguchil Y, Shinzato N, Meng X-Y, Enoki M, Kamagata Y. Application of pseudomurein endoisopeptidase to fluorescence in situ hybridization of methanogens within the family Methanobacteriaceae. Appl Environ Microbiol. 2006;72:6907–6913. doi: 10.1128/AEM.01499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okabe S, Oshiki M, Kamagata Y, et al. A great leap forward in microbial ecology. Microbes Environ. 2010;25:230–240. doi: 10.1264/jsme2.me10178. [DOI] [PubMed] [Google Scholar]
- 54.Orcutt B, Boetius A, Elvert M, Samarkin V, Joye S. Molecular biogeochemistry of sulfate reduction, methanogenesis and the anaerobic oxidation of methane at Gulf of Mexico cold seeps. Geochim. Cosmochim Acta. 2005;69:4267–4281. [Google Scholar]
- 55.Pavlekovic M, Schmid MC, Schmider-Poignee N, et al. Optimization of three FISH procedures for in situ detection of anaerobic ammonium oxidizing bacteria in biological wastewater treatment. J. Microbiol Methods. 2009;78:119–126. doi: 10.1016/j.mimet.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 56.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol. 2002;68:3094–3101. doi: 10.1128/AEM.68.6.3094-3101.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pernthaler A, Pernthaler J, Schattenhofer M, Amann R. Identification of DNA-synthesizing bacterial cells in coastal North Sea plankton. Appl Environ Microbiol. 2002;68:5728–5736. doi: 10.1128/AEM.68.11.5728-5736.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pernthaler A, Amann R. Simultaneous fluorescence in situ hybridization of mRNA and rRNA in environmental bacteria. Appl Environ Microbiol. 2004;70:5426–5433. doi: 10.1128/AEM.70.9.5426-5433.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pernthaler A, Pernthaler J. Simultaneous fluorescence in situ hybridization of mRNA and rRNA for the detection of gene expression in environmental microbes. Meth Enzymol. 2005;397:352–371. doi: 10.1016/S0076-6879(05)97021-3. [DOI] [PubMed] [Google Scholar]
- 60.Pernthaler A, Dekas AE, Brown CT, Goffredi SK, Embaye T, Orphan VJ. Diverse syntrophic partnerships from deep-sea methane vents revealed by direct cell capture and metagenomics. Proc Natl Acad Sci USA. 2008;105:7052–7057. doi: 10.1073/pnas.0711303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pernthaler J, Glockner F, Schönhuber W, Amann R. Fluorescence in situ hybridization (FISH) with rRNA-targeted oligonucleotide probes. Meth Microbiol. 2001;30:207–226. [Google Scholar]
- 62.Petersen JM, Zielinski FU, Pape T, et al. Hydrogen is an energy source for hydrothermal vent symbioses. Nature. 2011;476:176–180. doi: 10.1038/nature10325. [DOI] [PubMed] [Google Scholar]
- 63.Pfister P, Wasserfallen A, Stettler R, Leisinger T. Molecular analysis of Methanobacterium phage ΨM2. Mol Microbiol. 1998;30:233–244. doi: 10.1046/j.1365-2958.1998.01073.x. [DOI] [PubMed] [Google Scholar]
- 64.Pizzetti I, Gobet A, Fuchs B, Amann R, Fazi S. Abundance and diversity of Planctomycetes in a Tyrrhenian coastal system of central Italy. Aquat Microb Ecol. 2011;65:129–141. [Google Scholar]
- 65.Pratscher J, Dumont MG, Conrad R. Ammonia oxidation coupled to CO2fixation by archaea and bacteria in an agricultural soil. Proc Natl Acad Sci USA. 2011;108:4170–4175. doi: 10.1073/pnas.1010981108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Raap AK, van de Corput MP, Vervenne RA, van Gijlswijk RP, Tanke HJ, Wiegant J. Ultra-sensitive FISH using peroxidase-mediated deposition of biotin- or fluorochrome tyramides. Hum Mol Genet. 1995;4:529–534. doi: 10.1093/hmg/4.4.529. [DOI] [PubMed] [Google Scholar]
- 67.Schippers A, Neretin LN, Kallmeyer J, Ferdelman TG, Cragg BA, Parkes RJ, Jørgensen BB. Prokaryotic cells of the deep sub-seafloor biosphere identified as living bacteria. Nature. 2005;433:861–864. doi: 10.1038/nature03302. [DOI] [PubMed] [Google Scholar]
- 68.Schönhuber W, Fuchs BM, Juretschko S, Amann RI. Improved sensitivity of whole-cell hybridization by the combination of horseradish peroxidase-labeled oligonucleotides and tyramide signal amplification. Appl Environ Microbiol. 1997;63:3268–3273. doi: 10.1128/aem.63.8.3268-3273.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schönhuber W, Zarda B, Eix S, Rippka R, Herdman M, Ludwig W, Amann RI. In situ identification of cyanobacteria with horseradish peroxidase-labeled, rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1999;65:1259–1267. doi: 10.1128/aem.65.3.1259-1267.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schönhuber W, Le Bourhis G, Tremblay J, Amann RI, Kulakauskas S. Utilization of tmRNA sequences for bacterial identification. BMC Microbiol. 2001;1:20. doi: 10.1186/1471-2180-1-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schramm A, Fuchs BM, Nielsen JL, Tonolla M, Stahl DA. Fluorescence in situ hybridization of 16S rRNA gene clones (Clone-FISH) for probe validation and screening of clone libraries. Environ Microbiol. 2002;4:713–720. doi: 10.1046/j.1462-2920.2002.00364.x. [DOI] [PubMed] [Google Scholar]
- 72.Sekar R, Pernthaler A, Pernthaler J, Warnecke F, Posch T, Amann RI. An improved protocol for quantification of freshwater Actinobacteria by fluorescence in situ hybridization. Appl Environ Microbiol. 2003;69:2928–2935. doi: 10.1128/AEM.69.5.2928-2935.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sekar R, Fuchs BM, Amann RI, Pernthaler J. Flow sorting of marine bacterioplankton after fluorescence in situ hybridization. Appl Environ Microbiol. 2004;70:6210–6219. doi: 10.1128/AEM.70.10.6210-6219.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Steinberg LM, Regan JM. Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge. Appl Environ Microbiol. 2008;74:6663–6671. doi: 10.1128/AEM.00553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Teira E, Reinthaler T, Pernthaler A, Pernthaler J, Herndl GJ. Combining catalyzed reporter deposition-fluorescence in situ hybridization and microautoradiography to detect substrate utilization by bacteria and archaea in the deep ocean. Appl Environ Microbiol. 2004;70:4411–4414. doi: 10.1128/AEM.70.7.4411-4414.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tijssen P. Overview of principles of hybridization and the strategy of nucleic acid probe assays. In: van der Vliet PC, editor. Laboratory Techniques in Biochemistry and Molecular Biology, Hybridization with Nucleic Acid Probes Part I: Theory and Nucleic Acid Preparation. Elsevier; Amsterdam, London, New York, Tokyo: 1993. pp. 19–78. [Google Scholar]
- 77.Tischer K, Zeder M, Klug R, Pernthaler J, Schattenhofer M, Harms H, Wendeberg A. Fluorescence in situ hybridization (CARD-FISH) of microorganisms in hydrocarbon contaminated aquifer sediment samples. Syst. Appl. Microbiol. 2012 Mar 15; doi: 10.1016/j.syapm.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 78.Trebesius K, Amann RI, Ludwig W, Mühlegger K, Schleifer K. Identification of whole fixed bacterial cells with nonradioactive 23S rRNA-targeted polynucleotide probes. Appl Environ Microbiol. 1994;60:3228–3235. doi: 10.1128/aem.60.9.3228-3235.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tsunasawa S, Masaki T, Hirose M, Soejima M, Sakiyama F. The primary structure and structural characteristics of Achromobacter lyticus protease I, a lysine-specific serine protease. J Biol Chem. 1989;264:3832–3839. [PubMed] [Google Scholar]
- 80.Vaara M. Agents that increase the permeability of the outer membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van de Corput MP, Dirks RW, van Gijlswijk RP, van de Rijke FM, Raap AK. Fluorescence in situ hybridization using horseradish peroxidase-labeled oligodeoxynucleotides and tyramide signal amplification for sensitive DNA and mRNA detection. Histochem Cell Biol. 1998;110:431–437. doi: 10.1007/s004180050304. [DOI] [PubMed] [Google Scholar]
- 82.van Gijlswijk RP, Wiegant J, Raap AK, Tanke HJ. Improved localization of fluorescent tyramides for fluorescence in situ hybridization using dextran sulfate and polyvinyl alcohol. J Histochem Cytochem. 1996;44:389–392. doi: 10.1177/44.4.8601698. [DOI] [PubMed] [Google Scholar]
- 83.van Noorden CJ, Vogels IM. Polyvinyl alcohol and other tissue protectants in enzyme histochemistry: a consumer’s guide. Histochem J. 1989;21:373–379. doi: 10.1007/BF01789734. [DOI] [PubMed] [Google Scholar]
- 84.Visweswaran GRR, Dijkstra BW, Kok J. Two major archaeal pseudomurein endoisopeptidases: PeiW and PeiP. Archaea. 2010;2010:480492. doi: 10.1155/2010/480492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wagner M, Schmid M, Juretschko S, Trebesius KH, Bubert A, Goebel W, Schleifer KH. In situ detection of a virulence factor mRNA and 16S rRNA in Listeria monocytogenes. FEMS Microbiol Lett. 1998;160:159–168. doi: 10.1111/j.1574-6968.1998.tb12906.x. [DOI] [PubMed] [Google Scholar]
- 86.Wagner M, Hornt M, Daims H. Fluorescence in situ hybridisation for the identification and characterisation of prokaryotes. Curr Opin Microbiol. 2003;6:302–309. doi: 10.1016/s1369-5274(03)00054-7. [DOI] [PubMed] [Google Scholar]
- 87.Wagner M. Single-cell ecophysiology of microbes as revealed by Raman microspectroscopy or secondary ion mass spectrometry imaging. Annu Rev Microbiol. 2009;63:411–429. doi: 10.1146/annurev.micro.091208.073233. [DOI] [PubMed] [Google Scholar]
- 88.Wagner M, Haider S. New trends in fluorescence in situ hybridization for identification and functional analyses of microbes. Curr Opin Biotechnol. 2012;23:96–102. doi: 10.1016/j.copbio.2011.10.010. [DOI] [PubMed] [Google Scholar]
- 89.West NJ, Schönhuber WA, Fuller NJ, Amann RI, Rippka R, Post AF, Scanlan DJ. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology. 2001;147:1731–1744. doi: 10.1099/00221287-147-7-1731. [DOI] [PubMed] [Google Scholar]
- 90.Woebken D, Fuchs BM, Kuypers MMM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol. 2007;73:4648–4657. doi: 10.1128/AEM.02774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamaguchi N, Sasada M, Nasu M. Rapid detection of starved Escherichia coli with respiratory activity in potable water by signal-amplified in situ hybridization following formazan reduction. Microbes Environ. 2009;24:286–290. doi: 10.1264/jsme2.me09144. [DOI] [PubMed] [Google Scholar]
- 92.You Y, Moreira BG, Behlke MA, Owczarzy R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006;34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zarda B, Hahn D, Chatzinotas A, Schönhuber W, Neef A, Amann RI, Zeyer J. Analysis of bacterial community structure in bulk soil by in situ hybridization. Arch Microbiol. 1997;168:185–192. [Google Scholar]