Abstract
The Bayon temple in Angkor Thom, Cambodia has shown serious deterioration and is subject to the formation of various pigmented biofilms. Because biofilms are damaging the bas-reliefs, low reliefs engraved on the surface of sandstone, information about the microbial community within them is indispensable to control biofilm colonization. PCR-denaturing gradient gel electrophoresis (DGGE) analysis of biofilm samples from the pigmented sandstone surfaces showed that the bacterial community members in the biofilms differed clearly from those in the air and had low sequence similarity to database sequences. Non-destructive sampling of biofilm revealed novel bacterial groups of predominantly Rubrobacter in salmon pink biofilm, Cyanobacteria in chrome green biofilm, Cyanobacteria and Chloroflexi in signal violet biofilm, Chloroflexi in black gray biofilm, and Deinococcus-Thermus, Cyanobacteria, and Rubrobacter in blue green biofilm. Serial peeling-off of a thick biofilm by layers with adhesive sheets revealed a stratified structure: the blue–green biofilm, around which there was serious deterioration, was very rich in Cyanobacteria near the surface and Chloroflexi in deep layer below. Nitrate ion concentrations were high in the blue–green biofilm. The characteristic distribution of bacteria at different biofilm depths provides valuable information on not only the biofilm formation process but also the sandstone weathering process in the tropics.
Keywords: bacterial community, denaturing gradient gel electrophoresis, non-destructive sampling, cultural heritage, biodeterioration
A biofilm is an aggregation of microbial cells and extracellular polymer substances (EPSs) adhering to a solid substratum surface. Biofilms formed on stone monuments accelerate rock decay through physicochemical processes, namely the excretion of water-absorbing EPSs that contain surface-tension-reducing compounds and the uptake of capillary water followed by an increase in water-holding capacity (46). Biodeterioration caused by biofilm formation has been reported in many stone structures of cultural heritage (41, 47). In the Angkor monuments of Cambodia, there have been analyses of microorganisms in fresh and old biofilms by PCR amplification and clone libraries (21) and observations of colonizing cyanobacteria in green–brown and brown biofilms (9). Although the walls of the Bayon temple at Angkor Thom are covered with biofilms pigmented with various colors, there has been no comprehensive study on the bacterial communities in these biofilms.
Cambodia’s Angkor monuments are revered by the local people and have attracted visitors from around the world. In particular, the Bayon temple in the center of the historical city of Angkor Thom in Siem Reap is one of these great monuments. Its foundations are of laterite and its surface is of sandstone. On the walls of the inner and outer galleries that surround the central towers are engraved unfolding picture stories of Hindu myths and the life of the Khmer people as bas-reliefs. The temple has been deteriorating as a result of various factors, including water intrusion, mineral crystallization and microbial action, as is the case with many other historical stone monuments (41, 47). Exfoliate weathering, the flaking-off of fairly large and flat pieces of rock (14), has been observed at the base of sandstone pillars in Angkor Wat, where water intrusion is apparent. Previous studies showed that sulfur-oxidizing microorganisms play an important role in the decay of sandstone (23); sulfur-oxidizing Fusarium sp. and Mycobacterium spp. were isolated and their sulfur oxidation via chemolithoautotrophic process has been elucidated (19, 24).
In contrast, at the Bayon temple, biofilm formation on the sandstone walls is a more serious problem. Various pigmented biofilms are affecting not only the temple’s aesthetic appearance but also the integrity of its materials underneath (10, 27, 47) and the valuable bas-reliefs. Information about the structure of the microbial communities in the biofilms is therefore indispensable for understanding the microbial community involved and also controlling biofilm colonization of the historic bas-reliefs. To characterize the microorganisms in biofilms, we first compared bacteria in the air collected at the temple with bacteria found in the biofilms. The characteristics of the bacterial communities in variously pigmented biofilms and the stratified structure of bacteria in the biofilm were compared by using 16S rRNA-based PCR-denaturing gradient gel electrophoresis (DGGE) analyses among the bacterial communities after non-destructive collection of biofilm samples onto adhesive sheets. In addition, nitrate ions in the biofilms were also quantified as an indicator of microbial metabolism inside them due to ammonia/ammonium oxidation.
Materials and Methods
Sampling sites
The Bayon temple was built in the late 12th century. The climate of the area is governed by two monsoons: the northeastern monsoon resulting in dryness from November to April, and the southwestern monsoon brings the rainy season from May to October (25). Annual rainfall at the Angkor site is 1,300 to 1,500 mm (17) and the average temperature is 25°C (16). In the inner gallery of the Bayon the monthly mean temperature is between 25 and 30°C throughout the year, and the monthly mean relative humidity is between 60% and 80% in the dry season and 80% and 90% in the rainy season (39).
Airborne microbes and biofilms colonizing on the sandstone wall of sampling points (BA1 to BA4, BF1 to BF4, and BF6 to BF9, Fig. 1A) where the roof still stands were collected in September 2009. All other biofilm samples (Fig. 1A) were collected in September 2008, September 2009 or September 2011. Airborne microbes were collected onto an autoclaved polycarbonate membrane filter (pore size, 0.20 μm; disk diameter, 47 mm; Advantec, Tokyo, Japan), which was mounted in an open-face polycarbonate filter holder (Advantec) disinfected with 70% (v/v) ethanol. The filter holder was placed 2.5 m above the floor and 20 cm from the wall and connected to a diaphragm-type dry vacuum pump (model DA-30S; Ulvac Kiko, Kanagawa, Japan). Each air sample (1.4×104 L) was passed through the filter at 5 L min−1; this rate was controlled with a DMQ-V digital mass-flow controller (model MQV0050; Yamatake, Tokyo, Japan). Four samples (BA1 to BA4) were collected (once every 48 hours) over an 8-day period. After each sampling, the filters were put back into sterilized filter cases. To compare the bacterial communities of the air samples and the biofilms formed on the sandstone wall, biofilm samples were collected from the sandstone wall near the filter holder. Eight samples (BF1 to BF4 and BF6 to BF9) were collected by using adhesive sheets sterilized by gamma irradiation (adhesive area: 2.4×3.0 cm) (48). The colors of the biofilms sampled were brown for samples BF1, BF2, and BF9, brown and black for sample BF8, black for sample BF3, dark green for samples BF4 and BF6, and green for sample BF7. All samples were kept cold in an ice cooler in the field and stored at −20°C in a refrigerator of local office in Siem Reap, Cambodia, for a few days; they were then transported to our laboratory, where they were kept at −80°C until DNA extraction.
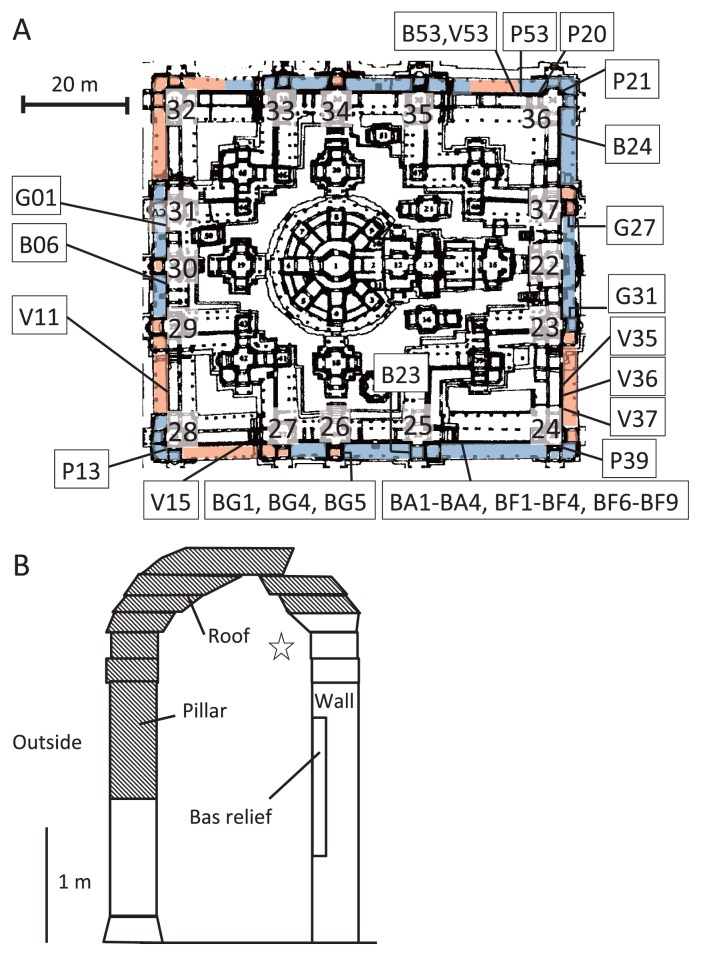
Fig. 1.
A. Map of the inner gallery of the Bayon, Angkor Thom, showing locations where biofilm and airborne microorganism samples were collected. Sample names are prefixed by the abbreviation of each biofilm color; sample identifications and biofilm characteristics are given in Table 1. The number next to the tower is the tower number. Blue area shows the area where the roof remained. Orange area shows the area where the roof was partly or completely missing. B. Side view of the inner gallery, which consisted of a sandstone wall on which bas-reliefs were engraved and a roof supported by pillars. Hatched areas show the parts of the pillars or roof that were partly or completely collapsed or missing. The area from which airborne bacterial samples were collected is shown by an asterisk.
To compare the bacterial communities of the various pigmented biofilms, adhesive sheets were used to take microorganisms from sandstone surface for routine analysis at 21 locations in the inner gallery (see Fig. 1A and Table 1 for sample numbers); the biofilms were grouped according to the five major colors observed on the gallery walls (Fig. 2A–C). The designated colors were based on a color chart (RAL Classic, German Institute for Quality Assurance and Certification, Sankt Augustin, Germany) as follows: salmon pink (P), RAL 3022; chrome green (G), RAL 6020; signal violet (V), RAL 4008; black–gray (B), RAL 7021; and blue–green (BG), RAL 6004. Microorganisms in the BG biofilm were collected from the surface to the deepest layer by a series of peeling-off the biofilm with adhesive sheets (23 sheets) at the same sampling position to analyze the stratified bacterial structure (sampling site BG4, Fig. 2B). This biofilm was chosen because it appeared thickest and seemed to be causing more serious deterioration of the biofilm underneath.
Table 1.
Colors and sampling locations of biofilms used for phylogenetic analysis in this study
| Sample | Color of biofilma | Sampling locationb | Roofc |
|---|---|---|---|
| P13 | Salmon pink (P) | Southwest area of tower 28 | + |
| P20 | North-facing wall between towers 35 and 36 | + | |
| P21 | Northeast area of tower 36 | + | |
| P39 | Southeast area of tower 24 | + | |
| P53 | North-facing wall in the northwest area of tower 36 | + | |
| G01 | Chrome green (G) | West-facing wall between towers 30 and 31 | + |
| G27 | South-facing wall in the southeast area of tower 37 | + | |
| G31 | North-facing wall in the northeast area of tower 23 | + | |
| V11 | Signal violet (V) | West-facing wall between towers 28 and 29 | − |
| V15 | South-facing wall between towers 27 and 28 | − | |
| V35 | East-facing wall between towers 23 and 24 | − | |
| V37 | East-facing wall between towers 23 and 24 | − | |
| V53 | North-facing wall between towers 35 and 36 | − | |
| B06 | Black gray (B) | West-facing wall between towers 29 and 30 | − |
| B23 | West-facing wall in the southwest area of tower 25 | + | |
| B24 | East-facing wall between towers 36 and 37 | + | |
| B36 | East-facing wall between towers 23 and 24 | − | |
| B53 | North-facing wall between towers 35 and 36 | − | |
| BG1 | Blue green (BG) | + | |
| BG4 | East-facing wall in the southeast area of tower 26 | + | |
| BG5 | + |
Samples P53, V53, B53, BG1, BG4, and BG5 were collected in September 2008. The other samples were collected in September 2009.
Letters in parentheses are abbreviations of colors.
All biofilms were obtained from the inner gallery of Bayon. Towers are numbered as described in Fig. 1A.
+, Roof indicated in Fig. 1B was present; −, roof was missing.
Fig. 2.
Various pigmented biofilms formed on the sandstone walls of the inner gallery of Bayon. Bar indicates 10 cm.
A. Salmon pink (P), black–gray (B), and signal violet (V) biofilms on the north-facing wall on the north side. Samples P53, B53, and V53 were obtained from the positions indicated “P”, “B”, and “V”, respectively.
B. Blue–green (BG) biofilm on the east-facing wall on the south side. Samples BG1, BG4, and BG5 were obtained from the position indicated “BG”, measuring 30 cm in circumference. Sample BG4 was used for analysis of bacterial stratified structure.
C. Chrome green (G) biofilm on the south-facing wall of the east side. Sample G27 was obtained from the position indicated by “G”.
To monitor the amounts of nitrate ion in the biofilms, sample No. 5 was taken from the wall near the BG; samples No. 1, 2, 9 and 10 were taken from the outer gallery where the roof was completely missing; samples No. 3, 4, 7 and 8 were taken from the inner gallery, where the roof was still present (Fig. 1B). The collected amount of biofilm from stone of no cultural importance was less than 1 g.
DNA extraction and PCR amplification
Samples were subjected to DNA extraction with a FastDNA spin kit for soil (MP Biomedicals, Illkirch, France). For DNA extraction, we performed one experiment on each sample because a tiny amount of sample was taken from the wall. Instead of replicates, we checked many samples of the similar colored biofilms. The adhesive side of the sheet used to sample the biofilm was placed in a tube, facing inward. DNA was extracted according to the kit manufacturer’s specifications.
To profile the bacterial community by DGGE, the V1 to V3 region of the bacterial 16S rRNA gene was amplified with the GC8F primer set, which was designed to attach a GC-clamp (cgcccgccgcgccccgcgcccgtcccgccgcccccgcccg) to the 5′ terminal of 8F (50), and with 520R (22). The PCR mixture contained 10 μL of 5× Phusion HF Buffer (Finnzymes, Espoo, Finland), 1 μL of 10 mM dNTPs (200 μM each), 1 μL of each primer (0.5 μM each), 5 μL of 0.1% bovine serum albumin, 1 μL of dimethyl sulfoxide, 0.5 μL of Phusion DNA polymerase (Finnzymes), 1 μL of Milli-Q water, and 1 μL of template DNA (0.1 to 20 ng) in a total volume of 50 μL. PCR amplification was performed in a thermal cycler (TP600, TaKaRa PCR thermal cycler dice; TaKaRa Bio, Otsu, Japan). The amplification conditions were as follows: 98°C for 30 s (initial denaturation), followed by 30 cycles of 98°C for 5 s, 50.7°C for 15 s, and 72°C for 60 s, with a final extension step of 72°C for 10 min. The amplified PCR products were checked on 1.5% agarose to verify amplification of the target region and purified with a GFX PCR DNA and gel band purification kit (GE Healthcare Life Sciences, Tokyo, Japan); they were then quantified with a UV spectrophotometer.
DGGE analysis
DGGE was performed as described by Muyzer et al. (29) using the DCode universal mutation detection system (Bio-Rad Laboratories, Hercules, CA, USA). DNA samples (150 ng) were loaded onto a 10% (w/v) polyacrylamide gel that contained a linear gradient of 30% to 70% denaturant (100% denaturant contained 7 M urea and 40% formamide). After electrophoresis at 60°C and 75 V for 14 h, the gel was soaked in SYBR Green I nucleic acid gel stain (Molecular Probes, Eugene, OR, USA) for 30 min, and then visualized and photographed under UV transillumination using a Printgraph (DT-20MP; Atto, Tokyo, Japan). DGGE marker I (Nippon Gene, Toyama, Japan) was used as a standard marker.
The relevant positions of each band on the DGGE gel were detected, and the peak intensities were converted to peak heights with CS Analyzer 3 (Atto). The bands were considered positive when the peak height as a percentage of the total peak height exceeded 5% in each lane in order to show the bacterial compositional characteristic more clearly. Bacterial compositions were calculated on the basis of the number of bands observed on the DGGE gels after analysis of the phylogenetic position as described below.
Sequence analysis of DGGE bands
Bands in the DGGE gel were excised from the gels with a Gel Cutting Tip (Axygen, CA, USA) and suspended in 50 μL of sterilized TE buffer (10 mM Tris-HCl, 0.5 M EDTA; pH 8.0). In order to check whether the target bands were obtained exactly, we compared with the DGGE gel picture before and after excising the bands. The DNA recovered after freeze-thawing the suspension was amplified under the same conditions as described above, except that as the forward primer we used 8F instead of GC8F. The size and amount of the PCR products were checked by 1.5% agarose gel electrophoresis for 30 min at 100 V. Cleaned PCR products were sequenced with a BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Carlsbad, CA, USA) by using the following reaction mixture: 0.5 μL of Ready Reaction Premix, 2 μL of 5×BigDye Sequencing Buffer, 0.32 μM of primer, cleaned PCR template, and Milli-Q water to a 10-μL total reaction volume. Amplification of cycle sequencing was as follows: 96°C for 1 min (initial denaturation) followed by 25 cycles of 96°C for 10 s, 50°C for 5 s, and 60°C for 4 min. Sequencing reactions were cleaned by ethanol precipitation, and sequencing was performed on an ABI 3130 Genetic Analyzer (Applied Biosystems). The closest relatives of the sequences of the DGGE bands were searched for in the GenBank database with a BLAST search program.
Phylogenetic analysis of 16S rRNA gene sequence
Each DNA sequence was compared with the sequences available in the database by using BLASTN 2.2.26 (51) from the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Potential DNA chimeric structures were detected by separately determining the species affiliation and the query coverage of the terminal 5′ and 3′ structures of each sequence. All of the sequences and their closest relatives were then aligned, and a phylogenetic tree was constructed with MEGA 5 (44) using the neighbor-joining method (37) with gaps and missing data removed. The presence of chimeric sequences was checked by comparison with the sequences in the Chimera-Checked database using BLASTN (1) from Greengenes (7; http://greengenes.lbl.gov/cgi-bin/nph-index.cgi).
Nitrate ion concentration in the biofilms
Biofilm samples collected in 2011 (0.1 g as wet weight) were suspended in 0.5 to 1.0 mL Milli-Q water and centrifuged at 5,000×g for 5 min at 4°C. The supernatant was used to measure pH with a compact pH meter (B-212; Horiba, Kyoto, Japan) and the concentration of nitrate ions with a compact NO3− meter (B-343; Horiba). These measurements were performed at the local office in Siem Reap. For nitrate ion measurements, we performed one experiment on each sample because a tiny amount of sample was taken from the wall. Instead of replicates, we checked many samples of the similar colored biofilms.
Nucleotide sequence accession numbers
All sequences of the 16S rRNA genes determined in this study have been submitted to the DNA Data Bank of Japan under accession numbers AB679757 to AB679829 and AB758608 to AB758622 (Tables S1 and S2).
Results and Discussion
Bacteria in wall biofilms and air
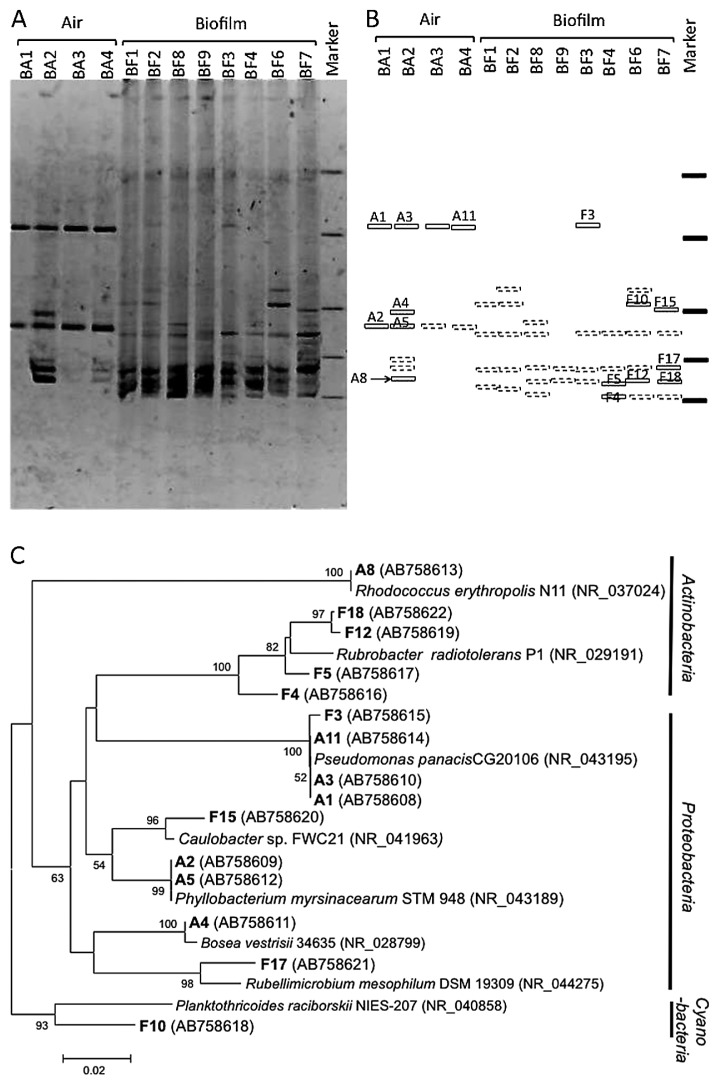
Bacteria in the wall biofilm samples BF1 to BF4 and BF6 to BF9 and the ambient air were compared by using DGGE to characterize the microorganisms in biofilms formed on the stone surface. The DNA banding patterns of PCR-amplified 16S rRNA genes, and hence the bacterial community structures, of the biofilm samples clearly differed from those of the bacteria found in the air samples (Fig. 3A). Two major DNA bands that were observed at the same position on the DGGE gels from all biofilm samples were Rubellimicrobium (band F17) and Rubrobacter (bands F5, F12, and F18) (Fig. 3A and B, Table S1), indicating that the major bacteria found in the biofilm were Proteobacteria and Actinobacteria (Fig. 3B and C). Except for band F3 and F15, the bacterial DNA sequences found in the biofilm had low levels of identity (91% to 94%) to the sequences in the database (Table S1). As already reported at many other historical sites (26, 40, 43), including Angkor sites (9, 21), the major bacteria in the biofilms colonizing the stone surfaces were characterized by the presence of unknown bacterial populations that were suited to growth in this environment.
Fig. 3.
Comparison based on partial 16S rRNA gene sequencing of bacterial communities in air collected in the inner gallery and in biofilms formed on the sandstone wall close to the air-sampling location.
A. Bacterial community fingerprints of air (BA) and biofilm (BF) samples by DGGE analysis. The partial 16S rRNA gene was amplified with primers GC8F and 520R.
B. Schematic diagram of the DGGE band patterns in A. Labeled bands correspond to 16S rRNA gene sequence types described in C and Table S1. Broken lines are DGGE bands that were not sequenced.
C. Phylogenetic relationships based on partial 16S rRNA gene sequences derived from air and biofilm samples by DGGE analysis. Sequences recovered in this study and corresponding to the band names in B are shown in bold type; “A” indicates bands derived from the air sample and “F” indicates bands derived from the biofilm sample. Numbers in parentheses are GenBank accession numbers. Neighbor-joining tree: bootstrap values based on 1,000 replicates are indicated for branches supported by 50% of trees. Scale bar represents 0.02 nucleotide changes per position.
In contrast to the bacteria found in the biofilms, the DNAs of only two major bacterial genera prevailed in the air samples (Fig. 3A): bands A1, A3, and A11 were affiliated with Pseudomonas panacis CG20106 with 100% identity, and bands A2 and A5 were affiliated with Phyllobacterium spp. with 98% or 100% identity (Table S1). The numbers of bands in the biofilm samples were higher than in the air samples (P<0.05) indicated that the bacterial diversity in the biofilm was higher than in the air. There has been a focus on airborne microorganisms at historical sites (33, 36, 45) because the atmosphere is a major vehicle for microorganisms (31). However, our results showed that the bacteria in the biofilm were unique and that major airborne bacteria seemed unable to establish communities under the extreme environmental conditions of exposure to severe sunlight and wind on the stone surface.
Bacterial communities in the pigmented biofilms
The P, G, and BG biofilms (Fig. 2) were frequently observed on the walls in places where the roof was still standing. The P biofilm was thin and dry. The BG biofilm was obviously thick (approx. >1 mm), comparatively wet, and easy to peel. The V biofilm was thin (approx. <1 mm) and was usually observed on the walls in places where the roof was missing (Fig. 1A). The stone surface under the BG biofilm exhibited exfoliation and serious deterioration. The G biofilm was comparatively thin (approx. <1 mm), hard, and difficult to peel off. The B biofilm was thin, easy to peel off, and was observed on the walls in places where the roof was either present or missing.
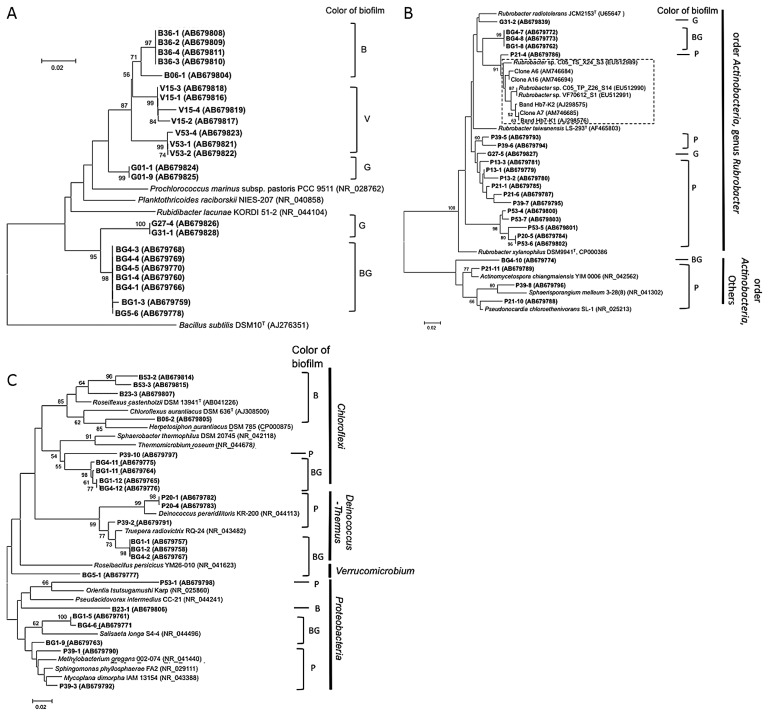
To compare the bacterial communities among these biofilms, three to five samples were collected from the top surface of each pigmented biofilm on four faces of the inner gallery (Fig. 1A) and subjected to 16S rRNA gene analyses. The numbers of DNA bands recovered from the DGGE gels were 36, 17, 24, 33, and 20 from the P, G, V, B, and BG biofilms, respectively. Of these, 24, 6, 7, 11, and 18, respectively, were successfully amplified, and then used for phylogenetic analyses after sequencing. We examined the phylogenetic relationships of these sequenced DNA bands with the DNA of members of the Cyanobacteria (Fig. 4A), Actinobacteria (Fig. 4B), and others (Fig. 4C).
Fig. 4.
Phylogenetic relationships based on partial 16S rRNA gene sequences recovered from salmon pink (P), chrome green (G), signal violet (V), black–gray (B), and blue–green (BG) biofilms derived from sandstone walls at Bayon by DGGE analysis. Sequences recovered in this study are shown in bold type. Numbers in parentheses are GenBank accession numbers Neighbor-joining tree: bootstrap values based on 1,000 replicates are indicated for branches supported by 50% of trees. Scale bar represents 0.02 nucleotide changes per position.
A. Phylogenetic affiliation of Cyanobacteria clade members. Bacillus subtilis DSM10T was used as an outgroup. Dashed box indicates 16S r RNA sequences derived from rosy discoloration of European historical stone monuments (15, 20, 40).
B, Phylogenetic affiliation of Actinobacteria clade members. Escherichia coli ATCC 11775T was used as an outgroup.
C, Phylogenetic affiliation of Chloroflexi, Proteobacteria, and Deinococcus-Thermus clade members.
Bacteria that were closely related to Cyanobacteria were found in many of the G, V, B, and BG biofilms, but none was found in the P biofilm (Fig. 4A). Actinobacteria-related bacteria prevailed in the P biofilms, especially in the genus Rubrobacter. In addition to the P biofilm, the G and BG biofilms included some bacteria closely related to the genus Rubrobacter (Fig. 4B).
Rubrobacter-related bacteria were observed in all pigmented biofilms collected at the Bayon and were especially rich in the P biofilm. The genus Rubrobacter is made up of Rubrobacter radiotolerans, Rubrobacter xylanophilus, and Rubrobacter taiwanensis. Rubrobacter radiotolerans has greater gamma radiation resistance than Deinococcus radiodurans (3, 28, 49). This radiation resistance might give Rubrobacter-related bacteria a growth advantage for colonizing the sandstone walls under strong sunlight at this tropical site. However, P biofilms from which the genus Rubrobacter were frequently detected were observed on the wall covered with a roof, suggesting colonization of uncultured bacteria that are clearly different from the Rubrobacter species so far known. Rubrobacter xylanophilus is also known to have a halotolerant phenotype. If a member of Rubrobacter-related bacteria possesses this trait, they will have an advantage for growing on the stone surface.
A relationship between Rubrobacter-related bacteria and rosy-pigmented biofilms has been reported in historical wall paintings or masonry in Europe (15, 20, 40), but to our knowledge our observation is the first of this type in Asia. Interestingly, most of the 16S rRNA sequences obtained from the P biofilm formed a cluster separate from most sequences derived from the rosy discoloration of wall paintings or masonry in Europe (15, 20, 40) (indicated by the shaded area in Fig. 4B). These results suggest that Rubrobacter-related bacteria colonize the rosy discoloration on historical stone monuments in Angkor Thom as well as in Europe, but their phylogenetic position differs slightly from those in Europe. Although we tried to isolate Rubrobacter-related bacteria by using the media described by Laiz et al. (20), we have not yet succeeded (data not shown).
The band sequences derived from the P, B, and BG biofilms were affiliated with members of the Chloroflexi, Proteobacteria, Acidobacteria, and Deinococcus-Thermus (Fig. 4C). Tolerance to irradiation, desiccation, and high temperature, which is known to occur in members of the phyla Chloroflexi and Deinococcus-Thermus and the genus Rubrobacter, appears advantageous for colonization of the sandstone walls.
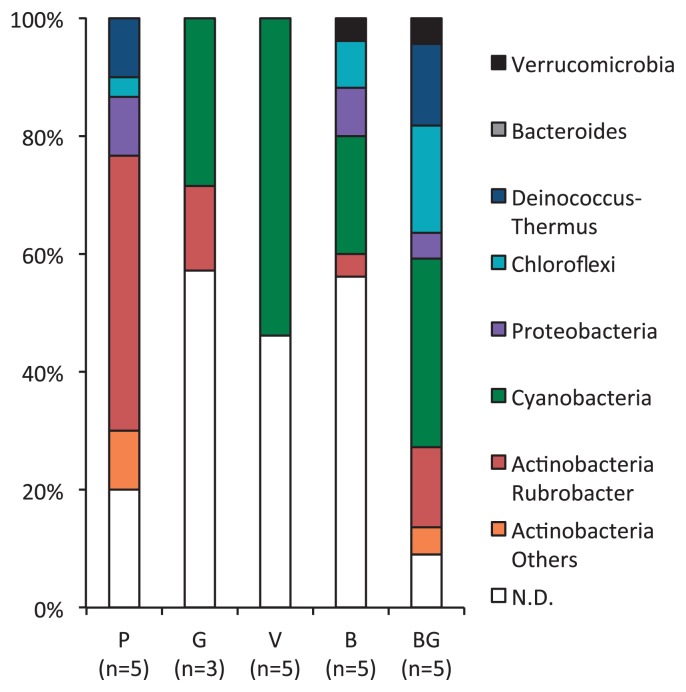
We calculated the bacterial compositions of each of the colored biofilms on the basis of the number of sequences obtained from the DGGE profiling followed by phylogenetic analysis (Fig. 5A). Bands at the same position in a gel were considered to represent the same taxon. Unsequenced bands were calculated as “not determined (N.D.)”. In P biofilms, members of the genus Rubrobacter was very rich (Fig. 5). In the V and B biofilms, bacteria belonging to the phylum Cyanobacteria or Chloroflexi had the highest species richness. In the G and BG biofilms, bacteria of the phylum Cyanobacteria or genus Rubrobacter showed high species richness. Bacteria of the phylum Deinococcus-Thermus were also observed in BG biofilms. In the G biofilm, although relatively few sequences were analyzed, bacteria grouped in the Chloroflexi, Cyanobacteria, and Rubrobacter were detected. These results suggested that the bacterial community composition of the biofilm surfaces had a considerable effect on their color. The color of the pigmentation in the biofilms could be a useful indicator of the bacterial communities present.
Fig. 5.
Comparison of bacterial compositions of pigmented biofilms formed on sandstone walls in Bayon. Sequences obtained from each biofilm sample were classified on the basis of the results of a BLAST search of Greengenes. Detailed affiliations are given in Table S2. P, salmon pink; B, black–gray; V, signal violet; G, chrome green; BG, blue–green; N.D., Not determined.
Members of the Cyanobacteria were observed in biofilms of all five colors. Some groups of Cyanobacteria have tolerance to extreme environmental conditions, such as radiation, desiccation, salinity, and high temperature (2, 4). This stress resistance can be advantageous for growth on sandstone walls under the severe conditions at the Bayon, as observed at a number of other historical monuments (6, 38). The Cyanobacteria-related bacteria detected in the biofilm samples may cause the accumulation of nitrogen nutrients in addition to organic compounds derived from photosynthesis, because some Cyanobacteria are known to have N2-fixing ability (4).
Spatial bacterial distribution in BG biofilm
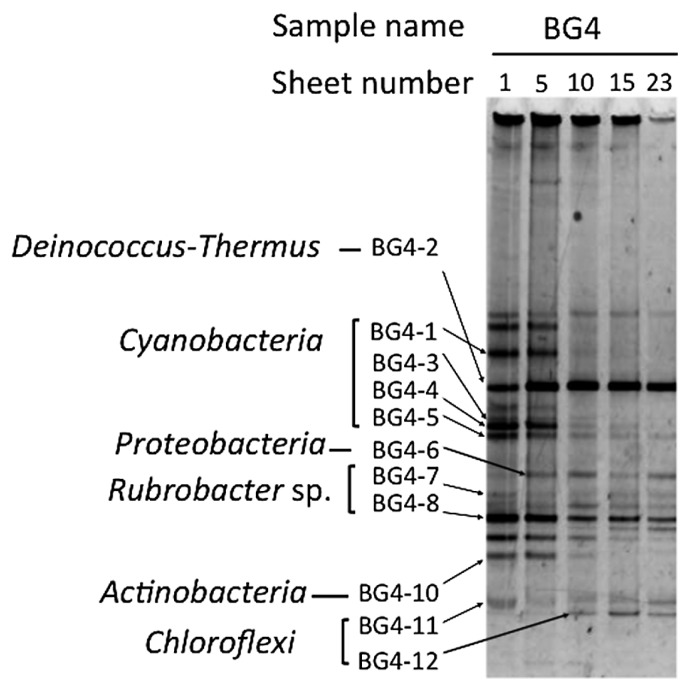
We examined the spatial bacterial distribution in the blue–green biofilm BG4 by using the DGGE profiles of 16S rRNA gene fragments (Fig. 6). Adhesive sheets with low numbering corresponded to the surface of the biofilm and those with higher numbering corresponded to the deepest layer of the biofilm. The banding pattern changed markedly from sheet numbers 5 to 10 of the 23 adhesive sheets. There were many kinds of bacteria, especially Cyanobacteria-related bacteria, in the surface layer of the biofilm, but there were fewer kinds of bacteria in the deeper layer. Two predominant bacteria were present throughout the layers. One was closely related to members of the phylum Deinococcus-Thermus, and the other was related to members of the genus Rubrobacter. In addition, a Chloroflexi-related bacterium, derived from band BG4-12, appeared only in the deeper layer of the biofilm. These results suggested that the bacterial habitats within each biofilm are very specific depending on the availability of light and organic nutrients and the oxygen requirement.
Fig. 6.
Changes in DGGE patterns of the blue–green (BG) biofilm. The sampling location, BG4, was on the south wall of a small room next to tower 26 on the south side of the inner gallery.
To examine in more detail the structure of the biofilm, samples were collected from the surface to the deepest part by using 23 adhesive sheets. Phylogenetic affiliations at the phylum level are shown next to the band names. Detailed affiliations of these bands are given in Table S2.
Spatial microbial distribution has been reported, especially in aquatic biofilms in experimental culture (5, 8, 32, 42) and in some environmental biofilms (13). However, there have been few reports of the biofilms on sandstone surfaces. McNamara et al. (26) reported that the endolithic bacterial community in terrestrial biofilms on limestone at a Mayan site differed from the epilithic community; their findings also suggest that a stratified bacterial structure forms in the biofilms on historical monuments. Because microbes living in the deepest part of the biofilm are likely to attack the stone more directly than those that live on the surface, an understanding of stratified microbial distribution is important.
A similar layered bacterial structure has already been reported in microbial mats in a hot spring. The surface layer was made up of Chloroflexus (phylum Chloroflexi) species and Cyanobacteria, and the deepest layer was composed of Roseiflexus castenholzii (phylum Chloroflexi) (13). The difference in the optical absorption spectra of the Chloroflexi and Cyanobacteria might enable them to live together in this terrestrial biofilm. The phylum Chloroflexi contains two orders of heterotrophic bacteria: the order Chloroflexales, a group of filamentous anoxygenic phototrophs, and the order Herpetosiphonales, which consists of non-phototrophic filamentous bacteria belonging to the genus Herpetosiphon. We found that a number of bacteria belonging to the phylum Chloroflexi grew on the sandstone wall, but most of them were uncultured members. This has been confirmed in a cloning analysis of biofilms of the Bayon temple (21). Although Chloroflexi bacteria have been detected at a few historical sites (26, 34), most have been detected in hot springs (11, 12, 35), in hypersaline mats (30), and in freshwater (18). The role of the organisms in the biofilms formed on the Bayon remains to be elucidated, and further study is required.
Accumulation of nitrate ions in biofilms
We examined the concentration of nitrate ions and the pH in the biofilms (Table 2). The concentration of NO3−-N in biofilm sample No. 5, which was derived from an area near samples BG1, BG4, and BG5, was 13 mg (g wet weight sample)−1. The concentrations of NO3−-N in the other samples derived from the roofed wall in the inner gallery ranged from 7.2 to 13 mg (g wet weight sample)−1. If this amount of nitrate ions were present as nitric acid, pH values would reach around 4. However, the pH values in these biofilm samples were from 6.5 to 7.5 (Table 2). This suggested that there was considerable accumulation of nitrate salts, not nitric acid, in the biofilm formed on the roofed wall. In contrast, the concentrations of nitrate ions in the biofilms where the roof were missing were in the range of 0.02 to 0.11 mg (g wet weight sample)−1.
Table 2.
Nitrate ion concentration in the biofilms
| Sample | Color of biofilm | Roofa | pHb | NO3−-N (mg [g wet weight sample]−1) | |
|---|---|---|---|---|---|
| No. 5 | Blue green (BG) | + | 7.0 | 13 | |
| No. 3 |
|
Yellow green | + | 7.5 | 12 |
| No. 4 | + | 6.5 | 7.2 | ||
| No. 7 |
|
Brown beige | + | 6.6 | 13 |
| No. 8 | + | 7.0 | 8.2 | ||
| No. 1 |

|
Purple violet | − | 6.4 | 0.11 |
| No. 2 | − | 7.2 | 0.04 | ||
| No. 10 | − | 6.3 | 0.02 | ||
| No. 9 | Brown green | − | 6.9 | 0.03 |
Samples were collected from the inner or outer gallery in September 2011.
+, Roof indicated in Fig. 1B was present; −, roof was missing.
Biofilms were diluted 1:4 with Milli-Q water and centrifuged; the supernatants were used for pH measurement in the field.
The presence of a roof over the wall can prevent direct exposure to rain and direct sunlight; however, at the same time, the presence of a roof might enhance the accumulation of nitrate ions in the biofilm because the roof would prevent the washing out of nitrate ions from the wall by rainwater. The accumulated nitrate ions could be crystallized in the sandstone, which would accelerate deterioration. Both acidification and bacterial growth will damage to the stone. Therefore, greenish-colored biofilms, including the BG biofilm, likely have the potential to do more severe damage to the stone than biofilms of other colors. Future work will attempt to determine how these microbial communities in each colored biofilm correlate to sandstone deterioration. Additional work will focus on the accumulation of nitrate ions and constructive differences as well as revealing the bacterial communities in upper and lower parts of biofilm.
Conclusions
The bacterial communities detected in the biofilms on this historical sandstone surface in Cambodia differed from those in the air. The biofilms were composed of more unknown bacteria, whereas the airborne bacteria consisted of cultured bacteria grouped mainly in the phylum Proteobacteria and Actinobacteria. Rubrobacter-related bacteria were retrieved especially from the salmon pink biofilms, whereas the black–gray, signal violet, chrome green, and blue–green biofilms were composed mainly of members of the phyla Cyanobacteria and Chloroflexi. Nitrate ion measurement suggested that the blue–green biofilm formed on the parts of the walls where the roof remained was more influential in causing stone deterioration than were the other pigmented biofilms.
Supplementary Information
Acknowledgements
We thank APSARA and the Ministry of the Environment, Kingdom of Cambodia, for permission to collect biofilm samples from the monuments and transport them to Japan. This experiment was performed under the regulation of the Plant Protection Station of the Ministry of Agriculture, Forestry, and Fisheries of Japan. Our research was supported by the Japanese Government Team for Safeguarding Angkor and by a Grant-in-aid for Scientific Research (No. 19251001) from the Ministry of Education, Culture, Sports, and Technology of Japan. We appreciate the advice and technical support given by Hiroshi Kobayashi, Faculty of Life and Environmental Sciences, University of Yamanashi, Takashi Someya, Faculty of Agriculture, Saga University, Ichita Shimoda, Tsukuba University, and Hiroshi Sakon, Yakult Honsha Co., Ltd.
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Billi D, Friedmann EI, Hofer KG, Caiola MG, Ocampo-Friedmann R. Ionizing-radiation resistance in the desiccation-tolerant cyanobacterium Chroococcidiopsis. Appl Environ Microbiol. 2000;66:1489–1492. doi: 10.1128/aem.66.4.1489-1492.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brooks BW, Murray RGE. Nomenclature for “Micrococcus radiodurans” and other radiation-resistant cocci: Deinococcaceae fam. nov. and Deinococcus gen. nov., including five species. Int J Syst Bacteriol. 1981;31:353–360. [Google Scholar]
- 4.Castenholz RW. Clus I. “Chlorofexi”. In: Boone DR, Castenholz RW, Garrity GM, editors. Bergey’s Manual of Systematic Bacteriology. 2nd ed. Vol. 1. Springer-Verlag; New York: 2001. p. 427. [Google Scholar]
- 5.Cole AC, Semmens MJ, LaPara TM. Stratification of activity and bacterial community structure in biofilms grown on membranes transferring oxygen. Appl Environ Microbiol. 2004;70:1982–1989. doi: 10.1128/AEM.70.4.1982-1989.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crispim CA, Gaylarde CC. Cyanobacteria and biodeterioration of cultural heritage: a review. Microb Ecol. 2005;49:1–9. doi: 10.1007/s00248-003-1052-5. [DOI] [PubMed] [Google Scholar]
- 7.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fdz-Polanco F, Méndez E, Urueña MA, Villaverde S, Garcìa PA. Spatial distribution of heterotrophs and nitrifiers in a submerged biofilter for nitrification. Wat Res. 2000;34:4081–4089. [Google Scholar]
- 9.Gaylarde CC, Rodríguez CH, Navarro-Noya YE, Ortega-Morales BO. Microbial biofilms on the sandstone monuments of the Angkor Wat complex, Cambodia. Curr Microbiol. 2012;64:85–92. doi: 10.1007/s00284-011-0034-y. [DOI] [PubMed] [Google Scholar]
- 10.Gu JD. Microbiological deterioration and degradation of synthetic polymeric materials: recent research advances. Int Biodeter Biodegr. 2003;52:69–91. [Google Scholar]
- 11.Hanada S, Hiraishi A, Shimada K, Matsuura K. Chloroflexus aggregans sp. nov., a filamentous phototrophic bacterium which forms dense cell aggregates by active gliding movement. Int J Syst Bacteriol. 1995;45:676–681. doi: 10.1099/00207713-45-4-676. [DOI] [PubMed] [Google Scholar]
- 12.Hanada S, Takaichi S, Matsuura K, Nakamura K. Roseiflexus castenholzii gen. nov., sp. nov., a thermophilic, filamentous, photosynthetic bacterium that lacks chlorosomes. Int J Syst Evol Microbiol. 2002;52:187–193. doi: 10.1099/00207713-52-1-187. [DOI] [PubMed] [Google Scholar]
- 13.Hanada S. Filamentous anoxygenic phototrophs in hot springs. Microbes Environ. 2003;18:51–61. [Google Scholar]
- 14.Hirsch P, Eckhardt FEW, Palmer RJ., Jr Methods for the study of rock-inhabiting microorganisms—a mini review. J Microbiol Meth. 1995;23:143–167. [Google Scholar]
- 15.Imperi F, Caneva G, Cancellieri L, Ricci MA, Sodo A, Visca P. The bacterial aetiology of rosy discoloration of ancient wall paintings. Environ Microbiol. 2007;9:2894–2902. doi: 10.1111/j.1462-2920.2007.01393.x. [DOI] [PubMed] [Google Scholar]
- 16.Iwasaki Y, Tsukuda E, Fukuda M. Geotechnology, geology, and environment. In: Nakagawa T, Fujiki Y, Iwasaki Y, Nishimura M, Uchida E, Narita T, editors. Annual Report on the Technical Survey of Angkor Monument 1995. Japan International Cooperation Center; Tokyo, Japan: 1995. pp. 191–309. (In Japanese) [Google Scholar]
- 17.Iwasaki Y. Geotechnology, geology, and environment survey (2) Water problems in Angkor. In: akagawa TN, Arai H, Iwasaki Y, Uchida E, Sakurada S, Akazawa Y, Shimizu N, Nishimoto S, Ota K, editors. Annual Report on the Technical Survey of Angkor Monument 2003. Japan International Cooperation Center; Tokyo, Japan: 2003. pp. 225–238. [Google Scholar]
- 18.Keppen OI, Tourova TP, Kuznetsov BB, Ivanovsky RN, Gorlenko VM. Proposal of Oscillochloridaceae fam. nov. on the basis of a phylogenetic analysis of the filamentous anoxygenic phototrophic bacteria, and emended description of Oscillochloris and Oscillochloris trichoides in comparison with further new isolates. Int J Syst Evol Microbiol. 2000;50:1529–1537. doi: 10.1099/00207713-50-4-1529. [DOI] [PubMed] [Google Scholar]
- 19.Kusumi A, Li XS, Katayama Y. Mycobacteria isolated from Angkor monument sandstones grow chemolithoautotrophically by oxidizing elemental sulfur. Front Microbiol. 2011 doi: 10.3389/fmicb.2011.00104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laiz L, Miller AZ, Jurado V, Akatova E, Sanchez-Moral S, Gonzalez JM, Dionísio A, Macedo MF, Saiz-Jimenez C. Isolation of five Rubrobacter strains from biodeteriorated monuments. Naturwissenschaften. 2009;96:71–79. doi: 10.1007/s00114-008-0452-2. [DOI] [PubMed] [Google Scholar]
- 21.Lan W, Li H, Wang W-D, Katayama Y, Gu J-D. Microbial community analysis of fresh and old microbial biofilms on Bayon temple sandstone of Angkor Thom, Cambodia. Microb Ecol. 2010;60:105–115. doi: 10.1007/s00248-010-9707-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic Acid Techniques in Bacterial Systematics. Wiley; New York: 1991. pp. 115–175. [Google Scholar]
- 23.Li X, Arai H, Shimoda I, Kuraishi H, Katayama Y. Enumeration of sulfur-oxidizing microorganisms on deteriorating stone of the Angkor monuments, Cambodia. Microbes Environ. 2008;23:293–298. doi: 10.1264/jsme2.me08521. [DOI] [PubMed] [Google Scholar]
- 24.Li XS, Sato T, Ooiwa Y, Kusumi A, Gu J-D, Katayama Y. Oxidation of elemental sulfur by Fusarium solani strain THIF01 harboring endobacterium Bradyrhizobium sp. Microb Ecol. 2010;60:96–104. doi: 10.1007/s00248-010-9699-1. [DOI] [PubMed] [Google Scholar]
- 25.Maxwell JF. Vegetation and vascular flora of the Mekong River, Kratie and Steung Treng Provinces, Cambodia. Maejo Int J Sci Technol. 2009;3:143–211. [Google Scholar]
- 26.McNamara CJ, Perry TD, IV, Bearce KA, Hernandez-Duque G, Mitchell R. Epilithic and endolithic bacterial communities in limestone from a Maya archaeological site. Microb Ecol. 2006;51:51–64. doi: 10.1007/s00248-005-0200-5. [DOI] [PubMed] [Google Scholar]
- 27.McNamara CJ, Konkol N, Mitchell R. Microbial deterioration of cultural heritage materials. In: Mitchell R, Gu JD, editors. Environmental Microbiology. 2nd ed. Wiley-Blackwell Hoboken; New Jersey: 2010. pp. 137–151. [Google Scholar]
- 28.Moseley BEB, Copland HJR. Isolation and properties of a recombination-deficient mutant of Micrococcus radiodurans. J Bacteriol. 1975;121:422–428. doi: 10.1128/jb.121.2.422-428.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer G, de Waal EC, Uitterkinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified gene coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nübel U, Bateson MM, Madigan MT, Kühl M, Ward DM. Diversity and distribution in hypersaline microbial mats of bacteria related to Chloroflexus spp. Appl Environ Microbiol. 2001;67:4365–4371. doi: 10.1128/AEM.67.9.4365-4371.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nugari MP, Realini M, Roccardi A. Contamination of mural paintings by indoor airborne fungal spores. Aerobiologia. 1993;9:131–139. [Google Scholar]
- 32.Okabe S, Hiratia K, Ozawa Y, Watanabe Y. Spatial microbial distributions of nitrifiers and heterotrophs in mixed-population biofilms. Biotechnol Bioeng. 1996;50:24–35. doi: 10.1002/(SICI)1097-0290(19960405)50:1<24::AID-BIT4>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 33.Petushkova J, Kandyba P. Aeromicrobiological studies in the Moscow cathedrals. Aerobiologia. 1999;15:193–201. [Google Scholar]
- 34.Portillo MC, Alloza R, Gonzalez JM. Three different phototrophic microbial communities colonizing a single natural shelter containing prehistoric paintings. Sci Total Environ. 2009;407:4876–4881. doi: 10.1016/j.scitotenv.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 35.Pierson BK, Giovannoni SJ, Stahl DA, Castenholz RW. Heliothrix oregonensis, gen. nov., sp. nov., a phototrophic filamentous gliding bacterium containing bacteriochlorophyll a. Arch Microbiol. 1985;142:164–167. doi: 10.1007/BF00447061. [DOI] [PubMed] [Google Scholar]
- 36.Saarela M, Alakomi H-L, Suihko M-L, Maunuksela L, Raaska L, Mattila-Sandholm T. Heterotrophic microorganisms in air and biofilm samples from Roman catacombs, with special emphasis on actinobacteria and fungi. Int Biodeter Biodegr. 2004;54:27–37. [Google Scholar]
- 37.Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 38.Saiz-Jimenez C. Microbial melanins in stone monuments. Sci Total Environ. 1995;167:273–289. [Google Scholar]
- 39.Sawada M, Matsui T, Inoue S, Ebisawa T. Japanese Government Team for Safeguarding Angkor, editor. Annual Technical Report on the Survey of Angkor Monument 2008. Angkor Project Office; Tokyo, Japan: 2009. 5.1 Weathering test and environmental survey of materials for the conservation and restoration of the bas-relief; pp. 149–154. [Google Scholar]
- 40.Schabereiter-Gurtner C, Piñar G, Vybiral D, Lubitz W, Roölleke S. Rubrobacter-related bacteria associated with rosy discolouration of masonry and lime wall paintings. Arch Microbiol. 2001;176:347–354. doi: 10.1007/s002030100333. [DOI] [PubMed] [Google Scholar]
- 41.Scheerer S, Ortega-Morales O, Gaylarde C. Microbial deterioration of stone monuments—an updated overview. Adv Appl Microbiol. 2009;66:97–139. doi: 10.1016/S0065-2164(08)00805-8. [DOI] [PubMed] [Google Scholar]
- 42.Stewart PS, Camper AK, Handran SD, Huang C-T, Warnecke M. Spatial distribution and coexistence of Klebsiella pneumoniae and Pseudomonas aeruginosa in biofilms. Microb Ecol. 1997;33:2–10. doi: 10.1007/s002489900002. [DOI] [PubMed] [Google Scholar]
- 43.Suihko M-L, Alakomi H-L, Gorbushina A, Fortune I, Marquardt J, Saarela M. Characterization of aerobic bacterial and fungal microbiota on surfaces of historic Scottish monuments. Syst Appl Microbiol. 2007;30:494–508. doi: 10.1016/j.syapm.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang W, Ma Y, Ma X, Wu F, Ma X, An L, Feng H. Diversity and seasonal dynamics of airborne bacteria in the Mogao Grottoes, Dunhuang, China. Aerobiologia. 2012;28:27–38. [Google Scholar]
- 46.Warscheid T, Oelting M, Krumbein WE. Physicochemical aspects of biodeterioration processes on rocks with special regard to organic pollutants. Int Biodeter. 1991;28:37–48. [Google Scholar]
- 47.Warscheid T, Braams J. Biodeterioration of stone: a review. Int Biodeter Biodegr. 2000;46:343–368. [Google Scholar]
- 48.Yamaguchi N, Ishidoshiro A, Yoshida Y, Saika T, Senda S, Nasu M. Development of an adhesive sheet for direct counting of bacteria on solid surfaces. J Microbiol Meth. 2003;53:405–410. doi: 10.1016/s0167-7012(02)00246-4. [DOI] [PubMed] [Google Scholar]
- 49.Yoshinaka T, Yano K, Yamaguchi H. Isolation of a highly radioresistant bacterium, Arthrobacter radiotolerans nov. sp. Agric Biol Chem. 1973;37:2269–2275. [Google Scholar]
- 50.Yuki N, Shimazaki T, Kushiro A, Watanabe K, Uchida K, Yuyama T, Morotomi M. Colonization of the stratified squamous epithelium of the non-secreting area of horse stomach by lactobacilli. Appl Environ Microbiol. 2000;66:5030–5034. doi: 10.1128/aem.66.11.5030-5034.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z, Schwartz S, Wagner L, Miller W. A greedy algorithm for aligning DNA sequences. J Comput Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.