Abstract
The tumor microenvironment plays an important role in regulating cell growth and metastasis. Recently, we developed an ex vivo lung cancer model (4D) that forms perfusable tumor nodules on a lung matrix that mimics human lung cancer histopathology and protease secretion pattern. We compared the gene expression profile (Human OneArray v5 chip) of A549 cells, a human lung cancer cell line, grown in a petri dish (2D), and of the same cells grown in the matrix of our ex vivo model (4D). Furthermore, we obtained gene expression data of A549 cells grown in a petri dish (2D) and matrigel (3D) from a previous study and compared the 3D expression profile with that of 4D. Expression array analysis showed 2,954 genes differentially expressed between 2D and 4D. Gene ontology (GO) analysis showed upregulation of several genes associated with extracellular matrix, polarity, and cell fate and development. Moreover, expression array analysis of 2D versus 3D showed 1006 genes that were most differentially expressed, with only 36 genes (4%) having similar expression patterns as observed between 2D and 4D. Finally, the differential gene expression signature of 4D cells (versus 2D) correlated significantly with poor survival in patients with lung cancer (n = 1,492), while the expression signature of 3D versus 2D correlated with better survival in lung cancer patients with lung cancer. Since patients with larger tumors have a worse rate of survival, the ex vivo 4D model may be a good mimic of natural progression of tumor growth in lung cancer patients.
Keywords: Lung cancer, matrigel, ex vivo 4D model, gene expression profile, survival
Introduction
The overall five-year survival rate for patients diagnosed with lung cancer in 2007 was 16%1. Because most patients with lung cancer present with distant disease and there are few successful treatments for patients with distant disease, overall survival is poor. For patients who would benefit from surgical resection of lung cancer, the major factor that contributes to a patient's survival is the pathologic stage at the time of the resection. For patients with non-small cell lung cancer, TNM staging system is used 2. The T, or tumor stage, is determined by the size, location, and degree of local invasion of the primary tumor. A higher T stage is correlated with greater metastatic disease to the lymph nodes and distant organs and leads to overall poor survival.
Recently, we have developed an ex vivo lung cancer 4D model (previously described as an ex vivo 3D model) that has been shown to produce growing perfusable lung nodules 3 that mimic the tumor growth or T stage of lung cancer in patients. Similar to the human condition, it allows formation of tumor nodules on a lung matrix from a collection of single tumor cells, which grow over time. The 4D model uses a natural matrix, which maintains its homology between species 4 and allows tumor cells from different species to grow in the model. However, the most important aspect of the 4D model is that it has an additional dimension of “continuous flow” in addition to allowing the tumor cells to grow in 3D space. It allows the tumor cells to grow with a constant continuous flow of media through the vascular space, which is separated from the epithelial space by a basement membrane 5. This aspect overcomes the limitations of other 3D models and allows for a more dynamic study of lung cancer growth. When we compared the growth of tumor cells growing in the 4D model to the petri dish (2D), we discovered significant differences in proliferation rates, cell death rates, and matrix metalloproteinase production 6. Moreover, the human lung cancer cells grown in the 4D model produced matrix metalloproteinases that are found in human lung cancer patients, not found from 2D culture 6.
The 4D model may be a better mimic of lung cancer growth than the 2D culture system, but it is unknown if the 4D model is a better mimic of the natural history of lung cancer growth in patients. In this study, we determined the differential gene expression profile between 2D and 4D as well as the differential gene expression profile from 2D and 3D of the A549 lung cancer cell line. We then determined the correlation between the differential gene expression profile and survival in patients with lung cancer. We demonstrated that the differential gene expression profile from the 4D model is correlated with poor survival in lung cancer patients, while the 3D model is correlated with better survival.
Materials and Methods
Animal Handling and Cell Lines
The protocols for animal experiments were approved by the Institutional Animal Care and Use Committee at the Methodist Hospital Research Institute (AUP-0910-0018). All the animal experiments were carried out in accordance with all applicable laws, regulations, guidelines, and policies governing the use of laboratory animals in research.
We used the human alveolar basal epithelial adenocarcinoma cell line A549, which was obtained from American Type Culture Collection (Manassas, VA, USA). The cells were grown in complete media made from RPMI 1640 medium (Hyclone, South Logan, UT, USA) supplemented with 10% fetal bovine serum (Lonza, Walkersville, MD, USA) and antibiotics (100 IU/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin; MP Biomedicals, Solon, OH, USA) at 37°C in 5% CO2. Once cells were 85% confluent, they were washed with phosphate buffered saline (PBS) and subjected to trypsinization using 0.25% trypsin (Cellgro, Manassas, VA, USA) to collect the cells from the flasks. The cells were washed with media and finally suspended in 50 mL of complete media.
4D Model
We created the 4D model (previously described as ex vivo 3D model) as previously described 3. Briefly, we used decellularized lung matrices from 6-week-old rats using 0.1% Sodium Dodecyl Sulfate and 1% Triton-X100. The acellular matrices were thoroughly washed with PBS supplemented with antibiotics (100 IU/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL amphotericin; MP Biomedicals, Solon, OH, USA). Next, we cannulated the trachea and placed 25 million A549 cells diluted in 50 ml of complete media through the trachea of the 4D model (n = 3). We collected the cells with media that passively came out of the model into a 500-mL container, we passed the media again through the trachea three times, and we incubated it at 37°C for 2 hours to allow for cell attachment. We then counted the cells in the container and determined the percentage of tumor cells that were seeded on the 4D model. We placed an additional 200 mL of complete media in the bioreactor previously described 3, and allowed the media to go through the pump at 6 ml per minute and then through the oxygenator and the pulmonary artery of the acellular lung matrix. The 6 ml per minute was used in the experiment since it provides perfusion pressure of 10-15 mm Hg7. This is in the range of mean pulmonary artery pressure in healthy humans of 14 ± 3.3 mm Hg8. Media in the bioreactor was replaced with fresh media every day for 15 days. After 15 days, we removed the tissue of the 4D lung model. We took a picture of the nodule and performed hematoxylin and eosin staining (H&E) as previously described 3 and stored in RNAzol for RNA extraction.
Immunohistochemistry (IHC)
Immunohistochemistry was performed in the Pathology Core Laboratory at The Methodist Hospital Research Institute as described previously6, 9. Briefly, for immunohistochemical analysis, slides were incubated with the primary antibodies overnight at 4°C (E-cadherin (Invitrogen, Camarillo, CA, USA) and CK7 (DAKO, Carpinteria, CA, USA)). Slides were washed thrice with Tris-Buffered NaCl Solution with Tween 20 (Dako), followed by incubation with the secondary antibody linked with horseradish peroxidase (Vector Laboratories, Burlingame, CA, USA) for 20 minutes. Slides were washed again and DAB (Dako) was used to develop the color for 5 minutes, followed by rinsing in distilled water. A counterstain of modified Mayer's Hematoxylin (Dako) was used for 20 seconds, and then the slides were rinsed sequentially with distilled water, 95% alcohol, 100% alcohol, and xylene. A permanent coverslip was mounted on the slide. Stained slides were examined by expert, board-certified pathologists, and images were captured using a microscope (Olympus, Center Valley, PA, USA).
2D Culture
We placed 25 million A549 cells in a 525-cm2 cell culture flask (BD) with 200 ml of complete media (n=3). We took a picture of the cell growing in the culture flask. We replaced the same amount of fresh media every day as media for the 4D model. After 15 days, the cells were trypsinized and fraction of pellet stored in RNAzol for RNA extraction and rest for protein analysis.
RNA Extraction and Quantification
Total RNA was extracted from monolayer cell cultures as well as recellularized lung matrix with Isol-RNA Lysis reagent (5 PRIME, Gaithersburg, MD, USA), followed by a SurePrep RNA cleanup and concentration kit (Fisher Scientific, Pittsburgh, PA, USA). RNA was treated with Ambion DNA-free kit (Applied Biosystems, Carlsbad, CA, USA) as per the manufacturer's instructions. RNA quality and quantity was determined using Nanodrop 1000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). Approximately 2 to 10 μg of RNA was overnight shipped to Phalanx Biotech service center, Belmont, California, for expression array. It was further processed for a quality check using Agilent RNA 6000 Nano assay to determine RIN score. RNA Samples with RIN score above 7 were used for further assay.
Expression Array of 2D vs 4D
The human genome expression array was carried out at the Phalanx Biotech Service Center (Belmont, CA, USA) using Human OneArray v5 chip (version HOA 6.1). The cDNA microarray consisted of 32,678 oligonucleotide probes, representing 19,118 unique, named human genes. Each group consisted of biological triplicate samples, each of which was processed in triplicate. Per the manufacturer's instructions, the target RNA was labeled with Cye5 dye and an OneArray slide is pre-hybridized to reduce the background signals and increase the performance of the microarray. Ten μg of dye-labeled RNA was further hybridized with an OneArray slide in a humidified oven with 2X SSPE buffer at 50°C for 14 to 16 hours. Following hybridization, microarray slides were washed and dried. Finally, the slides were scanned using Axon GenePix 4100 (Molecular Devices, CA, USA) at wavelengths 635 to 532 nm. The filtered data were log2 transformed and corrected using quantile normalization before their average ratios and significance values were calculated.
Quantitative PCR Validation
cDNA was prepared using a high-capacity cDNA Reverse Transcription kit (Applied Biosystems, NY, USA) with 200 ng of total RNA and real-time PCR assay was performed with SYBR green reagent (Applied Biosystems, NY, USA). We have used the published 18S rRNA as a housekeeping gene 10 and primers were designed for other target genes – COL4A1 (forward primer (F) 5′ GGCTCAAAGGTGACAAAGGA 3′, reverse primer (R) 5′ AGCAGGACCATATCCTG GAG 3′; 221 base pair (bp)), SOX2 (F 5′ AACCCCAAGATGCACAACTC 3′, R 5′ CGGGGCCGGTATTTATAATC 3′; 152 bp), E-cadherin (CDH1; F 5′ CTGGGTTATTCCTCCCATCA 3′, R 5′ GTGTATGTGGCAATGCGTTC 3′; 222 bp), N-cadherin (CDH2; F 5′ GTCAGCTGTCGGTGACAAAG 3′, R 5′ TGATCCCTCAGGAACTGTCC 3′; 191 bp), VIM (F 5′ GCAGGCTCAGATTCA GGAAC 3′; R 5′ GTGAGGGACTGCACCTGTCT 3′ ; 246 bp)11 and IL6 (F 5′ GGTCAGAAACCTGTCCACTG 3′; R 5′ CAAGAAATGATCTGGCT CTG 3′; 257 bp)12. A relative fold of gene expression was calculated using 2-(ΔΔCt) formula.
Western Analysis
Cells from monolayer culture and matrix tissues were homogenized on ice in a RIPA buffer (0.15 M NaCl, 1% deoxycholate sodium salt, 1% Triton X-100, 0.1% SDS, and 0.01 M Tris-HCl (pH 7.2)) containing complete protease inhibitor (Roche, Indianapolis, IN, USA). The cell lysate was centrifuged at 14,000 g for 15 min at 4°C and supernatant was collected for further analysis. Bio-Rad protein assay reagents were used to determine the protein concentration. Fifty μg of protein sample was mixed with sample buffer and separated on pre-set 4%-15% SDS-PAGE gels (Bio-Rad Life Science, Hercules, CA, USA) and transferred to nitrocellulose membrane (Bio-Rad, CA, USA) using semi-dry blotting system (Bio-Rad, CA, USA). Nitrocellulose membrane was blocked in 5% milk (Bio-Rad, CA, USA) for 1 hour at room temperature and incubated with primary antibodies against β-actin (1:1000), (1:500), (1:500), and Vimentin (VIM, 1:300) overnight at 4°C. The membrane was washed and incubated for 30 min at room temperature in a 1:2000 dilution of HRP-conjugated anti-mouse secondary antibodies (Santa Cruz Biotechnology, Dallas, TX, USA). The membrane was developed using the Lumi-Light Western blotting kit (Roche) that contains the chemiluminescent HR substrate, and exposed to X-ray film (Phenix Research Products, Candler, NC, USA). Furthermore, the band intensities were quantified using ImageJ 1.46r software (National Institutes of Health, Bethesda, MD, USA).
Statistical Analysis
Gene array data were processed and quantile normalized using Bioconductor. Array data have been deposited into the Gene Expression Omnibus (GEO GSE45142). Technical replicates were averaged together, and two-sided t-test and fold change (using log-transformed data) were used to compare gene expression between the 4D model and 2D cell cultures of the A549 lung cancer cell. Top genes were selected with < 0.5-fold or > 2-fold change at p-value less than 0.01. We used the method of Storey et al. 13 to estimate the false discovery rate from multiple hypothesis testing; of the ∼32,000 array probes in the entire dataset, 6,894 were nominally significant, with nominal p < 0.01 (no fold criteria), which yielded a false discovery rate of < 5%, indicating that the vast majority of changes would not be due to chance expected. Gene ontology analysis was performed using SigTerms 14 (Supplemental data file 1). JavaTreeview was used to visualize the expression patterns as heat maps 15. We also compared the gene expression between matrigel (3D) model and petri dish (2D) cell culture, using data obtained from GEO (Accession number GSE17347)16.
In order to examine the 4D and 3D gene signatures in human lung tumors, we assembled a “compendium” dataset of 11 published expression profiling datasets for human lung adenocarcinomas (n = 1,492 tumors), for which survival data (time to death) were available 17-27. Genes within each dataset were first normalized to standard deviations from the median; where multiple human array probe sets referenced the same gene, the probe set with the highest variation was used to represent the gene. In order to score each human lung tumor within the compendium dataset, for similarity to an experimentally derived gene signature of interest, we derived a “t-score” metric for each human tumor in relation to the experimental signature, similar to what we have done in previous analyses 28, 29; briefly, the t-score was defined for each external profile as the two-sided t-statistic comparing, within the profile, the average of the genes high in the signature with the average of the genes low in the signature. The significance of survival associations identified (using univariate Cox and log-rank tests) were further verified using simulations involving 1000 random gene signatures; for the 4D signature, no random signature had a univariate Cox significance level that exceeded that of the actual signature.
Standard t-test was performed to determine the difference between the relative expression of RT-PCR and relative intensity of Western blot analysis with p-value < 0.05 was considered significant.
Results
2D vs 4D
There was no formation of nodules in the 2D model (Fig 1A). In the 4D model, 97% (±1.9) of the A549 cells were seeded on the acellular scaffold. It formed a nodule on day 2 and the nodule grew over time on the 4D model (Fig 1B). The H&E of the nodule showed cell-cell and cell-matrix interaction on high power (Fig 1C) confirmed by cytokeratin (CK)-7 (Fig 1D) and E-cadherin (Fig 1E) immunohistochemistry staining.
Figure 1.
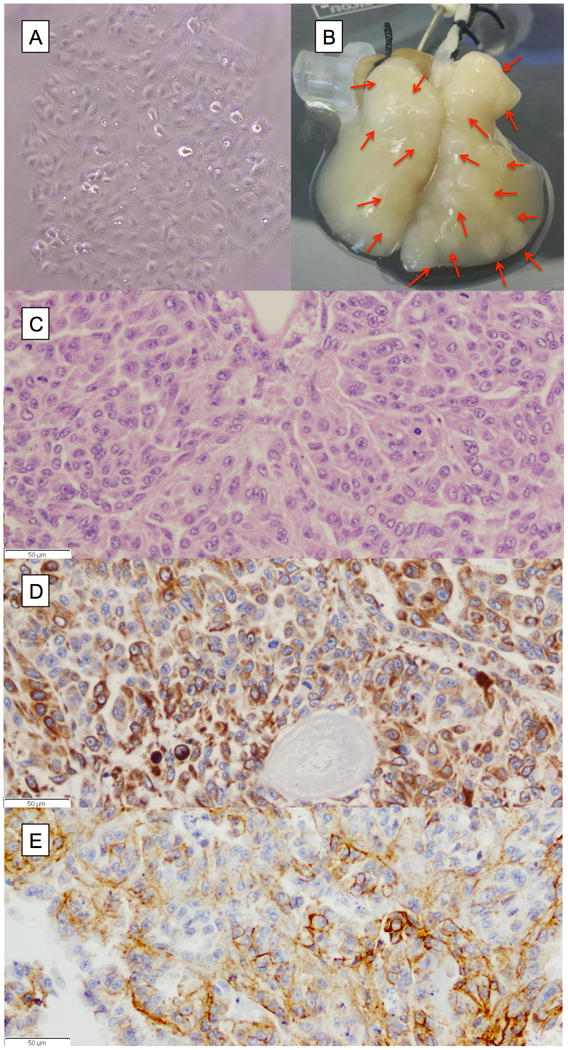
A549 cells grown on tissue of the 4D model and the 2D flask. (A) A549 cells growing in a culture flask in 2D fashion. (B) Photograph of tissue of the 4D model with multiple tumor nodules (red arrows). (C) The H&E stain (40X) showing cell-cell and cell-matrix interaction of cells growing in the 4D model supported by IHC staining of the cells with CK7 (D) and E-cadherin (E).
Gene Expression between 2D vs 4D
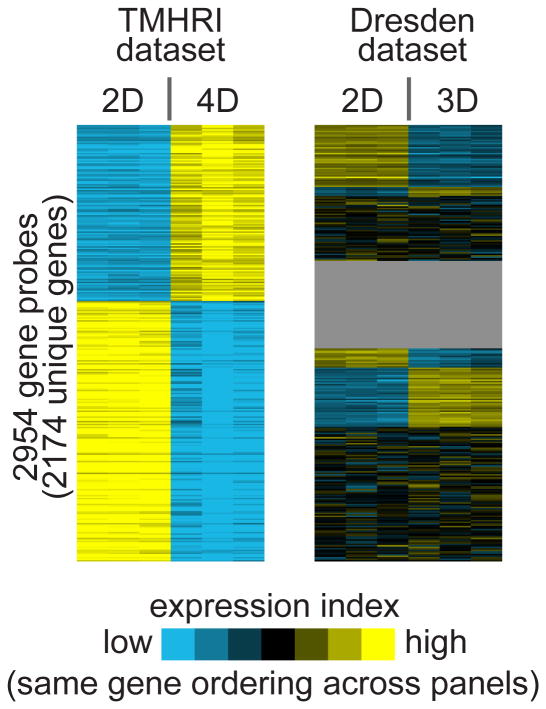
We compared the genome wide gene expression of growing A549 cells between our 4D model and monolayer 2D cultures, and found 2,954 genes probes significantly expressed with 2,174 unique genes. Out of the 2,954 gene probes, 1,194 gene probes were up-regulated and 1,760 gene probes were down-regulated in the 4D cultures (The Methodist Hospital Research Institute (TMHRI) dataset) (Fig 2). The global gene expression differences between 2D and 4D were therefore widespread, far exceeding chance expected (see Methods).
Figure 2.
Differential gene expression patterns associated with the 4D model. Heat map showing the top differential genes of the 2D vs the 4D model (TMHRI dataset), along with the corresponding patterns as observed in another profile dataset of the 2D vs the 3D model (Dresden dataset). Each row of the heat map represents a gene and each column represents a profiled sample. Relative gene expression is represented by a colorgram (yellow: high, blue: low). The ordering of the genes is the same between the two datasets. In the TMHRI dataset, comparison of the gene expression profiles of A549 cells grown in a petri dish (2D) and our ex vivo lung model (4D) defines the 2,954 unique gene probes differentially expressed (p < 0.01 and fold > 2). In the Dresden dataset, gene expression profiles were compared between A549 cells gown in a petri dish (2D) and A549 cells grown in a matrigel 3D culture system (gray, genes not represented).
Gene Ontology
Further gene ontology analysis of up-regulated genes in the 4D model showed that most of the genes belong to cell-matrix interaction (collagen type IV, collagen type V, and collagen fibril organization), cell-cell interaction (synapse assembly, actin binding, actomyosin structure organization, sarcomere, actin filament, actin cytoskeleton, and cell junction), polarity (positive regulation of the Wnt receptor signaling pathway, proximal/distal pattern formation, Notch binding, positive regulation of the Notch signaling pathway, basolateral plasma membrane, and apicolateral plasma membrane), migration (ectoderm and mesoderm interaction, positive regulation of filopodium assembly, and cell projection), tissue formation (organ regeneration, prostatic bud formation, and tissue regeneration) (Table 1). Most of the genes down-regulated in the 4D model belong to immune regulation (innate immune response, positive regulation of NF-κB transcription factor activity, positive regulation of chemokine production, immune response and complement activation, classic pathway) (Table 1).
Table 1. Selected Go Pathways for gene expression difference between A549 grown in 4D and 2D.
| No. of gene changed | p-value | z-score | |
|---|---|---|---|
| Up-regulated in 4D compared to 2D | |||
| Cell-Matrix Interaction | |||
| Collagen type IV | 3 | 0.0018 | 5.28 |
| Collagen type V | 2 | 0.0063 | 5.11 |
| Collagen fibril organization | 6 | 0.0010 | 4.47 |
| Cell-Cell Interaction | |||
| Synapse assembly | 7 | 0.0007 | 4.53 |
| Actin binding | 27 | 0.0002 | 4.04 |
| Actomyosin structure organization | 3 | 0.0018 | 5.28 |
| Sarcomere | 5 | 0.0074 | 3.43 |
| Actin filament | 5 | 0.0086 | 3.32 |
| Actin cytoskeleton | 13 | 0.0096 | 2.79 |
| Cell junction | 30 | 0.0044 | 2.94 |
| Polarity | |||
| Positive regulation of Wnt receptor signaling pathway | 4 | 0.0008 | 5.31 |
| Proximal/distal pattern formation | 5 | 0.0029 | 4.03 |
| Notch binding | 3 | 0.0068 | 4.09 |
| Positive regulation of Notch signaling pathway | 3 | 0.0030 | 4.80 |
| Basolateral plasma membrane | 11 | 0.0109 | 2.78 |
| Apicolateral plasma membrane | 2 | 0.0121 | 4.31 |
| Migration | |||
| Ectoderm and mesoderm interaction | 2 | 0.0022 | 6.41 |
| Positive regulation of filopodium assembly | 3 | 0.0030 | 4.80 |
| Cell projection | 11 | 0.0042 | 3.26 |
| Tissue formation | |||
| Organ regeneration | 6 | 0.0050 | 3.52 |
| Prostatic bud formation | 2 | 0.0063 | 5.11 |
| Tissue regeneration | 4 | 0.0123 | 3.26 |
| Down-regulated in 4D compared to 2D | |||
| Immune response | |||
| Innate immune response | 19 | 0.0001 | 4.40 |
| Positive regulation of NF-kappaB transcription factor activity | 11 | 0.0003 | 4.51 |
| Positive regulation of chemokine production | 5 | 0.0012 | 4.52 |
| Immune response | 35 | 0.0016 | 3.28 |
| Complement activation, classical pathway | 10 | 6.18E-06 | 6.22 |
Validation of Microarray Data by RT-PCR
The microarray data of gene expression between 2D and 4D was validated by RT-PCR for selected genes: collagen type IV alpha 1 (COL4A1), SRY (sex determining region Y)-box 2 (SOX2), interleukin 6 (IL6), CDH1, CDH2 and vimentin (VIM). We found that there were significantly higher levels of SOX2 (p < 0.001), COL4A1 (p = 0.008), CDH2 (p = 0.003), VIM (p = 0.03) in the 4D compared to 2D. Moreover, we found that there were significantly lower levels of IL6 (p = 0.002) and CDH1 (p = 0.02) (Fig 3). All of the genes validated the gene expression found between 2D and 4D. Both CDH1 and VIM had a similar pattern of expression in both gene expression array and RT-PCR, however, there was no significant difference in the gene expression analysis, while there was significant difference in RT-PCR due to the difference in the cutoff for significance of gene expression array of p < 0.01, while for the RT-PCR, we used p < 0.05.
Figure 3.
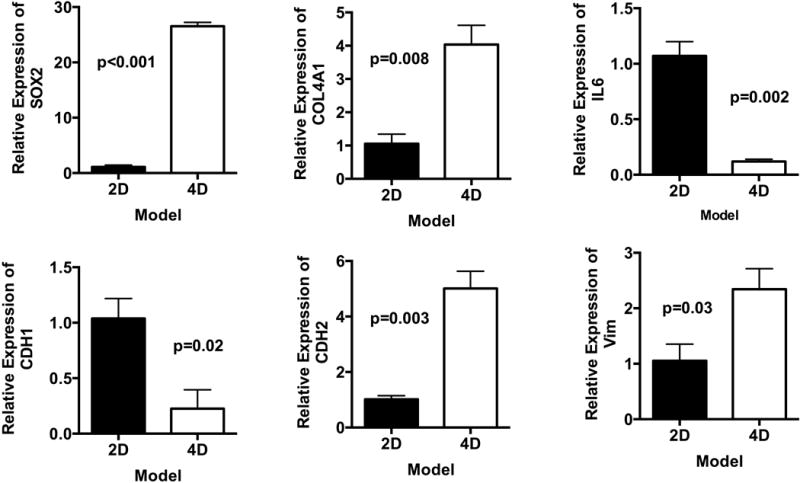
Validation of microarray data with qPCR. Relative gene expression of SOX2, COL4A1, IL-6, CDH1, CDH2 and VIM in A549 cells grown on our ex vivo 4D lung model (n=3) and 2D flask culture (n=3) on day 15. We have found significantly higher gene expression of SOX2, COL4A1, CDH2, and VIM in a 4D cell culture as compared to a 2D cell culture. We found significantly lower levels of IL-6 and CDH1 in the 4D models than in the 2D model. Error bar represents standard error of the mean.
Validation of Microarray Data by Western Blot Analysis
We performed Western Blot analysis on CDH1, CDH2, and VIM. We found that there was a good correlation between the protein level and the gene expression data except for that in CDH1 (Fig 4). Both CDH2 (p = 0.03) and VIM (p = 0.01) were significantly higher in the 4D model than in the 2D model. There was no significant difference in the CDH1 level between the 2D and 4D models due to one of the three samples in the 4D model having a higher expression of CDH1. That sample also had a significantly higher level of CDH2; thus we analyzed the CDH2 to CDH1 ratio, which showed trend toward significance (p = 0.06) between the 4D and 2D models.
Figure 4.
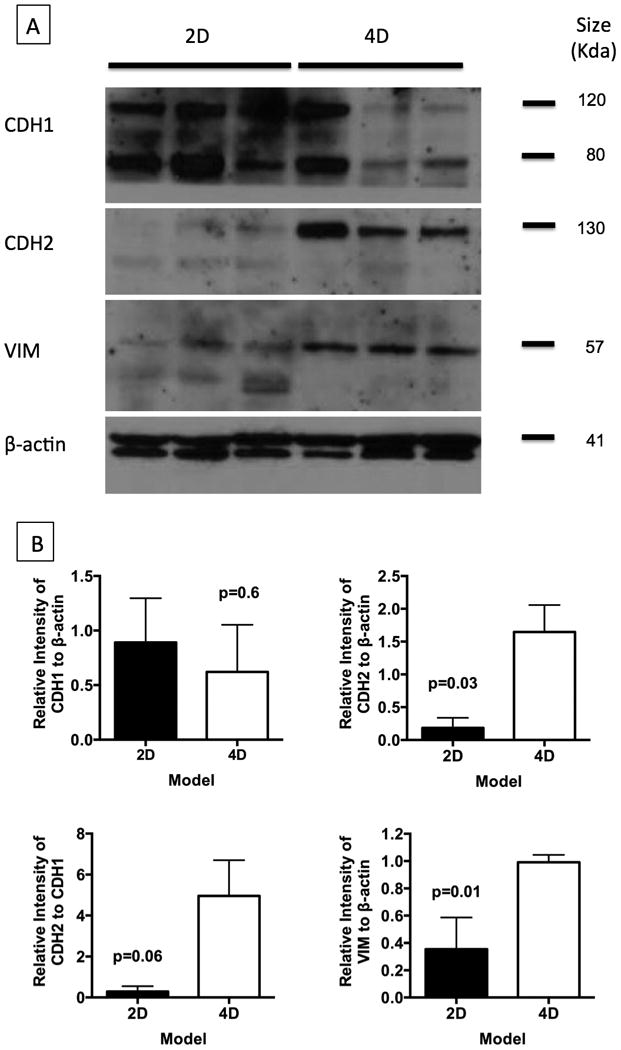
Validation of selected genes at protein level using Western blot analysis. (A) We have validated the CDH1 (80 & 120Kda), CDH2 (130KDa), and VIM (57KDa) genes, as they play an important role in tumor progression and metastasis. We used β-actin as the reference protein to compare the protein expression between the 2D (n = 3) and 4D (n = 3) models. (B) Western blots were further analyzed using imageJ software to normalize the protein expression of genes against β-actin. We found significantly higher levels of CDH2 and VIM in 4D tissues while there was no difference in CDH1 levels. It is well known that the CDH2/CDH1 ratio plays an important role epithelial to mesenchymal transition and tumor metastasis; therefore, we also calculated this value and found a trend of higher CDH2/CDH1 ratio in the 4D model than in the 2D model (p = 0.06). Error bar represents standard error of the mean.
Comparison between the Differential Expression Patterns of 3D versus 4D
Interestingly, the genes we identified as differentially expressed in our ex vivo 4D model, overall, did not show corresponding changes in a 3D matrigel model, based on analysis of public gene array data (Fig 2). For comparison with our data, we obtained gene expression data of A549 cells grown on petri dish (2D) and matrigel (3D) from a previous study by Zschenker et al. 16 (referred to here as the “Dresden” dataset). In this Dresden dataset, we identified 1006 top differential genes by our criteria (p < 0.01 with fold change > 2 in either direction); of these genes, 536 were up-regulated and 473 were down-regulated in the 3D versus 2D A549 cells (three genes being represented in both sets by different probes). However, among the Dresden 1006 unique genes, only 36 genes had similar expression patterns as the genes that were differentially expressed between 4D and 2D in our system (i.e. 36 Dresden genes were found within our top 4D genes, and were moving in an analogous direction). There were 10 genes that were up-regulated in both (GALNT5, COL4A6, EIF5, EPHA7, GRIK2, C13orfl5, MED31, ABCC8, KIAA1841, DOCK4) and 26 genes that were down-regulated in both (EDIL3, DHRS9, TRIOBP, ANXA10, CCDC85A, ANKRD22, LETM2, SGMS2, EMR1, AIM1, CLN8, FBN2, NID2, SYT11, HLX, AOX1, KCNS3, NOS1, PLAT, PMCH, XAF1, PAQR5, SNCA, WNT7B, CSDA, B3GALNT1, Supplement File 2). In fact, for the genes up-regulated in 4D in our data, many of these were down-regulated in the 3D cells in the Dresden data and vice versa (Fig 2).
Survival
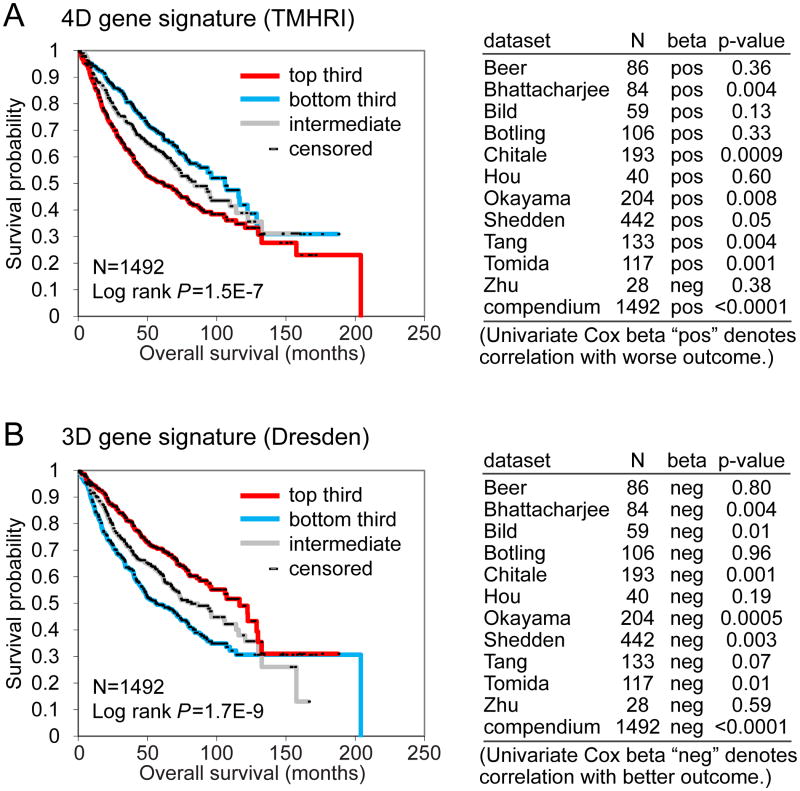
As the influence of the tumor microenvironment is thought to be a factor in tumor metastasis, we posited that our 4D transcriptomic signature would have prognostic value in patients, i.e. those human cancers that manifested a transcriptional program associated with microenvironment response would have a more aggressive phenotype. To investigate this question, we collected 11 independent cohorts of lung adenocarcinoma patients for which both gene expression and clinical outcome data were publically available 17-27. These tumor profiles (n = 1,492) were scored, based on the manifestation of our 4D gene signature (consisting of 2,174 unique human genes). We observed a significant correlation between the 4D signature and reduced survival duration, which reached significance in six of the individual cohorts and as a “compendium” of all 11 cohorts (Fig 5A, Log-rank p = 1.5E-7). Somewhat surprisingly, using the same analytical approach, the Dresden 3D signature (consisting of the top 1006 genes, p = 0.01, fold > 2) was significantly associated with better survival in the same compendium dataset (Fig 5B, Log-rank p = 1.7E-9), likely owing in part to the lack of overall concordance observed between the two signatures (Fig 2).
Figure 5.
Associations of 4D and 3D differential gene signatures with lung cancer patient survival. Gene signatures of both the 4D model system (TMHRI data) and the 3D culture (Dresden data) were probed in a large compendium of human lung tumor profile datasets (n = 1492). 4D gene signatures were associated with poor overall survival in patients with lung cancer, while 3D gene signatures were associated with better overall survival. (A) Survival analysis of lung cancer patients, comparing the differences in risk between tumors, according to the degree of manifestation of the TMHRI 4D gene signature. Kaplan-Meier plot compares top third (“strong manifestation”), bottom third (“weak manifestation”), and middle third (“intermediate”); table shows correlations by univariate Cox, for each individual array dataset examined, as well as for a “compendium” of all available datasets (featured in Kaplan-Meier plot). The top third manifestation (“strong manifestation”, red) of TMHRI 4D gene signature correlated with worse survival compared to the bottom third manifestation (“weak manifestation”, blue). (B) Same as with part A, but using the Dresden 3D signature. The top third manifestation (“strong manifestation”, red) of Dresden 3D gene signature correlated with better survival compared to the bottom third (“weak manifestation”, blue).
Discussion
Context plays an important role in cell behavior and function. A cell grown on a petri dish often grows in a 2D space with some cell-cell and cell-plastic interaction. This environment is very different from the environment of a tumor cell growing in a patient. This large discrepancy has recently been bridged with 3D culture systems 30, 31, which have grown the tumor cells in a natural and artificial matrix. The study of the tumor cells in this context have shown the importance of cell-matrix and cell-cell interaction in tumor growth and studies suggest that this is a better mimic of the natural condition than the cells grown on a petri dish. The major drawback of the 3D system is that it lacks vasculature. The tumors are grown in a matrix that is similar to the tumor growth in patients with lung cancer but it is different from the human condition due to the lack of flow of nutrients and removal of waste through the system. Recently, we developed a 4D model that had an additional dimension of flow to the 3D system 3, which is a better mimic of the natural condition than the cell growth on a 2D system.
The advantage of the 4D model is that it allows the tumor cells to form a perfusable nodule that grows over time. This phenotype is typically seen in patients with lung cancer. When a patient's lung cancer is observed over time, it grows in size. The size of the tumor has an important implication in the prognosis of the patient, since it is directly related to the metastatic lesions to the lymph nodes and other organs. The patients with small primary lung cancer rarely have metastatic lesions, while patients with large primary lung cancer often have metastatic disease. This may explain the reason for the differential expression between the 4D and 2D models, which compare a gene expression of a group of single cells (2D) to a large perfusable nodule (4D), correlates to poor survival.
When we analyzed the genes that were differentially expressed, we found the genes that were important in processing the extracellular matrix, such as matrix metalloproteinases as well as the production of different collagens, polarity and cell fate, and development. The most likely reason for the differential gene expression predicting poor survival is that the 4D culture may have more cells with mesenchymal features than the 2D culture. The analysis shows more up-regulation of the CDH2 and VIM in the 4D culture than the 2D culture. Since the hypothesis of tumor growth and metastasis is that as tumors form nodules, some cells break cell-cell and cell-matrix interaction and enter the circulation, going from an epithelial cell to a mesenchymal cell 32. The tumor cell in the mesenchymal phase moves in the lymphatic or vascular circulation and then enters the distant lymph node or organ and forms a metastatic disease. Thus, the 4D model may allow for tumor cells to undergo epithelial to mesenchymal transition.
The formation of this sub-population of cells that are undergoing epithelial to mesenchymal transition may explain the presence of contradictory markers in the 4D model. The GO analysis shows the presence of markers that enhance polarization such as cell junction markers and apical and basal membrane markers and markers that decrease polarization such as mesenchymal markers. The degree of the two sub-populations may account for the variation in the protein concentration of the E-cadherin in the 4D samples. The sample with higher concentration of polarized epithelial cells will likely have higher concentration of E-cadherin compared to a sample with higher concentration of less polarized mesenchymal cells which will have lower concentration of E-cadherin.
On the other hand, the differential gene expression between the 3D and 2D models shows the difference in expression due to the presence of the matrix without the vasculature. These cells grow without forming perfusable nodules. In an environment in which a tumor cell does not have an opportunity to break the cell-cell and cell-matrix interaction to enter a separate vascular space, there is no driver for the cells to undergo an epithelial to mesenchymal transition. Thus, the gene expression profile did not show any difference in the production of CDH1, CDH2, and VIM between the 3D and 2D conditions. Zschenker et al. 16, found 376 differentially expressed transcripts, using different cutoff for significant gene expression compared to our analysis, where they used the 3D extracellular matrix (ECM) scaffold for the 3D cell culture of the same A549 cell lines. Despite the difference in the expression pattern, the genes that were differentially expressed did predict a better survival in patients with lung cancer. The good prognostic indicator may be that the tumor cells grown on the matrigel may express genes that keep the cells in the epithelial phenotype, which may be less metastatic, thus providing better overall survival with this gene signature. This is supported by the fact that growing mouse lung cancer cells in matrigel induces expression of CDH1 and β-catenin, which are both epithelial markers 33. When human non-malignant normal bronchial cells and aggressive human lung cancer cell lines were plated on 3D lamina rich ECM (3D lrECM), there is a significant difference in the morphology of these tumor cells. In this 3D system, the non-malignant normal bronchial cells formed smooth spheroid agglomerates, while aggressive lung cancer cell lines formed a branched morphology 34. When A549 cells were grown in the 3D model, the cells formed a cluster of smooth cells, and the gene expression cell lines with smooth structure had a gene expression profile that had significantly improved survival compared to other lung cancer cell lines that formed a branch structure 35. The 3D model may stabilize the A549 cells to have similar morphology as non-malignant normal bronchial cells and provide overall better survival in analysis of patients with lung cancer.
In addition to the difference in the presence of vasculature in the 4D model compared to the 3D model, there are significant differences in the constitution of the matrix. The 3D culture system is formed by laminin-rich extracellular matrix (Matrigel) which is composed of primarily laminin, entactin and collagen IV36. The acellular lung matrix is composed of collagen I, collagen IV, laminin, fibronectin and elastin37. Differences in the composition of the matrix may have also led to differences in activity of tumor cells. However, since the matrix in the 4D model is derived from a natural source, the biology of the tumor cells grown in the 4D model may be a better mimic of the condition in patients with lung cancer compared to the artificial matrix of the 3D model.
There were 36 genes that had fold differences that were in similar direction for both 4D and 3D compared to 2D growth in gene expression analysis. Among the 36 genes, there were 10 genes that were up-regulated in both 4D and 3D compared to 2D. Two of those genes are related to cell-matrix and cell-cell interactions. In both 3D and 4D conditions, the collagen, type IV, alpha 6 (COL4A6) production was significantly higher compared to the 2D condition. Collagen type IV is an important structural element of basement membrane and regulator of smooth muscle differentiation38. Furthermore, in both 3D and 4D conditions, dedicator of cytokinesis 4 (DOCK4) was significantly elevated compared to the 2D condition. The DOCK4 produces proteins that are important in formation of cell-cell interaction39. The up-regulation of these genes is likely the result of presence of matrix in both conditions.
Further study needs to be done to elucidate how the environment plays a role in the gene expression of tumor cells. For example, future studies could include addition of other cellular components to the 4D model such as fibroblasts, monocytes, lymphocytes and endothelial cells, to determine if that leads to differential gene expression and differential tumor growth that may improve the current 4D model. Our study shows that the A549 cells grown on the 3D system predicts better survival in patients with lung cancer, while the same cells grown on the 4D system predict poor survival. This may be due to the 4D system with the addition of “continuous flow” to the matrix allowing the cells to undergo epithelial to mesenchymal transition and become more invasive. Moreover, use of natural matrix may allow the tumor cells to better mimic the activity in patients with lung cancer. Since lung cancer cells in patients form nodules and form metastatic lesions, our 4D model may be a good mimic of the tumor growth in patients with lung cancer.
Supplementary Material
Brief Description.
Tumor microenvironment plays an important role in tumor growth. Our 4D model with natural matrix and continuous flow of media leads to formation of tumor nodules and change in gene expression in A549 cells that correlates with poor survival in lung cancer patients. Since patients with larger tumors have a worse rate of survival, the ex vivo 4D model may be a good mimic of natural progression of tumor growth.
Acknowledgments
This work was supported in part by Cancer Prevention Research Institute of Texas Multi-investigator Research Award RP120713 (CJC) and P30 CA125123 (CJC). We thank Maria Kim for editing the manuscript.
Footnotes
Conflict of Interest: MPK has submitted a patent in regards to the 4D model.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA: a cancer journal for clinicians. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P. Staging manual in thoracic oncologyed. Orange Park, FL: Editorial Rx Press; 2009. International Association for the Study of Lung Cancer. [Google Scholar]
- 3.Mishra DK, Thrall MJ, Baird BN, Ott HC, Blackmon SH, Kurie JM, Kim MP. Human lung cancer cells grown on acellular rat lung matrix create perfusable tumor nodules. The Annals of thoracic surgery. 2012;93:1075–81. doi: 10.1016/j.athoracsur.2012.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kuttan R, Spall RD, Duhamel RC, Sipes IG, Meezan E, Brendel K. Preparation and composition of alveolar extracellular matrix and incorporated basement membrane. Lung. 1981;159:333–45. doi: 10.1007/BF02713933. [DOI] [PubMed] [Google Scholar]
- 5.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 6.Mishra DK, Sakamoto JH, Thrall MJ, Baird BN, Blackmon SH, Ferrari M, Kurie JM, Kim MP. Human lung cancer cells grown in an ex vivo 3D lung model produce matrix metalloproteinases not produced in 2D culture. PLoS One. 2012;7:e45308. doi: 10.1371/journal.pone.0045308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ott HC, Clippinger B, Conrad C, Schuetz C, Pomerantseva I, Ikonomou L, Kotton D, Vacanti JP. Regeneration and orthotopic transplantation of a bioartificial lung. Nat Med. 2010;16:927–33. doi: 10.1038/nm.2193. [DOI] [PubMed] [Google Scholar]
- 8.Kovacs G, Berghold A, Scheidl S, Olschewski H. Pulmonary arterial pressure during rest and exercise in healthy subjects: a systematic review. Eur Respir J. 2009;34:888–94. doi: 10.1183/09031936.00145608. [DOI] [PubMed] [Google Scholar]
- 9.Kiernan J. Histological and Histochemical Methods: Theory and Practice. 3rd. Oxford [U.K.]: 2008. p. 502. [Google Scholar]
- 10.Ohira T. WNT7a induces E-cadherin in lung cancer cells. Proceedings of the National Academy of Sciences. 2003;100:10429–34. doi: 10.1073/pnas.1734137100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JA. Primer3Plus, an enhanced web interface to Primer3. Nucleic acids research. 2007;35:W71–4. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y, Taveggia C, Melendez-Vasquez C, Einheber S, Raine CS, Salzer JL, Brosnan CF, John GR. Interleukin-11 potentiates oligodendrocyte survival and maturation, and myelin formation. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2006;26:12174–85. doi: 10.1523/JNEUROSCI.2289-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Creighton CJ, Nagaraja AK, Hanash SM, Matzuk MM, Gunaratne PH. A bioinformatics tool for linking gene expression profiling results with public databases of microRNA target predictions. RNA. 2008;14:2290–6. doi: 10.1261/rna.1188208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saldanha AJ. Java Treeview--extensible visualization of microarray data. Bioinformatics. 2004;20:3246–8. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 16.Zschenker O, Streichert T, Hehlgans S, Cordes N. Genome-wide gene expression analysis in cancer cells reveals 3D growth to affect ECM and processes associated with cell adhesion but not DNA repair. PloS one. 2012;7:e34279. doi: 10.1371/journal.pone.0034279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tomida S, Takeuchi T, Shimada Y, Arima C, Matsuo K, Mitsudomi T, Yatabe Y, Takahashi T. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:2793–9. doi: 10.1200/JCO.2008.19.7053. [DOI] [PubMed] [Google Scholar]
- 18.Chitale D, Gong Y, Taylor BS, Broderick S, Brennan C, Somwar R, Golas B, Wang L, Motoi N, Szoke J, Reinersman JM, Major J, et al. An integrated genomic analysis of lung cancer reveals loss of DUSP4 in EGFR-mutant tumors. Oncogene. 2009;28:2773–83. doi: 10.1038/onc.2009.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shedden K, Taylor JM, Enkemann SA, Tsao MS, Yeatman TJ, Gerald WL, Eschrich S, Jurisica I, Giordano TJ, Misek DE, Chang AC, Zhu CQ, et al. Gene expression-based survival prediction in lung adenocarcinoma: a multi-site, blinded validation study. Nature medicine. 2008;14:822–7. doi: 10.1038/nm.1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhattacharjee A, Richards WG, Staunton J, Li C, Monti S, Vasa P, Ladd C, Beheshti J, Bueno R, Gillette M, Loda M, Weber G, et al. Classification of human lung carcinomas by mRNA expression profiling reveals distinct adenocarcinoma subclasses. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:13790–5. doi: 10.1073/pnas.191502998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beer DG, Kardia SL, Huang CC, Giordano TJ, Levin AM, Misek DE, Lin L, Chen G, Gharib TG, Thomas DG, Lizyness ML, Kuick R, et al. Gene-expression profiles predict survival of patients with lung adenocarcinoma. Nature medicine. 2002;8:816–24. doi: 10.1038/nm733. [DOI] [PubMed] [Google Scholar]
- 22.Zhu CQ, Ding K, Strumpf D, Weir BA, Meyerson M, Pennell N, Thomas RK, Naoki K, Ladd-Acosta C, Liu N, Pintilie M, Der S, et al. Prognostic and predictive gene signature for adjuvant chemotherapy in resected non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:4417–24. doi: 10.1200/JCO.2009.26.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bild AH, Yao G, Chang JT, Wang Q, Potti A, Chasse D, Joshi MB, Harpole D, Lancaster JM, Berchuck A, Olson JA, Jr, Marks JR, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–7. doi: 10.1038/nature04296. [DOI] [PubMed] [Google Scholar]
- 24.Tang H, Xiao G, Behrens C, Schiller J, Allen J, Chow CW, Suraokar M, Corvalan A, Mao J, White MA, Wistuba II, Minna JD, et al. A 12-Gene Set Predicts Survival Benefits from Adjuvant Chemotherapy in Non-Small Cell Lung Cancer Patients. Clin Cancer Res. 2013 doi: 10.1158/1078-0432.CCR-12-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Okayama H, Kohno T, Ishii Y, Shimada Y, Shiraishi K, Iwakawa R, Furuta K, Tsuta K, Shibata T, Yamamoto S, Watanabe S, Sakamoto H, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer research. 2012;72:100–11. doi: 10.1158/0008-5472.CAN-11-1403. [DOI] [PubMed] [Google Scholar]
- 26.Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Ponten F, Konig A, Fernandes O, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clinical cancer research : an official journal of the American Association for Cancer Research. 2013;19:194–204. doi: 10.1158/1078-0432.CCR-12-1139. [DOI] [PubMed] [Google Scholar]
- 27.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, Grosveld F, Philipsen S. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PloS one. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibbons DL, Lin W, Creighton CJ, Zheng S, Berel D, Yang Y, Raso MG, Liu DD, Wistuba II, Lozano G, Kurie JM. Expression signatures of metastatic capacity in a genetic mouse model of lung adenocarcinoma. PLoS One. 2009;4:e5401. doi: 10.1371/journal.pone.0005401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–15. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cukierman E, Pankov R, Stevens DR, Yamada KM. Taking cell-matrix adhesions to the third dimension. Science. 2001;294:1708–12. doi: 10.1126/science.1064829. [DOI] [PubMed] [Google Scholar]
- 31.Pampaloni F, Reynaud EG, Stelzer EH. The third dimension bridges the gap between cell culture and live tissue. Nature reviews Molecular cell biology. 2007;8:839–45. doi: 10.1038/nrm2236. [DOI] [PubMed] [Google Scholar]
- 32.Thiery JP. Epithelial-mesenchymal transitions in tumour progression. Nature reviews Cancer. 2002;2:442–54. doi: 10.1038/nrc822. [DOI] [PubMed] [Google Scholar]
- 33.Gibbons DL, Lin W, Creighton CJ, Rizvi ZH, Gregory PA, Goodall GJ, Thilaganathan N, Du L, Zhang Y, Pertsemlidis A, Kurie JM. Contextual extracellular cues promote tumor cell EMT and metastasis by regulating miR-200 family expression. Genes Dev. 2009;23:2140–51. doi: 10.1101/gad.1820209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Batran SE, Astner ST, Supthut M, Gamarra F, Brueckner K, Welsch U, Knuechel R, Huber RM. Three-dimensional in vitro cocultivation of lung carcinoma cells with human bronchial organ culture as a model for bronchial carcinoma. Am J Respir Cell Mol Biol. 1999;21:200–8. doi: 10.1165/ajrcmb.21.2.3205. [DOI] [PubMed] [Google Scholar]
- 35.Cichon MA, Gainullin VG, Zhang Y, Radisky DC. Growth of lung cancer cells in three-dimensional microenvironments reveals key features of tumor malignancy. Integr Biol (Camb) 2012;4:440–8. doi: 10.1039/c1ib00090j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes CS, Postovit LM, Lajoie GA. Matrigel: a complex protein mixture required for optimal growth of cell culture. Proteomics. 2010;10:1886–90. doi: 10.1002/pmic.200900758. [DOI] [PubMed] [Google Scholar]
- 37.Wallis JM, Borg ZD, Daly AB, Deng B, Ballif BA, Allen GB, Jaworski DM, Weiss DJ. Comparative assessment of detergent-based protocols for mouse lung de-cellularization and re-cellularization. Tissue Eng Part C Methods. 2012;18:420–32. doi: 10.1089/ten.tec.2011.0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhou J, Mochizuki T, Smeets H, Antignac C, Laurila P, de Paepe A, Tryggvason K, Reeders ST. Deletion of the paired alpha 5(IV) and alpha 6(IV) collagen genes in inherited smooth muscle tumors. Science. 1993;261:1167–9. doi: 10.1126/science.8356449. [DOI] [PubMed] [Google Scholar]
- 39.Yajnik V, Paulding C, Sordella R, McClatchey AI, Saito M, Wahrer DC, Reynolds P, Bell DW, Lake R, van den Heuvel S, Settleman J, Haber DA. DOCK4, a GTPase activator, is disrupted during tumorigenesis. Cell. 2003;112:673–84. doi: 10.1016/s0092-8674(03)00155-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.