ABSTRACT
The mammalian X chromosome contains a large number of multicopy genes that are expressed during spermatogenesis. The roles of these genes during germ cell development and the functional significance of gene multiplication remain mostly unexplored, as the presence of multicopy gene families poses a challenge for genetic studies. Here we report the deletion of a 1.1-Mb segment of the mouse X chromosome that is syntenic with the human Xq22.1 region and contains 20 genes that are expressed predominantly in testis and brain, including three members of the nuclear export factor gene family (Nxf2, Nxf3, and Nxf7) and five copies of preferentially expressed antigen in melanoma-like 3 (Pramel3). We have shown that germline-specific Cre/loxP-mediated deletion of this 1.1-Mb segment is efficient and causes defective chromosomal synapsis, meiotic arrest, and sterility in male mice. Our results demonstrate that this 1.1-Mb region contains one or more novel X-linked factors that are essential for male meiosis.
Keywords: male fertility, male infertility, meiosis, meiotic arrest, segmental deletion, spermatogenesis, X chromosome
A 1.1-Mb segment between Nxf2 and Nxf3 on the X chromosome is essential for meiosis in male mice.
INTRODUCTION
Sex chromosomes have evolved from a pair of autosomes; their unique status, however, has shaped specific gene content during evolution. In mammalian males, the hemizygous state of the X chromosome exerts immediate selection for or against new X-linked alleles encoding sexually dimorphic traits [1]. The X chromosome also encodes a large number of genes that are highly expressed in the brain, and mutations in more than 100 of these genes have been associated with mental retardation syndromes in males [2, 3]. During male meiosis, the X and the Y chromosome are transcriptionally silenced in a process termed meiotic sex chromosome inactivation (MSCI), whereas autosomes remain transcriptionally active [4, 5]. In postmeiotic germ cells, the transcription of sex chromosome-linked genes remains substantially repressed [6, 7]. Consistent with these changes in transcriptional activity during spermatogenesis, the X chromosome is enriched for genes that are expressed prior to meiosis but depleted of genes that play roles during or after meiosis [8, 9]. Many X-linked micro-RNAs, however, escape from MSCI and are present during the pachytene stage of male meiosis [10]. Genetic studies in the mouse have shown that several X-linked genes regulate male meiosis and play critical roles during germ cell maturation [11].
Families of multiple or multicopy genes can arise from gene duplication, retrotransposition, or from segmental duplications. Approximately 5% of the human genome consists of large interspersed segmental duplications with high sequence identity, producing recombination hot spots that can result in genomic inversions, amplifications, and deletions [12]. Segmental duplications also pose a challenge for genome assembly due to their repetitive nature, leaving gaps in the “finished” genome [13]. The mouse X chromosome contains 22 ampliconic regions enriched for identical or nearly identical multicopy genes that are expressed in the testis [14]. Thirty-three such multicopy gene families are expressed preferentially in postmeiotic germ cells, suggesting that this increase in gene copy numbers evolved to counteract the transcriptional repression of the X chromosome in postmeiotic germ cells. Ampliconic regions make up 19.4 Mb of genomic sequence and approximately 12% of the X chromosome; however, little is known about their functional relevance in the male germline.
The International Knockout Mouse Consortium (IKMC) aims to mutate all protein-coding genes in the mouse using the Cre/loxP targeting approach, among others; however, multicopy genes are not amenable to genetic studies on a one-gene-at-a-time basis. A transgenic RNAi approach has been successfully used to knock down the X-linked multicopy Slx/Slxl1 genes and the Y-linked multicopy Sly genes [15, 16]. Cre/loxP-mediated deletion of entire genomic regions could be an effective alternative strategy for studying multicopy genes. Meiotic recombination in mice with independent conditional null mutant alleles flanked by loxP sites (floxed) can generate alleles in which large genomic regions are flanked by loxP sites. Such floxed genomic segments can then be deleted using Cre recombinase, provided that the loxP sites are in the same orientation. The continually expanding number of conditional mutants available from the IKMC will eventually render this strategy feasible for many locations [17]. Here, we have generated a floxed allele for a 1.1-Mb segment of the murine X chromosome, an ampliconic region between Nxf2 and Nxf3 that contains 20 genes. We report that germline-specific deletion of this large genomic segment causes male sterility, revealing that it contains factors that are essential for meiotic progression in males.
MATERIALS AND METHODS
Isolation of Mouse Spermatogenic Cells
Cell populations that are highly enriched for specific spermatogenic cell types were isolated from the testes of male CD-1 mice (Charles River Laboratories) using the Sta Put method as described previously [18, 19]. Isolation of each of the spermatogenic cell types was done once [20]. The purity of each of the spermatogenic cell types was reported previously: >95% for pachytene spermatocytes and round spermatids, respectively, and >85% for all other cell types [20].
RNA Extraction and RT-PCR
Transcript levels were assessed using a semiquantitative RT-PCR method. Total RNA was extracted from adult mouse tissues, E13.5 fetuses, placenta, isolated spermatogenic cells, and testes from mice of various ages (1, 2, 4, and 8 wk or 4 mo) using TRIzol reagent (Invitrogen). Poly (A)+ RNA was isolated using a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech); 0.5 μg of total RNA from tissues or 70 ng of poly (A)+ RNA from germ cells were used for reverse transcription primed with random hexamers or oligo (dT) 18. Bulk cDNA was diluted and used for PCR reactions as previously described [20]. To avoid saturation of PCR reactions, aliquots of products were removed after various cycles of PCR and examined by agarose gel electrophoresis. Each PCR reaction was performed at least twice.
Mice
Nxf2fl and Nxf3fl mice were on mixed genetic backgrounds (129xC57BL/6). Nxf2fl and Nxf3fl mice were intercrossed to generate Nxf2fl/+ Nxf3+/fl females, which carried the Nxf2fl and Nxf3fl alleles on separate X chromosomes [21, 22]. Among 49 offspring of Nxf2fl/+ Nxf3+/fl females mated to wild-type males, we identified one Nxf2fl/Y Nxf3fl/Y male that carried both Nxf2fl and Nxf3fl on the same X chromosome due to meiotic recombination between these two alleles in the Nxf2fl/+ Nxf3+/fl female. This new Nxf2 Nxf3 floxed allele is referred to as Nxffl in this study; the approved symbol for the allele after Cre recombinase-mediated excision of the Nxf fragment is Del (XNxf2-Nxf3)1Jw (MGI ID: 5297602; see Supplemental Fig. S2). Nxffl/+ females (backcrossed to C57BL/6J for two generations) were crossed with Ddx4-Cre mice (Jackson Laboratory; stock no. Ddx4-Cre, 006954) to produce Nxffl/Y Ddx4-Cre males [23]. Genotyping was performed on genomic DNA isolated from tails. Mice were maintained and used in accordance with standards approved by the Institutional Animal Care and Use Committee of the University of Pennsylvania.
Generation of Antibodies
A carboxy-terminal segment of 200 aa (263–462) of the mouse PRAMEL3 protein was expressed as a GST fusion protein in Escherichia coli using the pGEX4T-1 vector. After affinity purification with glutathione sepharose, the GST-PRAMEL3 fusion protein was used to immunize rabbits (Cocalico Biologicals Inc.). Two antisera, UP2340 and UP2341, were obtained, from which antibodies were affinity purified by immunoblotting [24]. Western blot analysis was performed on 20 μg of testicular extract as previously described [22].
Sperm Count and Mating Tests
Cauda epididymides were dissected. Epidydimal sperm were fixed in 4% formaldehyde and counted using a hematocytometer. Sperm count was performed as previously described [25]. For mating tests, four wild-type and four Nxffl/Y Ddx4-Cre males (2 mo of age) were individually housed with two wild-type virgin females (2–3 mo of age) for 2 mo, with daily inspection of cages. The wild-type males sired an average of 2.5 litters per male, with 8.6 pups per litter. The four Nxffl/Y Ddx4-Cre males produced no offspring.
Histological Analysis
Testes were fixed in Bouin solution, embedded in paraffin, and sectioned, and slide-mounted sections were stained with hematoxylin and eosin. To quantify the seminiferous tubules, 100 cross sections of seminiferous tubules per mouse were examined and categorized into tubules with meiotic arrest (absence of spermatids) and tubules without meiotic arrest (presence of spermatids). Testes from five 8-wk-old and six 4-mo-old Nxffl/Y Ddx4-Cre males were analyzed. The data were presented as average ± standard deviation.
Immunofluorescence, TUNEL, and Nuclear Spread Analyses
For immunofluorescence, testes were fixed in 4% paraformaldehyde overnight at 4°C, dehydrated in 30% sucrose overnight, frozen in dry ice/95% ethanol, and sectioned at −20°C. TUNEL assays were performed on frozen sections using ApopTag Fluorescein In Situ Apoptosis Detection Kit (Chemicon) combined with immunostaining with anti-H1t antibodies. Surface spreads of spermatocyte nuclei were prepared as previously described [26, 27]. The following primary antibodies were used for immunofluorescence: anti-PRAMEL3 (this study), antihistone H1t (gift from Mary Ann Handel) [28], CREST antiserum (gift from Bill Brinkley) [29], anti-SYCP1 (Abcam, ab15090), and anti-SYCP2 [30]. Secondary antibodies were FITC- and Texas red-conjugated. Images were captured on an Axioskop 40 fluorescence microscope using a digital camera (Evolution QEi, MediaCybernetics).
RESULTS
A 1.1-Mb Nxf2–Nxf3 Segment on the X Chromosome Is Enriched for Testis and Brain-Expressed Genes
A 1.1-Mb segment of the murine X chromosome, located between the Nxf2 and Nxf3 genes, encodes 20 genes with testis- and brain-specific expression patterns (Fig. 1 and Supplemental Table S1; all Supplemental Data available online at www.biolreprod.org). This region, here termed Nxf segment, is syntenic with the human Xq22.1 region and maps to the cytogenetic bands XF1 (18 genes: 3632454L22Rik to Nxf3) and XE3 (two genes: Nxf2 and Zmat1; Fig. 1). Supplemental Table S1 summarizes the known or putative functions of the 20 genes, of which so far only Nxf2, Nxf3, and G protein-coupled receptor associated sorting protein 1 (Gprasp1) have been subject to targeted inactivation in the mouse. Nxf2 and Nxf3 are expressed in germ cells and Sertoli cells of the testis, respectively [21, 22]. Inactivation of Nxf2 impairs male fertility [21], whereas Nxf3-deficient mice are grossly normal and fertile [22]. A double mutant of Nxf2 and Nxf3 lacks compound effects [22]. GPRASP1 has been implicated in regulating dopamine and cannabinoid receptor signaling, and Gprasp1 null-mutant mice exhibit impaired striatum-dependent responses [31, 32]. The Nxf segment also contains a cluster of genes belonging to the Prame (preferentially expressed antigen in melanoma) family of genes that are expressed in testis and several types of cancers (cancer-testis genes): Pramel, Pramel3 (five nearly identical copies), and Prame; two genes (Tceal6 and Bhlhb9) encode transcription factors; Gm6215 (three nearly identical copies) and 3632454L22Rik are long intergenic noncoding RNA (lincRNA) genes; and Mir1970 is a micro-RNA gene. In addition, the fragment contains three predicted pseudogenes (Gm6207, Gm6228, and Gm15016), for which RNA reference sequences are lacking.
FIG. 1.

Relative chromosomal locations and tissue-specific expression of genes contained within the 1.1-Mb Nxf fragment on the X chromosome. Expression was assayed by RT-PCR analysis of RNA isolated from adult mouse tissues and from E13.5 whole embryo and placenta. Chromosomal locations shown are based on the NCBI mouse reference genome. Almost identical gene copies of Pramel3 (five copies) and Gm6215 (three copies) are marked by multiple lines. Primer sequences are listed in Supplemental Table S2. Gapdh served as a positive control; reactions without reverse transcriptase were negative (data not shown).
We determined expression patterns of 18 genes encoded within the Nxf fragment in diverse tissues by semiquantitative RT-PCR analysis (Fig. 1) and found that eight of these were specifically or preferentially expressed in testis: Nxf2, Pramel, Pramel3, Gm6215, Prame, Tcp11l3, Tmsb15a, and Nxf3. Nxf7 transcript levels were highest in placenta but detectable in other tissues such as testis and brain, whereas Tceal6 expression was restricted to brain. Transcripts of the remaining eight genes (Zmat1, 3632454L22Rik, Armcx5, Gprasp1, Gprasp2, Bhlhb9, Arxes1/2, and Bex2) were detected in a broad range of tissues, including testis and brain. The Nxf segment therefore includes both genes with highly tissue-specific expression patterns, restricted predominantly to testis and brain, and broadly expressed genes that may play roles in multiple tissues.
X-Linked Genes Exhibit Stage-Specific Expression Patterns During Spermatogenesis
For 16 genes in the Nxf fragment that were expressed in testis, we next characterized expression profiles throughout spermatogenesis by assessing transcript levels in seven specific subpopulations of spermatogenic cells isolated from testis (Fig. 2). These highly enriched cell populations included three types of spermatogonia, that is, mitotically dividing diploid germ cells: primitive type A, which contain a subpopulation of self-renewing spermatogonial stem cells; type A, which proliferate and differentiate into intermediate spermatogonia, which then divide to become type B spermatogonia. Type B spermatogonia enter meiosis to become Prophase I spermatocytes (preleptotene, leptotene, zygotene, pachytene, and diplotene). Round spermatids are postmeiotic haploid germ cells that undergo a complex differentiation process to develop into mature spermatozoa. Eight of the 16 genes (Zmat1, Armcx5, Gprasp1, Gprasp2, Bhlhb9, Arxes1, Arxes2, and Bex2) were expressed in all spermatogenic subpopulations and in XXY* testis (Fig. 2), which are germ cell deficient [33], revealing activity of these genes in germ cells throughout spermatogenesis and in somatic cells of the testis. In addition to Nxf2 and Pramel3 [8], four genes (Nxf7, Prame, Tcp11l3, and Tmsb15a) were found to be specifically expressed in germ cells of the testis, lacking expression in germ cell-deficient XXY* testis. These genes also exhibited differential expression in spermatogenic subpopulations: transcripts were found in spermatocytes and round spermatids but not in spermatogonia. During male meiosis, sex chromosome-linked genes undergo transcriptional inactivation (MSCI) at the pachytene stage [4, 7]. We observed reduced expression of many of the 16 genes analyzed in pachytene spermatocytes, providing evidence that these X-linked genes were subject to MSCI.
FIG. 2.
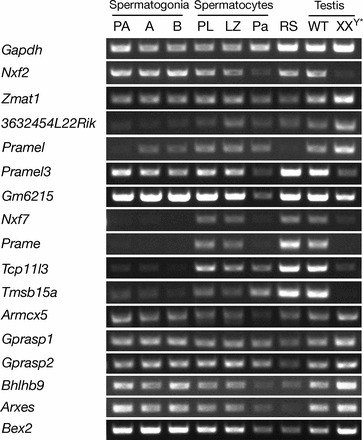
Expression profiles of 16 genes of the 1.1-Mb Nxf segment during spermatogenesis. Relative transcript levels were determined by RT-PCR, with Gapdh as positive control for ubiquitous and Nxf2 and Pramel3 for testis-specific gene expression, respectively [20]. Nxf3 is specifically expressed in Sertoli cells [22] and was therefore not included. Testes from XXY* mice lack germ cells and served to identify genes expressed in somatic cells. Abbreviations for spermatogenic cell types: PA, primitive type A spermatogonia; A, type A spermatogonia; B, type B spermatogonia; PL, preleptotene spermatocytes; LZ, leptotene plus zygotene spermatocytes; Pa, adult pachytene spermatocytes; RS: round spermatids. WT: wild type. Gene designations are as described in Figure 1. Primer sequences are listed in Supplemental Table S2.
Dynamic Expression and Subcellular Localization of PRAMEL3 During Spermatogenesis
Pramel3 is a cancer-testis gene of unknown physiological functions. Transcript analysis suggested that PRAMEL3 is restricted to germ cells of the testis and absent from somatic cells (Fig. 2). To confirm this finding on the protein level and to characterize PRAMEL3 protein localization during spermatogenesis, we raised polyclonal antibodies for PRAMEL3. Immunostaining of testis sections revealed a dynamic expression pattern of PRAMEL3 during spermatogenesis (see Supplemental Fig. S1): consistent with analyses of transcript distribution (Fig. 2), PRAMEL3 protein was detected in spermatogonia, early spermatocytes (preleptotene, leptotene, and zygotene), and round spermatids but absent from pachytene spermatocytes, metaphase spermatocytes, and elongated spermatids. In spermatogonia and round spermatids, PRAMEL3 was evenly distributed across cytoplasm and nuclei (see Supplemental Fig. S1A), whereas leptotene and zygotene spermatocytes exhibited higher protein levels in the cytoplasm (see Supplemental Fig. S1B, C). Absence of PRAMEL3 in pachytene spermatocytes suggests that Pramel3, like other X-linked genes, is subject to MSCI. Although most X-linked genes remain suppressed after meiosis and are not active in round spermatids, multicopy genes escape from postmeiotic silencing by an increase in copy number, thereby compensating for a low expression level per copy [14]. The Pramel3 gene is present in five copies (Fig. 1). Consistent with the notion on the postmeiotic expression of multicopy genes, PRAMEL3 transcript and protein are abundantly expressed in round spermatids (Fig. 2 and see Supplemental Fig. S1).
Germline-Specific Segmental Deletion Causes Male Sterility and Meiotic Defects
To ascertain the consequences of the loss of a chromosomal segment enriched in testis-expressed genes, including multicopy genes (Fig. 1), we generated a conditional allele of the 1.1-Mb Nxf fragment, permitting its tissue-specific deletion. From intercrosses of mice with loxP-flanked alleles for Nxf2 (Nxf2fl) and Nxf3 (Nxf3fl), we obtained offspring with both Nxf2fl and Nxf3fl alleles on the same X chromosome as a consequence of meiotic recombination [21, 22]. In this new conditional allele (approved designation: Del [XNxf2-Nxf3] 1Jw [MGI:5297602]; here abbreviated Nxffl), Nxf2 and Nxf3 are flanked by loxP sites of the same orientation (see Supplemental Fig. S2). Ubiquitous deletion of the Nxf segment in males using Actb-Cre causes neonatal lethality. To obtain germ-cell specific deletion of this allele, we generated Nxffl/Y Ddx4-Cre males. Ddx4-Cre (previously termed Vasa-Cre) expression is restricted to the germline; expression begins in embryonic germ cells at Embryonic Day 15 (E15) and continues throughout germ cell maturation, in males until the postmeiotic spermatid stage [23]. The Ddx4-Cre has been used to efficiently disrupt several genes specifically in male germ cells [34–36]. Nxffl/Y Ddx4-Cre males were sterile, producing no offspring in mating tests. Testes from 8-wk-old Nxffl/Y Ddx4-Cre males were significantly reduced in size, at 40% of the weight of wild-type testes (Fig. 3, A and B). The sperm count of Nxffl/Y Ddx4-Cre males was less than 8% that of wild-type males (Fig. 3C). Therefore, deletion of the Nxf2-Nxf3 segment on the X chromosome causes male sterility.
FIG. 3.
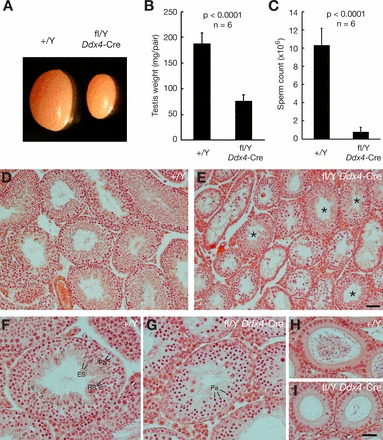
Germline-specific segmental deletion causes meiotic failure and male sterility. A) Significant size reduction in 8-wk-old Nxffl/Y Ddx4-Cre testis. B and C) Significantly reduced testis weight and sperm production in 8-wk-old Nxffl/Y Ddx4-Cre mice. Data were analyzed by Student t-test. D and E) Hematoxylin and eosin stained sections of testes from 8-wk-old wild-type and Nxffl/Y Ddx4-Cre mice (low magnification). Asterisks mark tubules of the mutant testes that contain postmeiotic germ cells (E). F) Wild-type tubule contains a full spectrum of germ cells. G) Mutant seminiferous tubule exhibiting meiotic arrest at the pachytene stage. H) Wild-type epididymal tubule filled with sperm. I) Epididymal tubules from Nxffl/Y Ddx4-Cre mice, devoid of sperm. Pa, pachytene spermatocytes; RS, round spermatids; ES, elongating spermatids. Bars = 50 μm (D and E) and 25 μm (F–I).
Histological analysis of testes from Nxffl/Y Ddx4-Cre males revealed meiotic defects. Seminiferous tubules from wild-type adult testes contained a full spectrum of spermatogenic cells at different stages, including spermatocytes and spermatids (Fig. 3, D and F). In contrast, most seminiferous tubules (57 ± 10%) from 8-wk-old Nxffl/Y Ddx4-Cre males contained spermatocytes but lacked postmeiotic germ cells, indicating meiotic arrest. The remaining tubules contained postmeiotic germ cells, such as round spermatids and/or elongated spermatids (Fig. 3E). We assume that the subset of tubules with apparent normal spermatogenesis failed to undergo complete Cre-mediated deletion of the Nxf segment due to its large size (1.1 Mb). In tubules with meiotic arrest, the most advanced germ cells were pachytene spermatocytes (Fig. 3G). Consistent with testis histology and sperm count data, epididymal tubules from Nxffl/Y Ddx4-Cre males lacked sperm or contained visibly fewer sperm compared to those of wild type mice (Fig. 3, H and I).
As an approach to assess the efficacy of Ddx4-Cre-mediated deletion of the Nxf segment and to determine if the presence of spermatid-containing tubules in Nxffl/Y Ddx4-Cre testes (Fig. 3E) was due to incomplete Cre-mediated deletion, we examined expression of PRAMEL3 protein (Fig. 4). Western blot analysis confirmed the presence of PRAMEL3 protein in wild-type but not germ cell-depleted XXY* testes (Fig. 4A). In testes from Nxffl/Y Ddx4-Cre males, PRAMEL3 was not detectable by Western blot, suggesting that Cre-mediated deletion of the Nxf fragment was overall efficient (Fig. 4B). Immunofluorescent analysis confirmed that in Nxffl/Y Ddx4-Cre mutant tubules with meiotic arrest, germ cells of all stages lacked detectable PRAMEL3 protein (Fig. 4D). In Nxffl/Y Ddx4-Cre mutant tubules that contained spermatids, PRAMEL3 was expressed in the majority of round spermatids (Fig. 4E), while other round spermatids of the same tubule lacked PRAMEL3 protein (Fig. 4E, arrowheads). These observations suggest incomplete Ddx4-Cre-mediated deletion of the Nxf fragment in spermatogonia, possibly due to its large size, permitting germ cell maturation in some seminiferous tubules (Fig. 3E). In some of these cells, Ddx4-Cre may mediate deletion of the Nxf fragment at postmeiotic stages.
FIG. 4.

Cre-mediated recombination of the Nxf fragment in germ cells assayed by the expression of PRAMEL3 protein. A) PRAMEL3 is restricted to germ cells and absent in in germ cell-deficient XXY* testes. B) Testes from Nxffl/Y Ddx4-Cre lack detectable PRAMEL3 protein. ACTB serves as a loading control. C–E) Expression of PRAMEL3 in testicular germ cells from 8-wk-old wild-type (C) and Nxffl/Y Ddx4-Cre (D and E) mice. Testis sections were immunostained with anti-PRAMEL3 antibody (green). Chromatin stained with DAPI (blue) is also shown in black and white to visualize nuclear morphology. PRAMEL3 is absent in mutant tubules with meiotic arrest (D). In the mutant tubule with spermatids (E), PRAMEL3 is present in some round spermatids but absent in other round spermatids (arrowheads), demonstrating partial Cre-mediated deletion of the Nxf genomic fragment. Bar = 25 μm.
The ongoing expression of Ddx4-Cre in spermatogonia of Nxffl/Y Ddx4-Cre males may lead to an increase in Cre-mediated deletion of the Nxf fragment over time. Consistent with this, the testis weight of mutant males was further reduced at 4 mo (see Supplemental Fig. S3). We observed that, in testes from 4-mo-old Nxffl/Y Ddx4-Cre males, the majority of tubules (93 ± 2%; n = 6 mice) exhibited complete meiotic arrest (see Supplemental Fig. S3), compared to 57 ± 10% (n = 5 mice) in 8-wk-old mutant testes (Fig. 3E).
We monitored the Ddx4-Cre-mediated deletion of the Nxf segment by analyzing the expression of genes within this segment in testes at different times after birth from 1 wk to 4 mo (see Supplemental Fig. S4). The expression of Pramel3 is most informative, as it is germ cell-specific and is expressed in spermatogonia. The lower or absent expression of Pramel3 in Nxffl/Y Ddx4-Cre testes showed that the deletion of the Nxf segment was substantial (in spermatogonia) beginning at 1 wk after birth but was not complete until 4 mo. This conclusion was further supported by the expression profiles of other genes, such as Gm6215, Prame, Tcp11l3, and Tmsb15a. The genes that are highly expressed in testicular somatic cells, such as Zmat1, Gprasp2, and Bhlhb9 (Fig. 2), appeared to be expressed at a higher level in Nxffl/Y Ddx4-Cre testes than wild type at 4 wk and beyond (see Supplemental Fig. S4) due to the reduced percentage of germ cells in the mutant testes caused by meiotic arrest. In addition, we examined the expression of three genes on either side of the Nxf segment, respectively. The levels of their expression were either unchanged (Bex4) or higher (Armcx6) in Nxffl/Y Ddx4-Cre testes compared to wild-type controls, mostly likely due to their ubiquitous expression or predominant expression in testicular somatic cells.
Defective Chromosomal Synapsis in Nxffl/Y Ddx4-Cre Males
Seminiferous tubules from Nxffl/Y Ddx4-Cre testes contained substantially more apoptotic cells than tubules from wild-type testes (Fig. 5, A and B), suggesting effective elimination of abnormal spermatocytes through the pachytene checkpoint that operates at the midpachytene stage [5, 37]. Supporting this hypothesis, we found that prophase I spermatocytes from 20-day-old Nxffl/Y Ddx4-Cre males contained proportionally fewer mid- to late (H1t-positive) pachytene cells [28] than early (histone H1t-negative) stages (Fig. 5C).
FIG. 5.
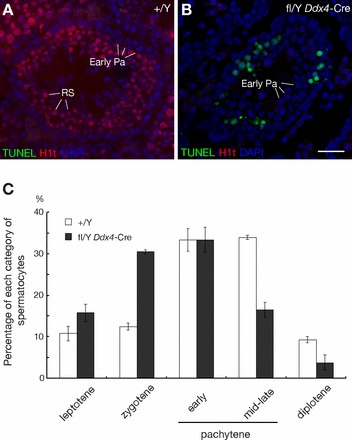
Apoptosis of spermatocytes in Nxffl/Y Ddx4-Cre testes. A and B) Spermatocytes in Nxffl/Y Ddx4-Cre testis undergo apoptosis. TUNEL assay of frozen testis sections from 8-wk-old wild-type and Nxffl/Y Ddx4-Cre males, combined with immunostaining for H1t. Chromatin is visualized with DAPI. RS, round spermatids (H1t-positive); Early Pa, early pachytene spermatocytes (H1t-negative). Bar = 25 μm. C) Reduced proportion of mid-late pachytene spermatocytes among prophase I spermatocytes from Nxffl/Y Ddx4-Cre males. Surface spreads of spermatocytes from 20-day-old mice were immunostained with antibodies against SYCP2 and histone H1t; 200 spermatocytes from each mouse (three mice per genotype) were counted.
To determine the cause of meiotic arrest in Nxffl/Y Ddx4-Cre germ cells, we examined the formation of the synaptonemal complex (SC), which mediates the alignment and synapsis of homologous chromosomes during meiosis. Immunostaining for SYCP2, a component of the axial/lateral elements of the SC (Fig. 6A) [30, 38, 39], showed that SC lateral elements were present in spermatocytes from Nxffl/Y Ddx4-Cre males (Fig. 6, B–D). However, visualization of SYCP1, a component of SC transverse filaments that localizes to the synapsed regions of SC [40, 41], revealed defects in chromosomal synapsis in pachytene spermatocytes from Nxffl/Y Ddx4-Cre testes (Fig. 6E). In most (90%) wild-type pachytene spermatocytes, the 19 pairs of autosomes were fully synapsed, and the XY chromosomes synapsed at the pseudoautosomal regions (Fig. 6, A and E). In contrast, 46% of mutant spermatocytes from 20-day-old testes exhibited synaptic defects: 8% displayed asynapsis of only the XY chromosomes (Fig. 6B), 13% of cells contained incompletely synapsed autosomes with split ends (Fig. 6C), and in 25% of cells both sex chromosomes and autosomes were affected (Fig. 6D).
FIG. 6.
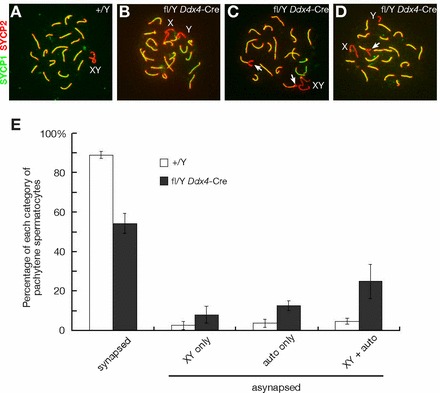
Defective chromosomal synapsis in germ cells from Nxffl/Y Ddx4-Cre males. A) Wild-type pachynema; 19 pairs of autosomes are fully synapsed. XY chromosomes are synapsed only in the pseudoautosomal regions. B) Mutant pachynema with asynapsed XY chromosomes, autosomes are not affected. C) Mutant pachynema with partially asynapsed autosomes (white arrows). D) Mutant pachynema with synaptic defects affecting both autosomes (white arrow) and sex chromosomes. E) Analysis of chromosomal synapsis in pachytene spermatocytes from 20-day-old wild-type and Nxffl/Y Ddx4-Cre testes. Testes from three wild-type and three Nxffl/Y Ddx4-Cre mice were analyzed by surface nuclear spread analysis using anti-SYCP1 and anti-SYCP2 antibodies; 100 pachynema were analyzed per mouse, and grouped into the categories depicted in A–D.
Mouse chromosomes are telocentric, and their centromeres are the last regions to synapse [42, 43]. Combined immunostaining with anti-SYCP2 antibodies and the centromere-specific CREST antiserum [44] showed that among 33 pachytene spermatocytes from Nxffl/Y Ddx4-Cre testes with unsynapsed autosomal ends, 22 cells exhibited asynapsis only at some centromeric ends, three cells displayed asynapsis only at some noncentromeric ends, and in eight cells asynapsis affected both centromeric and noncentromeric ends, showing that Nxf deletion could affect synapsis at either chromosomal end. In summary, these data suggest the presence of a single or multiple X-linked factors in the Nxf2-Nxf3 segment that are important for chromosomal synapsis.
DISCUSSION
The 1.1-Mb Nxf2-Nxf3 genomic region on the X chromosome is enriched for testis-specific single-copy and multicopy genes. Deletion of this segment in the male germline causes male sterility, revealing that this region contains new factors that are essential for male meiosis. This finding is intriguing considering that the mammalian X chromosome is transcriptionally inactivated during male meiosis due to MSCI [4, 7]. We previously reported that Tex11, an X-linked germ cell-specific gene, is essential for male meiosis [45]. TEX11 interacts with SYCP2 and regulates both chromosomal synapsis and meiotic recombination. TBP-associated factor 7 like (Taf7l), which is also X-linked, functions synergistically with Tex11 in the regulation of male meiosis [11]. These X-linked genes are transcriptionally silenced at the pachytene stage of meiosis as a consequence of MSCI. However, continued availability of their respective protein products is essential for meiotic progression and may rely on accumulation of transcripts prior to MSCI.
The question remains as to which gene in the 1.1-Mb Nxf region is responsible for meiotic defects. It might be a single gene or more than one gene. The testis-specific genes (Pramel, Pramel3, Prame, Tcp11l3, and Tmsb15a) are strong candidates, as they are expressed in early spermatocytes. However, we cannot exclude the broadly expressed genes. Future genetic studies are needed to further narrow down the genomic region and identify the gene or genes critical for male meiosis. It is also possible that the synaptic abnormality observed in the Nxffl Ddx4-Cre mice is a downstream effect of other deficiencies in germ cells. Recently, aneuploidy (loss or gain of a chromosome) causes genomic instability and proteotoxic stress in yeast mitotic cells [46–48]. The deletion of the 1.1-Mb Nxf region on the X chromosome could have a more global effect on the transcriptome or proteome or even on the expression of the adjacent genes on either side of this segment. Notably, there is abnormal synapsis between X and Y in the In (X)1H male mice harboring a segmental inversion on the X chromosome [49]. Because the loxP sites are located within the introns of Nxf2 and Nxf3 genes (Supplemental Fig. S2), the 1.1-Mb Nxf segmental deletion does not delete the promoters of the neighboring genes. However, it remains possible that the deletion may interfere with the expression of genes outside the deleted region. Our attempt to address this possibility using testis samples was inconclusive due to the wide expression patterns of the neighboring genes (Supplemental Fig. S4).
Chromosomal segmental deletion is a frequent cause of human disease. Such chromosomal microdeletion regions span a few Mb genomic DNA sequences and contain multiple genes. Deletion of multiple genes often leads to haploinsufficiency syndromes. The 22q11 deletion syndrome, typically caused by a 3-Mb deletion, is the most common human microdeletion syndrome, occurring in 1:4000 live births [50, 51]. Medical cytogenetics and array comparative genomic hybridization have identified and defined the deletion intervals for a number of human deletion syndromes, including 1p36 [52, 53], 2q37 [54], 3p [55], 9p [56, 57], 13q [58], 18q23 [59], and Yq [60]. While studies of human patients are important for understanding the pathogenesis of these complex syndromes, there is a pressing need for generating animal models of human deletion syndromes. The mouse model of the 22q11 deletion syndrome has been valuable for characterizing this human syndrome [61–63].
In a recent report, a familial 1.1-Mb deletion in human chromosome Xq22.1 was found in heterozygous female patients with epilepsy, mental retardation, and other developmental defects [64]. Our 1.1-Mb deletion region (Nxf2-Nxf3) on the mouse X chromosome is syntenic to the 1.1-Mb deletion region in human Xq22.1. We have generated Nxf+/− heterozygous females using a ubiquitously expressed Actb-Cre [65]. The characterization of Nxf+/− heterozygous females will be published in a separate study. Therefore, our Nxf segmental deletion mouse mutant is an animal model for this new human syndrome: the Xq22.1 deletion syndrome.
Supplementary Material
ACKNOWLEDGMENT
We are grateful to Mary Ann Handel for H1t antibodies, Bill R. Brinkley for CREST antiserum, Sigrid Eckardt for help with manuscript preparation, and Seth Kasowitz and Qi Fu for comments on the manuscript.
Footnotes
Supported by NIH grants R01GM089893 and R01GM076327 to P.J.W.
REFERENCES
- Rice WR. Sexually antagonistic genes: experimental evidence. Science 1992; 256: 1436 1439. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. High expression of the mammalian X chromosome in brain. Brain Res 2006; 1126: 46 49. [DOI] [PubMed] [Google Scholar]
- Lubs HA, Stevenson RE, Schwartz CE, Fragile X. and X-linked intellectual disability: four decades of discovery. Am J Hum Genet 2012; 90: 579 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development 2007; 134: 1823 1831. [DOI] [PubMed] [Google Scholar]
- Handel MA, Schimenti JC. Genetics of mammalian meiosis: regulation, dynamics and impact on fertility. Nat Rev Genet 2010; 11: 124 136. [DOI] [PubMed] [Google Scholar]
- Namekawa SH, Park PJ, Zhang LF, Shima JE, McCarrey JR, Griswold MD, Lee JT. Postmeiotic sex chromatin in the male germline of mice. Curr Biol 2006; 16: 660 667. [DOI] [PubMed] [Google Scholar]
- Turner JM, Mahadevaiah SK, Ellis PJ, Mitchell MJ, Burgoyne PS. Pachytene asynapsis drives meiotic sex chromosome inactivation and leads to substantial postmeiotic repression in spermatids. Dev Cell 2006; 10: 521 529. [DOI] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 2001; 27: 422 426. [DOI] [PubMed] [Google Scholar]
- Khil PP, Smirnova NA, Romanienko PJ, Camerini-Otero RD. The mouse X chromosome is enriched for sex-biased genes not subject to selection by meiotic sex chromosome inactivation. Nat Genet 2004; 36: 642 646. [DOI] [PubMed] [Google Scholar]
- Song R, Ro S, Michaels JD, Park C, McCarrey JR, Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat Genet 2009; 41: 488 493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Yang F, Wang PJ. Regulation of male fertility by X-linked genes. J Androl 2010; 31: 79 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Gu Z, Clark RA, Reinert K, Samonte RV, Schwartz S, Adams MD, Myers EW, Li PW, Eichler EE. Recent segmental duplications in the human genome. Science 2002; 297: 1003 1007. [DOI] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet 2004; 5: 345 354. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JM. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat Genet 2008; 40: 794 799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Mahadevaiah SK, Affara NA, Ward MA, Burgoyne PS. The multicopy gene Sly represses the sex chromosomes in the male mouse germline after meiosis. PLoS Biol 2009; 7: e1000244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Yamauchi Y, Riel JM, Karacs TP, Rattigan A, Ojarikre OA, Affara NA, Ward MA, Burgoyne PS. Deficiency in the multicopy Sycp3-like X-linked genes Slx and Slxl1 causes major defects in spermatid differentiation. Mol Biol Cell 2010; 21: 3497 3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan C, Ye C, Yang X, Gao J. A review of current large-scale mouse knockout efforts. Genesis 2010; 48: 73 85. [DOI] [PubMed] [Google Scholar]
- Bellve AR. Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 1993; 225: 84 113. [DOI] [PubMed] [Google Scholar]
- La Salle S, Sun F, Handel MA. Isolation and short-term culture of mouse spermatocytes for analysis of meiosis. Methods Mol Biol 2009; 558: 279 297. [DOI] [PubMed] [Google Scholar]
- Wang PJ, Page DC, McCarrey JR. Differential expression of sex-linked and autosomal germ-cell-specific genes during spermatogenesis in the mouse. Hum Mol Genet 2005; 14: 2911 2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Eckardt S, Leu NA, Buffone MG, Zhou J, Gerton GL, McLaughlin KJ, Wang PJ. Inactivation of Nxf2 causes defects in male meiosis and age-dependent depletion of spermatogonia. Dev Biol 2009; 330: 167 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Pan J, Eckardt S, Leu NA, McLaughlin KJ, Wang PJ. Nxf3 is expressed in Sertoli cells, but is dispensable for spermatogenesis. Mol Reprod Dev 2011; 78: 241 249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 2007; 45: 413 417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E, Lane D. Using Antibodies: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1998. [Google Scholar]
- Cheng Y, Buffone MG, Kouadio M, Goodheart M, Page DC, Gerton GL, Davidson I, Wang PJ. Abnormal sperm in mice lacking the Taf7l gene. Mol Cell Biol 2007; 27: 2582 2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters AH, Plug AW, van Vugt MJ, de Boer P. A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 1997; 5: 66 68. [DOI] [PubMed] [Google Scholar]
- Kolas NK, Marcon E, Crackower MA, Hoog C, Penninger JM, Spyropoulos B, Moens PB. Mutant meiotic chromosome core components in mice can cause apparent sexual dimorphic endpoints at prophase or X-Y defective male-specific sterility. Chromosoma 2005; 114: 92 102. [DOI] [PubMed] [Google Scholar]
- Cobb J, Cargile B, Handel MA. Acquisition of competence to condense metaphase I chromosomes during spermatogenesis. Dev Biol 1999; 205: 49 64. [DOI] [PubMed] [Google Scholar]
- Brenner S, Pepper D, Berns MW, Tan E, Brinkley BR. Kinetochore structure, duplication, and distribution in mammalian cells: analysis by human autoantibodies from scleroderma patients. J Cell Biol 1981; 91: 95 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol 2006; 173: 497 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeuf J, Trigo JM, Moreau PH, Lecourtier L, Vogel E, Cassel JC, Mathis C, Klosen P, Maldonado R, Simonin F. Attenuated behavioural responses to acute and chronic cocaine in GASP-1-deficient mice. Eur J Neurosci 2009; 30: 860 868. [DOI] [PubMed] [Google Scholar]
- Mathis C, Bott JB, Candusso MP, Simonin F, Cassel JC. Impaired striatum-dependent behavior in GASP-1-knock-out mice. Genes Brain Behav 2011; 10: 299 308. [DOI] [PubMed] [Google Scholar]
- Hunt PA, Eicher EM. Fertile male mice with three sex chromosomes: evidence that infertility in XYY male mice is an effect of two Y chromosomes. Chromosoma 1991; 100: 293 299. [DOI] [PubMed] [Google Scholar]
- Yang F, Wei Q, Adelstein RS, Wang PJ. Non-muscle myosin IIB is essential for cytokinesis during male meiotic cell divisions. Dev Biol 2012; 369: 356 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audouard C, Christians E. Hsp90beta1 knockout targeted to male germline: a mouse model for globozoospermia. Fertil Steril 2011; 95: 1475 1477. [DOI] [PubMed] [Google Scholar]
- Liu D, Li L, Fu H, Li S, Li J. Inactivation of Dicer1 has a severe cumulative impact on the formation of mature germ cells in mouse testes. Biochem Biophys Res Commun 2012; 422: 114 120. [DOI] [PubMed] [Google Scholar]
- Roeder GS, Bailis JM. The pachytene checkpoint. Trends Genet 2000; 16: 395 403. [DOI] [PubMed] [Google Scholar]
- Offenberg HH, Schalk JA, Meuwissen RL, van Aalderen M, Kester HA, Dietrich AJ, Heyting C. SCP2: a major protein component of the axial elements of synaptonemal complexes of the rat. Nucleic Acids Res 1998; 26: 2572 2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schalk JA, Dietrich AJ, Vink AC, Offenberg HH, van Aalderen M, Heyting C. Localization of SCP2 and SCP3 protein molecules within synaptonemal complexes of the rat. Chromosoma 1998; 107: 540 548. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci 1994; 107 (pt 10): 2749 2760. [DOI] [PubMed] [Google Scholar]
- de Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, Yuan L, Liu JG, van Zeeland AA, Heyting C, Pastink A. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev 2005; 19: 1376 1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisig CG, Guiraldelli MF, Kouznetsova A, Scherthan H, Hoog C, Dawson DS, Pezza RJ. Synaptonemal complex components persist at centromeres and are required for homologous centromere pairing in mouse spermatocytes. PLoS Genet 2012; 8: e1002701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao H, Chen JK, Reynolds A, Hoog C, Paddy M, Hunter N. Interplay between synaptonemal complex, homologous recombination, and centromeres during mammalian meiosis. PLoS Genet 2012; 8: e1002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Spyropoulos B. Immunocytology of chiasmata and chromosomal disjunction at mouse meiosis. Chromosoma 1995; 104: 175 182. [DOI] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Hoog C, McLaughlin KJ, Wang PJ. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 2008; 22: 682 691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oromendia AB, Dodgson SE, Amon A. Aneuploidy causes proteotoxic stress in yeast. Genes Dev 2012; 26: 2696 2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Blank HM, Pfau SJ, Tange Y, George BM, Humpton TJ, Brito IL, Hiraoka Y, Niwa O, Amon A. Aneuploidy drives genomic instability in yeast. Science 2011; 333: 1026 1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci U S A 2012; 109: 12644 12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley T. Nonhomologous synapsis of the XY during early pachynema in In(X)1H male mice. Genetica 1987; 72: 81 84. [DOI] [PubMed] [Google Scholar]
- Saitta SC, Harris SE, Gaeth AP, Driscoll DA, McDonald-McGinn DM, Maisenbacher MK, Yersak JM, Chakraborty PK, Hacker AM, Zackai EH, Ashley T, Emanuel BS. Aberrant interchromosomal exchanges are the predominant cause of the 22q11.2 deletion. Hum Mol Genet 2004; 13: 417 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devriendt K, Fryns JP, Mortier G, van Thienen MN, Keymolen K. The annual incidence of DiGeorge/velocardiofacial syndrome. J Med Genet 1998; 35: 789 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YQ, Heilstedt HA, Bedell JA, May KM, Starkey DE, McPherson JD, Shapira SK, Shaffer LG. Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Hum Mol Genet 1999; 8: 313 321. [DOI] [PubMed] [Google Scholar]
- Battaglia A. Del 1p36 syndrome: a newly emerging clinical entity. Brain Dev 2005; 27: 358 361. [DOI] [PubMed] [Google Scholar]
- Doherty ES, Lacbawan F. 2q37 Microdeletion Syndrome : Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP. (eds.), GeneReviews, Seattle, WA: University of Washington Press; 1993. [PubMed] [Google Scholar]
- Malmgren H, Sahlen S, Wide K, Lundvall M, Blennow E. Distal 3p deletion syndrome: detailed molecular cytogenetic and clinical characterization of three small distal deletions and review. Am J Med Genet A 2007; 143A: 2143 2149. [DOI] [PubMed] [Google Scholar]
- Alfi O, Donnell GN, Crandall BF, Derencsenyi A, Menon R. Deletion of the short arm of chromosome no.9 (46,9p-): a new deletion syndrome. Ann Genet 1973; 16: 17 22. [PubMed] [Google Scholar]
- Onesimo R, Orteschi D, Scalzone M, Rossodivita A, Nanni L, Zannoni GF, Marrocco G, Battaglia D, Fundaro C, Neri G. Chromosome 9p deletion syndrome and sex reversal: novel findings and redefinition of the critically deleted regions. Am J Med Genet A 2012; 158A: 2266 2271. [DOI] [PubMed] [Google Scholar]
- Allderdice PW, Davis JG, Miller OJ, Klinger HP, Warburton D, Miller DA, Allen FH, Jr,, Abrams CA, McGilvray E. The 13q-deletion syndrome. Am J Hum Genet 1969; 21: 499 512. [PMC free article] [PubMed] [Google Scholar]
- Margarit E, Morales C, Rodriguez-Revenga L, Monne R, Badenas C, Soler A, Clusellas N, Mademont I, Sanchez A. Familial 4.8 MB deletion on 18q23 associated with growth hormone insufficiency and phenotypic variability. Am J Med Genet A 2012; 158A: 611 616. [DOI] [PubMed] [Google Scholar]
- Reijo R, Lee TY, Salo P, Alagappan R, Brown LG, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page DC. Diverse spermatogenic defects in humans caused by Y chromosome deletions encompassing a novel RNA-binding protein gene. Nat Genet 1995; 10: 383 393. [DOI] [PubMed] [Google Scholar]
- Paylor R, McIlwain KL, McAninch R, Nellis A, Yuva-Paylor LA, Baldini A, Lindsay EA. Mice deleted for the DiGeorge/velocardiofacial syndrome region show abnormal sensorimotor gating and learning and memory impairments. Hum Mol Genet 2001; 10: 2645 2650. [DOI] [PubMed] [Google Scholar]
- Mukai J, Dhilla A, Drew LJ, Stark KL, Cao L, MacDermott AB, Karayiorgou M, Gogos JA. Palmitoylation-dependent neurodevelopmental deficits in a mouse model of 22q11 microdeletion. Nat Neurosci 2008; 11: 1302 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark KL, Xu B, Bagchi A, Lai WS, Liu H, Hsu R, Wan X, Pavlidis P, Mills AA, Karayiorgou M, Gogos JA. Altered brain microRNA biogenesis contributes to phenotypic deficits in a 22q11-deletion mouse model. Nat Genet 2008; 40: 751 760. [DOI] [PubMed] [Google Scholar]
- Grillo L, Reitano S, Belfiore G, Spalletta A, Amata S, Bottitta M, Barone C, Falco M, Fichera M, Romano C. Familial 1.1 Mb deletion in chromosome Xq22.1 associated with mental retardation and behavioural disorders in female patients. Eur J Med Genet 2010; 53: 113 116. [DOI] [PubMed] [Google Scholar]
- Lewandoski M, Meyers EN, Martin GR. Analysis of Fgf8 gene function in vertebrate development. Cold Spring Harb Symp Quant Biol 1997; 62: 159 168. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


