SUMMARY
Anchorage of tissue cells to their physical environment is an obligate requirement for survival which is lost in mature hematopoietic and in transformed epithelial cells. Here we find that a lymphocyte lineage-restricted transcription factor, Aiolos, is frequently expressed in lung cancers and predicts markedly reduced patient survival. Aiolos decreases expression of a large set of adhesion-related genes, disrupting cell-cell and cell-matrix interactions. Aiolos also reconfigures chromatin structure within the SHC1 gene, causing isoform-specific silencing of the anchorage reporter p66Shc and blocking anoikis in vitro and in vivo. In lung cancer tissues and single cells, p66Shc expression inversely correlates with that of Aiolos. Together, these findings suggest that Aiolos functions as an epigenetic driver of lymphocyte mimicry in metastatic epithelial cancers.
INTRODUCTION
Epigenetic dysregulation and instability occur at the earliest steps of tumorigenesis and accompany every stage of cancer progression. Consequent activation and/or silencing of tumor related genes confers premalignant epithelial cells with the capacity for unrestrained proliferation, resistance to cell death, evasion from immune destruction, and progression to frank malignancy (Mehlen and Puisieux, 2006; Timp and Feinberg, 2013; Valastyan and Weinberg, 2011). Despite its importance, specific epigenetic mechanisms by which evolving tumor cells acquire metastatic potential are poorly understood. In particular, processes by which widespread transcriptional reprogramming endows epithelial cells with hematopoietic characteristics allowing metastatic behavior are not clear.
Aiolos (encoded by IKZF3) is a member of the Ikaros zinc finger family whose expression is normally restricted to lymphoid cells (Morgan et al., 1997). Aiolos exerts broad changes in gene expression patterns, in large part through the recruitment of chromatin modifiers such as the nucleosome remodeling and deacetylase (NuRD) complex (Kim et al., 1999; Koipally et al., 1999). During lymphocyte development, Aiolos is first detected in pro-B and pro-T cells and is upregulated as these cells progress to pre-B and double positive CD4/CD8 cells, reaching maximum expression in mature, activated B and T cells (Wang et al., 1998). Aiolos facilitates the transition from pre-BCR-driven proliferation to differentiation and immunoglobulin light chain rearrangement, and plays important roles in late stages of B and T lymphocyte development (Ma et al., 2008; Mandal et al., 2009; Quintana et al., 2012; Reynaud et al., 2008; Thompson et al., 2007). Notably, this developmental stage also coincides with a sustained loss of lymphocyte adhesion to its matrix-rich microenvironment which is required for lymphocyte entry into the blood system and subsequent circulation to other organs (Glodek et al., 2003).
Aiolos expression has also been noted to be altered in both solid and liquid tumors. Aiolos is highly expressed and may promote survival in B-cell leukemia and certain lymphomas; conversely, Aiolos is absent in acute lymphoblastic leukemia (Antica et al., 2008; Billot et al., 2011; Duhamel et al., 2008; Holmfeldt et al., 2013; Niebuhr et al., 2013; Romero et al., 1999). Further, Aiolos has been detected in multiple malignant solid tumor cell lines including MCF-7, SW480, HEK, PC3 and HeLa (Billot et al., 2010). In the breast cancer cell line BT474, ectopic expression of Aiolos results from a fusion event leading to its transcriptional control by the VABP promoter (Edgren et al., 2011). In addition, a broad bioinformatic analysis of the human transcriptome in normal and abnormal tissues revealed upregulation of Aiolos in some breast cancers (Kilpinen et al., 2008). These reports suggest that Aiolos may be aberrantly expressed in some solid tumors; however, the functional consequence of Aiolos expression in carcinomas is completely unknown. Given the involvement of Aiolos in hematopoietic cell development, we hypothesized that Aiolos may promote the ability of cancer cells to survive in an unanchored state. Here, we study the molecular and clinical consequences of Aiolos expression by lung cancers.
RESULTS
Aiolos Expression Correlates with Poor Prognosisin Human Lung Cancer
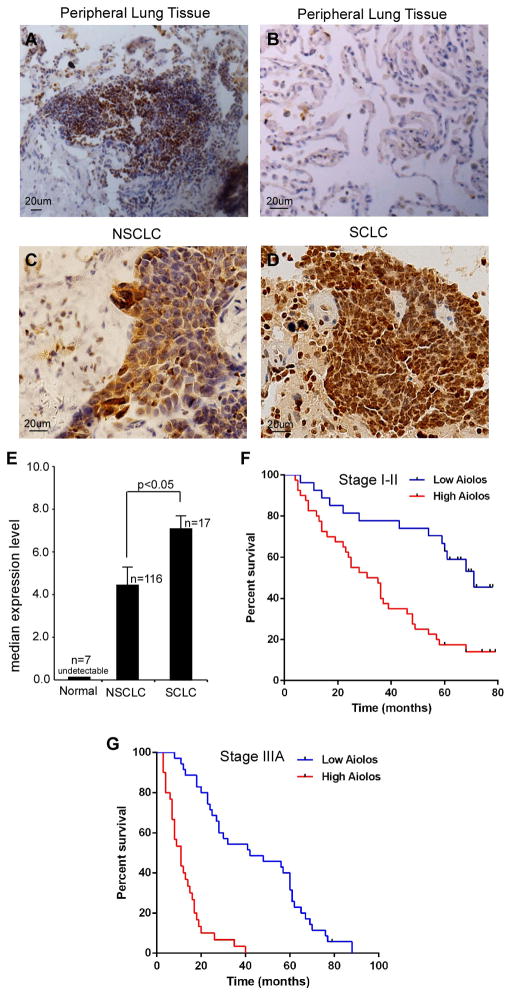
We studied Aiolos expression in 116 non-small cell lung cancers (NSCLC), 17 small cell lung cancers (SCLC), and 7 tumor-adjacent normal lung tissues, using immunohistochemical (IHC) analyses. Normal lung epithelial cells in tumor-adjacent lung tissues and stromal cells in lung cancer tissues did not express Aiolos. Lymphocytes in folliculi lymphaticus exhibited strong Aiolos staining (Figure 1A), consistent with previous reports (Wang et al., 1998), supporting antibody specificity. Both NSCLC (adenocarcinomas and squamous cell carcinomas) and SCLC cells expressed Aiolos to varying degrees; many Aiolos-positive NSCLC cells and all Aiolos-positive SCLC cells exhibited nuclear localization of Aiolos (Figures 1B–D). Quantification of staining on a scale of 0 to 8.0 revealed significantly higher expression of Aiolos in SCLCs than NSCLCs (Figure 1E). Of note, SCLC is associated with an extremely poor prognosis (median survival <2 yr) owing to its strong tendency to disseminate early and present with established metastatic foci (Fischer and Arcaro, 2008; Jackman and Johnson, 2005). Indeed, all 13 SCLC subjects with available survival data had high Aiolos staining scores (>4.0) and low survival rates (median survival 15 mo, Table S1). Thus the expression level of Aiolos correlates inversely with overall prognosis between histologic subtypes of lung cancers.
Figure 1.
Aiolos is expressed in lung cancer cells and predicts mortality. A–D. IHC staining with anti-Aiolos was performed on 7 normal lung tumor adjacent tissues, 116 NSCLCs and 17 SCLCs specimens. Scale bars are 20 μm. Representative fields show positive Aiolos staining in B lymphocytes in folliculi lymphaticus(A), lack of Aiolos staining in normal lung epithelial cells (B), moderate Aiolos staining in NSCLC cells (C) and strong Aiolos staining in SCLC cells (D). E. Semi-quantitative scoring of Aiolos staining showed significantly stronger staining in SCLC than NSCLC (p < 0.05). Error bars indicate SD. F, G. Kaplan-Meier survival rates for 67 stage I–II and 65 stage IIIA NSCLC subjects with low (0–4.0 staining scores, blue lines, n=27 for stage I–II, n=35 for stage IIIA), versus high (4.1–8.0 staining scores, red lines, n=40 for stage I–II, n=30 for stage IIIA) Aiolos expression were compared. Median survivals were 71 mo (low Aiolos) vs 33 mo (high Aiolos) for stage I–II (p=0.0003) and 42 mo (low Aiolos) vs 11 mo (high Aiolos) for stage IIIA subjects (p<0.001). See also TablesS1 –4.
To more closely assess the prognostic significance of Aiolos, we examined expression levels in resected NSCLC tumors from subjects with known clinical outcomes. Early stage (stage I–II, n=67) subjects whose tumors had low Aiolos expression levels (staining scores 0–4.0, n=27) had strikingly longer survival times than those whose tumors had high expression levels (staining scores 4.1–8.0, n=40), with median survivals of 71 months (low Aiolos) vs. 33 months (high Aiolos, p=0.0003). Of 65 stage IIIA subjects, median survivals were 42 mo with low Aiolos staining scores (n=35) and 11 mo with high Aiolos scores (n=30, p<0.0001). The survival rate of the high Aiolos expressors is poor and similar to that for extensive-stage disease SCLC. In a Kaplan-Meier model, Aiolos protein expression was a strong predictor of lung cancer patient survival rates for both stage I–II patients (Hazard ratio 2.95, CI 1.63–5.32, Figure 1F) and stage IIIA patients (Hazard ratio 11.26, CI 5.53–22.90, Figure 1G). Aiolos staining intensity did not correlate with age, TNM status, smoking history, or sex (Tables S2–S4). These results indicate that the hematopoietic lineage protein Aiolos is frequently expressed in lung cancers and its high expression level correlates with extremely poor survival rates in human lung cancer.
Aiolos Downregulates Cell Adhesion Genes
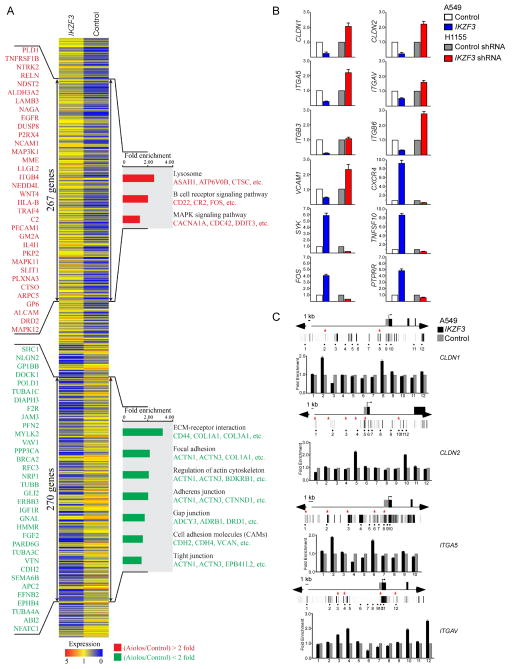
To explore the effect of Aiolos on established cancer cells, we expressed Aiolos in A549 cells, a lung carcinoma cell line which does not express endogenous Aiolos, and analyzed the resultant expression profile using a microarray platform. Functional classification of differentially expressed genes was performed, revealing ontologic gene groupings with high fold-enrichment and low false discovery rates. Within these enriched functional groups, two gene trees that significantly changed expression levels (>2-fold in either direction) after Aiolos/IKZF3 overexpression were identified. Tree 1 included 270 downregulated genes and tree 2 contained 267 upregulated genes upon Aiolos overexpression (Figure 2A). Functional groups most commonly downregulated represented genes which control cellular adhesion, including extracellular matrix-receptor interacting proteins, focal adhesion proteins, actin cytoskeleton regulating proteins, and cell adhesion molecules, with significant downregulation of adherens, tight, and gap junction proteins, indicating a broad repression of cell-matrix and cell-cell adhesion machinery. Individual Aiolos-regulated genes and subgroups of different molecular pathways are shown in TablesS5 –S7.
Figure 2.
Aiolos expression downregulates cell adhesion genes. A. Functional profiling of genes differentially expressed between control A549 cells and Aiolos/IKZF3-expressing A549 cells. 267 genes upregulated and 270 genes downregulated by Aiolos are marked by double-headed arrows. Representative Aiolos-induced (red) and Aiolos-repressed genes (green) are listed vertically (left) and under each molecular pathway (right). B. Quantitative RT–PCR was performed to confirm the transcriptional change of indicated genes identified in the microarray. RNA was purified from control or Aiolos/IKZF3 expressing A549 cells and control or Aiolos/IKZF3 shRNA expressing H1155 cells. Relative expression is shown as fold differences relative to GAPDH. C. ChIP was performed in A549 cells transfected with Aiolos/IKZF3 or vector. Gene names are indicated at right. Vertical lines indicate DNase I hypersensitive sites, red stars mark sites with Aiolos consensus sequences. Bar graphs show fold enrichment of Aiolos binding. Mean ± SD of 3 determinations is shown. See also TablesS5 –7.
We confirmed Aiolos-dependent differential transcription of 12 genes identified in the microarray analysis including downregulation of CLDN1, CLDN2, ITGB3, ITGB6, ITGAV, ITGA5, and VCAM1, which play important roles in mediating cell-matrix and cell-cell adherence, and upregulation of SYK, which participates in pre-B signaling and promotes tumor metastasis upon ectopic expression (Sung et al., 2009), and TNFSF10, CXCR4, FOS and PTPRR, which also promote cancer metastasis (Figure 2B). To confirm the role of Aiolos in gene regulation, we studied H1155 cells, a NSCLC cell line which expresses endogenous Aiolos. Knockdown of Aiolos in these cells caused reciprocal changes in expression levels of these genes compared with expression of Aiolos in A549 cells (Figure 2B).
To further study whether adhesion related genes are regulated directly by Aiolos, we determined the in vivo binding status of Aiolos within genomic loci of four Aiolos-repressed genes (CLDN1, CLDN2, ITGA5 and ITGAV) using ChIP. Cis-regulatory elements identified by DNase I hypersensitive sites, as reported in the ENCODE database, and potential Aiolos binding consensus sites were analyzed for Aiolos association (Figure 2C). ChIP was performed in A549 cells expressing Aiolos-FLAG. All four genes analyzed contained Aiolos consensus sites which were occupied by Aiolos (Figure 2C). These data suggest that Aiolos directly regulates key cell-matrix and cell-cell adhesion-related genes.
Aiolos Disrupts Cell-Cell and Cell-Matrix Adhesion
The shift in epithelial gene expression profile induced by Aiolos is consistent with its physiologic role in lymphocytes preparing for release into the circulation. Notably, we observed major differences in A549 cell morphology between control cells and Aiolos-expressing cells in regular 2D culture. While control cells spread well and grew as clusters, Aiolos expressing cells appeared rounded and exhibited less cell-cell interactions (Figure 3A). This morphological change is consistent with the decrease in expression of tight junctional proteins including Claudin 1, Claudin 2, and Occludin following Aiolos expression. Expression of E-cadherin was not affected; however, E-cadherin redistributed away from the cell periphery, indicating disruption of adherens junctions (Figure 3B). These data demonstrate that Aiolos may disrupt intercellular junctions through specific repression of key adhesion proteins.
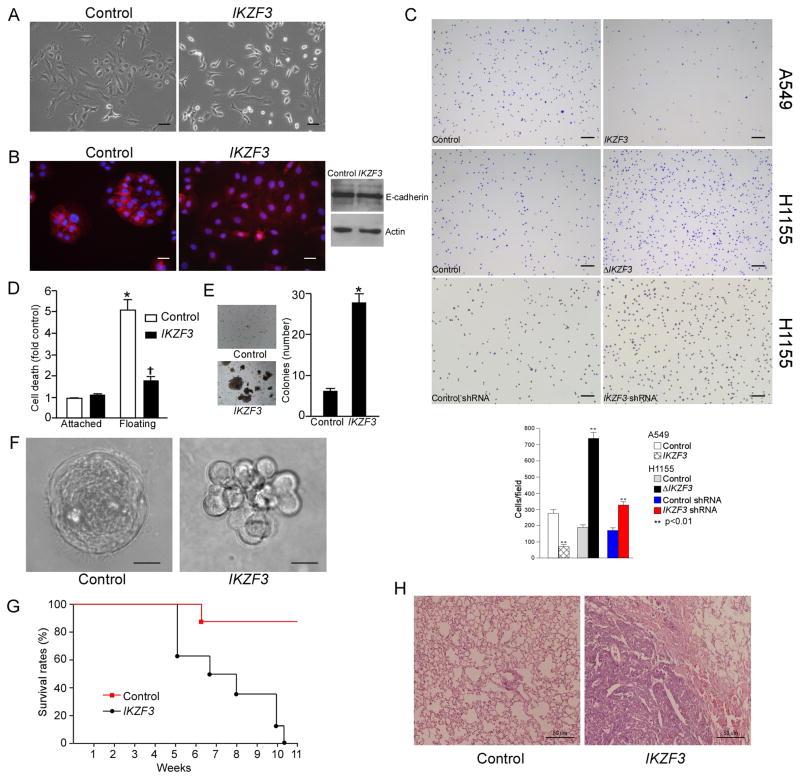
Figure 3.
Aiolos promotes cell detachment, anchorage independence, and lung metastasis. A. Phase-contrast micrographs of A549 cells expressing vector or Aiolos. Scale bars are 40 μm. B. Immunofluorescence and immunoblot were performed to examine the localization of E-cadherin (left two panels) and expression level (right panel) in Aiolos-expressing A549 cells. Scale bars are 20 μm. C. Aiolos/IKZF3 expressing A549 cells, Aiolos/IKZF3 shRNA expressing H1155 cells, or dominant negative Aiolos (ΔIKZF3) expressing H1155 cells were plated on fibronectin coated plates. After 15 min, attached cells were counted. Scale bars are 200 μm. Bar graph below shows number of adherent cells. Mean ± SD of 3 duplications is shown. D. HUVEC cells were transduced with Aiolos/IKZF3. Cell death was assessed after 16 h under attached or floating conditions. *p<0.05 from vector attached conditions; †p<0.05 from vector floating conditions. Error bars indicate SEM. E. A549 cells were transduced as indicated, allowed to grow in soft agar for 4 weeks, and colonies counted. Error bars indicate SD (n=3). F. Phase-contrast micrographs of control and Aiolos/IKZF3-expressing MCF10A acini cultured on Matrigel for 18 days. Scale bars are 15 μm. G. A549 cells expressing empty vector or Aiolos/IKZF3 were injected into the tail vein of 6-week-old female BALB/C nude mice. Survival curve is shown; survival between groups was different, p<0.001. H. Representative hematoxylin and eosin stain of lungs from mice injected with vector control and Aiolos/IKZF3-expressing A549 cells. Scale bars are 50 μm. See also Figure S1 and Table S8.
We then tested the effect of Aiolos expression on adherence of lung cancer cells to fibronectin, a major extracellular matrix component in lung and tumor stroma. In replating experiments, Aiolos expression dramatically decreased the number of A549 cells adherent to fibronectin (Figure 3C). Conversely, knockdown of Aiolos or overexpression of a dominant negative form of Aiolos (ΔAiolos/IKZF3, lacking the DNA-binding domain (Caballero et al., 2007))significantly increased the adhesion of H1155 cells to fibronectin (Figure 3C). These data are consistent with the downregulation by Aiolos of genes involved in at least three major fibronectin receptors, α4β1, α5β1, and αvβ3 (Table S5). However, SCLC cells (H209, H69), which float in culture, do not adhere following Aiolos knockdown (data not shown) despite increases in integrin and claudin gene expression (Figure S1A). Thus SCLC cells have a more profound adhesion defect and may lack other proteins not under Aiolos control but which are necessary for firm anchorage. Aiolos expression had no effect on cell proliferation or invasiveness and caused a slight decrease in cell motility, as evaluated by BrdU incorporation, Boyden chamber and wound healing assays(Figure S1B–D).
Aiolos Promotes Anchorage Independence
Similar to hematopoietic cells, metastatic cancer cells are able to survive in suspension and thus bypass anoikis, a form of apoptosis triggered by loss of anchorage to substratum. This led us to ask whether Aiolos is able to relax cellular anchorage requirements. We found that transient expression of Aiolos by primary endothelial cells, which are highly sensitive to anoikis, effectively blocked anoikis (Figure 3D). We then examined the effect of Aiolos on anchorage-independent proliferation. We expressed Aiolos in A549 cells and found that Aiolos significantly increased anchorage-independent growth in soft agar (Figure 3E).
As a third test of anchorage sensing, we studied the ability of MCF10A cells to form acinus-like spheroids in three dimensional basement membrane gels, which requires loss of inner cell mass through anoikis (Debnath et al., 2002; Han et al., 2008). Whereas control cells developed into well-formed hollow spheroids, Aiolos expression resulted in disrupted acinus formation with luminal filling and lack of polarity (Figure 3F). These results indicate that Aiolos promotes anchorage independence in primary endothelial cells, immortalized epithelial cells, and transformed cancer cells. Thus besides decreasing adhesion mechanisms, Aiolos protects cells from anoikis following loss of anchorage.
Anchorage independence is essential for survival of circulating cancer cells during metastasis (Valastyan and Weinberg, 2011). To examine the effect of Aiolos expression on metastatic capacity in vivo, we injected A549 cells into the tail vein of BALB/C nude mice. Aiolos-expressing A549 cells caused lethality in 8/8 mice by 77 days, with a median survival of 48 days (Figures 3G, H). In all cases, death was accompanied by a heavy lung metastatic burden, verified by histologic examination (Figure 3H). Mice injected with control A549 cells had longer survival times, with only 1/8 mice dying from lung metastases by 77 days. Therefore in parallel with loss of anoikis in vitro, ectopic expression of Aiolos promotes metastatic capacity of lung cancer cells in vivo.
We expanded our analysis to include solid tumor cell lines from breast, liver, and colon, and found Aiolos to be expressed in 5/10 cell lines examined (Figure S1E). Further, Aiolos expression correlated with the reported ability of these cells to metastasize in nude mice with only one exception (Table S8). Consistently, knock down of Aiolos in these Aiolos-expressing cells also lead to increased adhesion (Figure S1F). To further correlate these findings with human data, The Cancer Genome Atlas (TCGA) was interrogated for breast cancer outcomes. Analysis of this dataset revealed that high Aiolos/IKZF3 expression correlates with poor survival rates in breast cancer (Figure S1G). In total, these data suggest that Aiolos may be of general significance to anchorage mechanisms in tumors other than lung cancers.
Expression of Aiolos Negatively Correlates with p66Shcin Human Lung Cancers
Of particular note given the prominent effects of Aiolos in promoting anchorage independence, we observed that the adaptor protein Shc was downregulated 2.3-fold by Aiolos expression. SHC1 contains tandem promoters which are thought to independently control p52Shc and p66Shc transcription (Figure 4A), though very little is known about control of either promoter (Ventura et al., 2002). p66Shc is required for anoikis and functions as a powerful metastasis suppressor in mice, whereas the shorter p52Shc does not (Ma et al., 2010). We found that expression of Aiolos in A549 cells selectively repressed p66Shc but not p52Shc expression (Figure 4A). Transient expression of Aiolos also selectively decreased p66Shc protein levels in A549, HepG2, and MDA-MB-231 cells, confirming an inhibitory effect of Aiolos on p66Shc expression (Figure 4B). However, knockdown of endogenous Aiolos did not restore p66Shc expression in SCLC (H209, H69, H82) as well as NSCLC (H1155) or non-lung carcinoma (Hep3B, MCF7) cell lines (Figure S2A). Accordingly, knockdown of Aiolos did not increase sensitivity to anoikis in H209, H69, and H82 lung cancer cells (Figure S2B). Thus while Aiolos is sufficient to repress p66Shc, it does not appear to be necessary in established cancer cells. To study this effect further, A549 cells were stably transfected with an inducible Aiolos transgene. Upon induction of Aiolos by doxycycline treatment for 8h, p66Shc protein levels markedly decreased; however, removal of doxycycline for eight subsequent days caused early loss of Aiolos expression but no recovery of p66Shc levels (Figure S2C). These results indicate that Aiolos may be necessary for the initiation but not the maintenance of p66Shcsilencing.
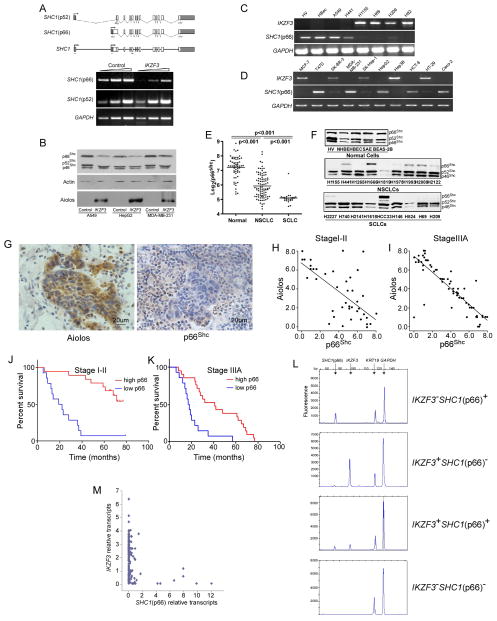
Figure 4.
Aiolos negatively correlates with p66Shcexpression. A. Semi-quantitative RT-PCR was performed for p52Shc and p66Shc on control or Aiolos/IKZF3-expressing A549 cells. Upper panel shows splicing diagram of two separate mRNA transcripts for SHC1. Shaded areas are 5′ and 3′ untranslated sequences. Arrows show location of PCR primers. Lower panel shows PCR products of serial threefold dilutions of cDNA. B. Transient expression of Aiolos/IKZF in A549, MDA-MB-231 and HepG2 cells downregulated p66Shc expression by immunoblot. C, D. Expression of Aiolos/IKZF3 and p66Shc is shown for normal (HV, HBec) and lung cancer (C) or breast, liver, and colon cancer (D) cell lines as indicated. E. Expression of p66Shc was assessed from a probe specific for the Shc CH2 domain. Log base 2 of the signal strength is shown. Expression of p66Shc was lower (p < 0.001) in NSCLC than normal bronchial epithelial cells and lower (p < 0.001) in SCLC than NSCLC or normal cells. F. Immunoblot of Shc in normal (top panel), NSCLC (middle panel), and SCLC (lower panel) cells showing relative expression of 66, 52, and 46 kDa forms. G. IHC for Aiolos and p66Shc was performed on 105 lung specimens. Representative fields show lack of Aiolos staining in lung parenchymal cells with variable staining in intra-alveolar mononuclear cells, compared with strong staining in malignant cells. Antibodies to p66Shc variably stain alveolar septal wall cells; malignant cells in the center of the field are negative for p66Shc staining. H, I. Semi-quantitative scoring was performed and Aiolos scores were correlated with p66Shc scores in stage I–II (panel H, r2 = 0.451, p < 0.0001) or stage IIIA (panel I, r2 = 0.719, p < 0.0001) tumors. J, K. Kaplan-Meier survival curves based on p66Shc staining for stage I–II (J) and stage IIIA (K) subjects. Median survivals in stage I–II subjects were 22 mo low and >80 mo for high p66Shc staining (p< 0.0001). Median survivals in Stage IIIA subjects were 11 mo for low and 42 mo for high p66Shc staining (p <0.0001). L. Representative capillary electropherograms showing PCR products of Aiolos/IKZF3, p66Shc, GAPDH and KRT19. M. Quantitative analysis was performed to show the correlation of Aiolos/IKZF3 and p66Shc. Each dot represents one single cancer cell. See also Figure S2 and Table S9.
In strong support of p66Shc repression by endogenously expressed Aiolos, we noted in a panel of 18 human cell lines of diverse origin a strict correlation between the expression of Aiolos and repression of p66Shc (Figure 4C, D) but not p52Shc (data not shown). In contrast, all cells without detectable Aiolos expression retained p66Shcexpression (Figure 4C, D).
To assess whether p66Shc is commonly repressed in lung cancer, we screened 207 human lung epithelial cell lines for expression of p66Shc. Normal lung epithelium consisted of human bronchial epithelial cells (HBEC, n=30) and small airway epithelial cells (HSAEC, n=29). Expression of p66Shc was robust and not different between small and large airway epithelium (p > 0.05). Cells derived from lung cancers (n=148), however, had significantly reduced expression compared with normal lung epithelium (p<0.001, Figure 4E). Notably, p66Shc expression was significantly lower in SCLC than in NSCLC, and each group was decreased compared to non-malignant human bronchial epithelium (p<0.001, Figure 4E). Expression at the level of protein in a subset of cells confirmed consistently robust levels of p66Shc in normal endothelial or lung epithelial cells (Figure 4F). In contrast, p66Shc protein levels were diminished in most NSCLC and SCLC cell lines, despite persistence of p52Shc expression. This expression pattern for p66Shc is opposite from that which we found for Aiolos and suggests that repression by Aiolos is biologically relevant.
To define the correlation between Aiolos and p66Shc in individual primary tumors, we used IHC to examine the expression levels of Aiolos and p66Shc in serial sections of the previously-characterized NSCLC lung cancer samples. Tumors with significant levels of staining for Aiolos showed low levels of p66Shc staining; conversely, surrounding lung parenchyma was negative for Aiolos but positive for p66Shc (Figure 4G). When intratumoral staining was quantified, a highly significant negative correlation was found between Aiolos and p66Shc expression in both stage I–II (r2=0.451, p<0.0001) and stage IIIA (r2=0.719, p<0.0001) tumors (Figure 4H, I). Accordingly, low p66Shc expression correlated with worse survival rates in both stage I/II and stage IIIA groups (Figure 4J, K). Median survivals in stage I–II subjects were 22 mo for low and >80 mo for high p66Shc staining (p<0.0001). Median survivals in stage IIIA subjects were 11 mo for low and 42 mo for high p66Shcstaining (p<0.0001).
Because of limitations associated with serial section analysis and cell-to-cell heterogeneity in protein expression within tumors, we sought more definitive evidence for biologically relevant repression of p66Shc by Aiolos in human lung cancers through single cell analysis. Single cancer cells were isolated from four freshly dispersed human NSCLC tumors for single cell RT-PCR. Expression of Aiolos/IKZF3, p66Shc, KRT19 (cytokeratin 19, a biomarker for NSCLC cells) and GAPDH were determined in a single PCR reaction. Figure 4L shows examples of capillary electropherograms for the four possible qualitative patterns (±Aiolos/IKZF3, ±p66Shc). A total of 198 cells isolated from 4 human lung cancer tissues expressing KRT19 and GAPDH were further analyzed. Cancer cells purified from tumor 1 expressed p66Shc but not Aiolos/IKZF3, whereas the majority of cancer cells isolated from the other three tumors expressed Aiolos/IKZF3 but repressed p66Shc (Table S9). Quantification of transcript levels revealed a negative correlation between Aiolos/IKZF3 and p66Shc expression (r2= −0.239, p<0.001) (Figure 4M). This correlation appeared biphasic, suggesting that at the single cell level Aiolos controls a binary decision, strongly repressing p66Shc when expressed above a certain threshold level and becoming permissive below this level. Altogether, these results indicate that expression of endogenous Aiolos strongly represses p66Shc in multiple carcinomas in vitro and in tumors and single cancer cells in vivo.
Aiolos Confers Anoikis Resistance Throughp66 Shc Repression
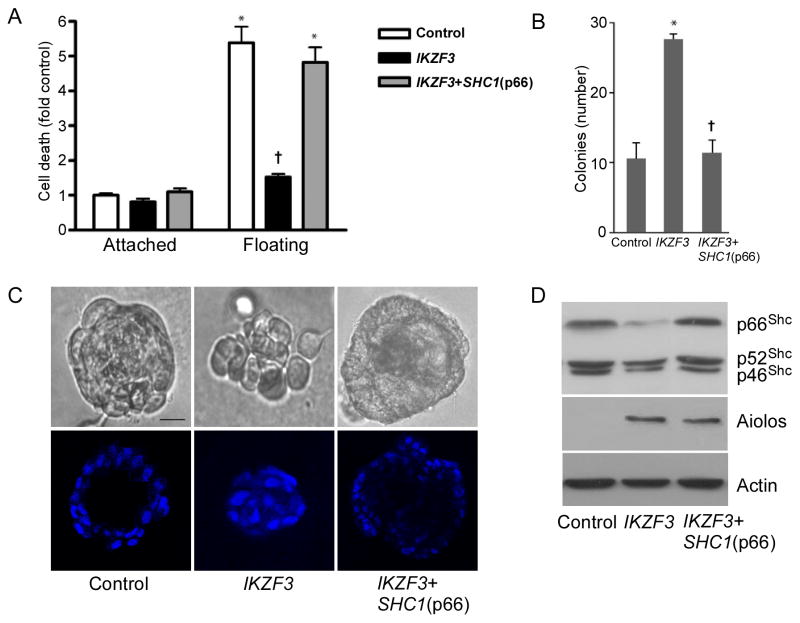
To assess whether Aiolos abrogates anchorage dependence through p66Shc repression, we re-expressed p66Shc in Aiolos-transfected primary endothelial cells, A549 cells and MCF10A cells. Indeed, re-expression of p66Shc completely restored anoikis in primary endothelial cells (Figure 5A), decreased anchorage independent growth of A549 cells in soft agar to baseline levels (Figure 5B), and restored the ability of MCF10A to form differentiated spheroids (Figures 5C, D). These results demonstrate that Aiolos represses p66Shc expression and thus promotes anchorage independent behavior, offering an explanation for the poor prognostic association of Aiolos with lung cancer mortality rates.
Figure 5.
Aiolos confers anchorage independence through p66Shc silencing. A. HUVEC were transduced with Aiolos/IKZF3 or both Aiolos and p66Shc as indicated. Cell death was assessed after 16 h under attached or floating conditions. *p < 0.05 from vector attached conditions; †p < 0.05 from vector floating conditions. Error bars indicate SEM. B. A549 cells were transduced as indicated, allowed to grow in soft agar for 4 weeks, and colonies counted. Error bars indicate SD (n=3). C. MCF10A cells were transduced as indicated and plated in Matrigel. Phase contrast and confocal midpoint slices of DAPI stained acini are shown. Scale bars are 15 μm. F. Immunoblots of MCF10A cells for Shc, Aiolos, and actin under conditions shown as in E. See also Figure S3.
To assess the specificity of this effect for Aiolos, we tested the expression of other Ikaros family members including Ikaros (IKZF1), Helios (IKZF2), Eos (IKZF4), and Pegasus (IKZF5) in normal endothelial and epithelial cells and multiple cancer cell lines. Surprisingly, Helios, Pegasus, and Eos were expressed in all 18 cell lines analyzed (Figure S3). The significance of this broad expression pattern is not clear, though we did not find any correlation with p66Shc repression. Ikaros is structurally most similar to Aiolos, and was expressed in only four cell lines (Figure S3). Three of these cell lines also expressed Aiolos and repressed p66Shc, and one (HCT-8) expressed Ikaros but not Aiolos, with weaker but distinct expression of p66Shc (Figure 4D). While it is possible that Ikaros has repressive effects on p66Shc similar to Aiolos, Aiolos appears to be more commonly expressed and correlates more strongly with p66Shc repression across all cell lines.
Aiolos Represses p66Shc Transcription by Disrupting Long Range Enhancer-Promoter Interactions
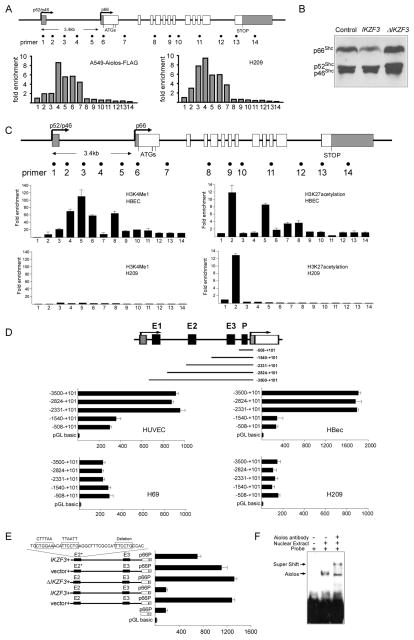
We then explored the mechanism by which Aiolos silences p66Shc. Using bisulfite sequencing, we detected variable DNA methylation of the p66Shc promoter region in SCLC cells (Figures S4A, B). ChIP analyses to scan 14 regions along the SHC1 gene for histone marks revealed decreased association of activating histones (H3K9acetyl, H3K4me2 and H3K4me3) and enrichment of repressing histones (H3K9me2) in the p66Shc promoter region in SCLC cells, indicating epigenetic silencing of p66Shc (Figure S4C, D). Of note, repressive histones were enriched primarily in a 3.5 kb region between the two promoters in SCLC cells, suggesting a chromatin domain specifically controlling p66Shc expression. Further ChIP studies revealed the occupancy of this region, including the p66Shc promoter, by endogenous Aiolos in SCLC cells, as well as by transfected Aiolos in A549 cells (Figure 6A). Transient expression of ΔAiolos/IKZF3 failed to repress p66Shc transcription (Figure 6B), indicating that Aiolos requires direct DNA binding, possibly to this upstream cis-regulatory element, to silence p66Shctranscription.
Figure 6.
Aiolos directly regulates p66Shctranscription. A. ChIP demonstrates association of Aiolos-Flag in transfected A549 cells with regions 4–7, indicated in the upper diagram. Right panel shows similar association of native Aiolos with regions 2–7 in H209 SCLC cells. B. p66Shc expression was examined in A549 cells expressing wild type Aiolos/IKZF3 or ΔAiolos/IKZF3. C. ChIP shows distribution of H3K4me1 and H3K27ac histone modifications in HBEC (p66Shc-expressing) and H209 (p66Shc-nonexpressing) cells. Error bars are SD of three independent chromatin preparations. D. Diagram shows serial deletions of the luciferase reporter upstream of the p66Shc TSS; bar graphs show luciferase activity. E. Luciferase reporter studies with the −2,824– +101 segment containing E2 and E3. Transient expression of Aiolos/IKZF3, but not ΔAiolos/IKZF3, abrogated p66Shc promoter enhancement. p66P denotes the p66Shc basal promoter. Triple mutation of three Aiolos consensus sites within E2 impaired Aiolos-induced loss of enhancer activity (top bar). Normalized signals for D and E represent mean ± SD of triplicates. F. EMSA shows mobility shift of probe with E2 sequence containing the first two Aiolos consensus sites, with supershift following anti-Aiolos treatment. See also Figure S4.
Alignment of human, mouse and rat p66Shc genes revealed a 113 bp conserved sequence residing within the noncoding region 2.1 kb upstream from the p66Shc transcriptional start site (TSS), marking one potential cis-regulatory enhancer (site 4, Figure 6C). As a second method to screen for enhancers, we used ChIP to scan the SHC1 gene for H3K27ac and H3K4me1, modified histones which are frequently associated with human enhancers (Heintzman et al., 2007; Rada-Iglesias et al., 2011). We identified two regions enriched with H3K27ac in normal bronchial epithelium, again within the putative p66Shc-regulatory domain (sites 2 and 5, Figure 6C). Site 5 but not site 2 was co-occupied by H3K4me1. In H209 SCLC cells by contrast, only the H3K27ac modification was recovered from site 2 and neither modification from sites 4 or 5 (Figure 6C).
We then tested the function of sites 2, 4, and 5 (renamed E1, −3,474– −3,311; E2, −2,132– −2,019; and E3, −779– −610 from the p66Shc TSS) as potential enhancers of the p66Shc promoter. Luciferase reporters transfected into both HUVEC and HBEC, which express p66Shc, showed basal promoter activity contained within the −508– +101 fragment, as expected (Figure 6D). Serial extensions of this fragment demonstrated that inclusion of E2+E3 but not E3 alone significantly enhanced basal promoter activity, and inclusion of E1 had no further effect. Of significance, the basal promoter had equivalent activity when transfected into H69 and H209 SCLC cells, but no enhancement was observed upon inclusion of E1, E2, or E3 (Figure 6D). As these cells express no detectable p66Shc, enhancer rather than promoter activity appears to determine functional p66Shc expression. Cotransfection of wild type but not ΔAiolos/IKZF3 with the luciferase reporter fully repressed enhancer activity (Figure 6E), indicating that DNA binding is necessary for Aiolos to inhibit enhancer activity. Three Aiolos consensus sites were identified within E2 and one within the p66Shc promoter. While mutation of each of these single binding sites in a luciferase reporter had no effect (Figure S4E), mutation of all three sites within E2 significantly blocked Aiolos-dependent inhibition of enhancer activity (Figure 6E). Further, EMSA with supershift showed direct binding of Aiolos to this sequence (Figure 6F). These data support a model whereby Aiolos suppresses p66Shc transcription through DNA binding within a cis-regulatory upstream region with consequent deactivation of p66Shcenhancer function.
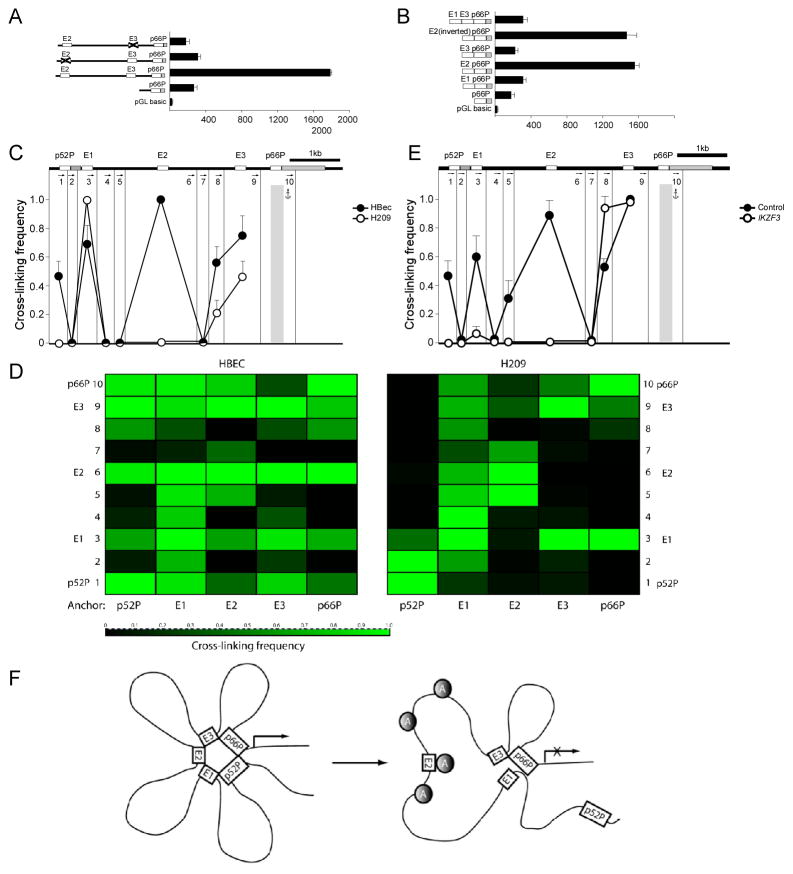
Further luciferase studies revealed that in their native positions, both E2 and E3 were necessary for enhancer activity, whereas deletion of either abrogated enhancement (Figure 7A). In contrast, ectopic placement of E2 just upstream of the promoter, in forward or inverted orientation, restored full enhancer activity, whereas E1 and E3, alone or in combination, had no effect (Figure 7B). Thus E2 harbors the principal enhancer activity for the p66Shc promoter. As relocalization of E2 to a position adjacent to the promoter obviated the requirement for E3, E3 may promote physical interactions between E2 and the p66Shc promoter which are required for promoter activation.
Figure 7.
Aiolos disrupts E2-p66Shc promoter interactions. A. Luciferase reporter activity is shown with deletion of either E2 or E3. B. Ectopic placement of the candidate enhancers adjacent to the p66Shc promoter was performed as shown. C–E. 3C was used to calculate cross linking frequency between chromatin segments to assess proximity in HBEC (p66Shc-expressing, solid circles) and H209 (p66Shc-nonexpressing, open circles) cells. Vertical lines represent Dpn II restriction sites; arrows indicate PCR primer sites and direction. Anchor symbols mark anchoring primer for each data set. Cross linking frequency between the p66Shc promoter and other Dpn II-defined segments is shown in C. Mean ± SD of 3 independent chromatin preparations is shown. D. Heatmaps of cross-linking frequency in HBEC and H209 cells. Anchoring primer is listed below, Dpn II-defined segments numbered as in C. E. Cross linking frequency is shown using the p66Shc promoter as anchor in A549 cells transiently expressing Aiolos (open circles) or vector control (solid circles). Mean ± SD of 3 independent experiments is shown. F. Schematic showing formation of a transcriptionally active complex containing p52Shc /p66Shc promoters and E1–E3 regions. Upon binding of Aiolos (shaded circles) to E2 and surrounding regions, the complex is disrupted and p66Shcis silenced. See also Figure S5.
To more directly study the importance of intragenic chromatin interactions in p66Shc regulation, we employed chromosome conformation capture (3C) to examine the physical proximity of enhancer and promoter elements. In brief, cross-linked chromatin was digested with Dpn II, diluted, re-ligated, and long-range association frequencies assessed with PCR. When the p66Shc promoter fragment was used as a PCR anchor, very strong associations were seen between the p66Shc promoter and the E1, E2, E3, and p52Shc promoter regions in p66Shc-expressing normal human bronchial epithelial cells (HBEC) (Figure 7C). With the E1, E2, E3 and p52Shc promoter fragments as anchors, strong interactions were confirmed between all three candidate enhancers and both p66Shc and p52Shc promoter regions, and between the two promoters in HBEC (Figures S5A–D). Such extensive interactions between the five cis-acting sites are seen in a heat-map representation of cross-linking frequencies across chromatin domains (Figure 7D). Thus in p66Shc-expressing cells, we detect a higher-order complex containing both promoters and all three candidate enhancers.
In H209 SCLC cells, repression of p66Shc was associated with persistence of interactions between E1 and the p66Shc promoter (Figure 7C), and between E3 and E1 (Figures 7D, S5A–B), consistent with the enrichment of H3K27ac in E1 in H209 cells (Figure 6C). However, interactions between E2, the p52Shc promoter, and the p66Shc promoter were lost (Figures 7C–D, S5C–D). Importantly, transient expression of Aiolos by A549 cells caused delocalization of E1, E2, and the p52Shc promoter from the p66Shc promoter (Figure 7E), leading to a chromatin structure resembling that seen in H209 cells. We next studied the persistence of SHC1 structural remodeling following transient Aiolos expression using the doxycycline-inducible model. Eight days after doxycycline withdrawal and the disappearance of Aiolos, E2 remained strongly dissociated from the p66Shc promoter, while the dislocation of E1 from the p66Shc promoter was reversed (Figure S5E). The stability of this chromatin reconfiguration with respect to the E2 enhancer matches the durable silencing of p66Shc after Aiolos knockdown. These data support a model whereby p66Shc transcription requires long-range physical interaction between the primary enhancer E2 and the p66Shc promoter. Aiolos associates with multiple upstream sites including an E2-containing chromatin domain, altering higher order chromatin structure, disrupting enhancer-promoter interactions, and silencingp66 Shc transcription (Figure 7F).
DISCUSSION
Metastatic spread of tumors accounts for the vast majority of cancer-related deaths; here, we describe an integrated cellular program which enables transformed epithelium to gain metastatic properties through lymphocyte mimicry. This process requires cells to downregulate proteins which constitute cell-cell and cell-matrix adhesion complexes and in addition, to acquire anchorage independence. In part, this cellular behavior resembles normal lymphocyte development as pro- and pre-B cell progenitors prepare to leave the hematopoietic niche. During this transition, B lymphocytes lose their ability to form sustained adhesions to bone marrow matrix, and are capable of only transient adhesions as circulating mature cells (Glodek et al., 2003). Accordingly, this lymphocyte maturation phase is accompanied by broad shifts in the expression of integrins and other adhesion proteins, with a preponderance of downregulated adhesion-related genes moving from the early B through the pro- and pre-B cell stages to immature B lymphocytes (Hystad et al., 2007).
Aiolos expression in lymphocytes parallels this progression and peaks in mature B and T cells. Consequently, loss of Aiolos results in an increased pre-B population within the bone marrow environment with diminished recirculating B lymphocytes (Wang et al., 1998). While temporally correlated with changes in lymphocyte adhesion properties, it is not clear whether Aiolos expression directly modulates adhesion-related genes in lymphocytes. However, it is particularly striking that a major effect of ectopic Aiolos expression in lung cancer cells is the repression of a large number of adhesion-related genes. This shift in expression profile suggests that Aiolos may generally control a broad transcriptional program that allows hematopoietic, or in an abnormal context, epithelial cell departure from a developmental niche to gain entry into the circulation.
Beyond the loss of durable cell-matrix adhesion, Aiolos may confer other hematopoietic properties important for the acquisition of metastatic ability by epithelial cells. First, Aiolos downregulates claudins and occludins and disrupts cell-cell junctions. Epithelial polarity and differentiation are maintained by intact tight and adherens junctions, and disruption of such intercellular junctions induces epithelial-to-mesenchymal transition and metastatic behavior (Derksen et al., 2006; Onder et al., 2008). The mechanisms linking such loss of polarity to oncogenic transformation involves changes in gene expression patterns induced by alterations in Wnt, TGF-β, Hippo, and TBX2 pathways (Frisch et al., 2013; Kumar et al., 2011).
Second, while release from a solid matrix environment is a prerequisite for metastatic spread, circulating tumor cells must also be able to survive such a loss of anchorage, which normally triggers anoikis (Frisch and Francis, 1994). Accordingly, loss of junctional integrity has also been shown to suppress anoikis, which is critical for metastatic capacity (Derksen et al., 2006; Kumar et al., 2011; Onder et al., 2008). Of importance, Aiolos also silences p66Shc, a Shc isoform not expressed by hematopoietic cells. SHC1 gives rise to both p66Shc and p52Shc, proteins with divergent cellular functions. p52Shc is recruited to integrins and couples mitogenic stimulation of Ras and Rac1 to integrin ligation, thus providing anchorage context to proliferative signaling pathways (Mainiero et al., 1997; Wary et al., 1996). In contrast, p66Shc inhibits growth factor signaling, restrains Ras and Rac1, and induces anoikis (Giorgio et al., 2005; Ma et al., 2010; Ma et al., 2007; Xi et al., 2010). Whereas p52Shc permits anchorage-independent growth (Pelicci et al., 1992), p66Shc enforces anchorage dependence and prevents metastatic behavior in mice (Ma et al., 2010; Ma et al., 2007). Within the SHC1 gene, we find that Aiolos selectively silences p66Shc but not p52Shc. Thus p66Shc represents a critical target for Aiolos-related silencing in metastatic cancers, confirmed by the ability of p66Shc to reverse Aiolos-induced anchorage independent behavior in vitro. However, upon Aiolos knockdown, we also find that cell-matrix adhesion and the expression of adhesion-related genes increase, whereas p66Shc remains deeply silenced. This implies that Aiolos exerts both p66Shc-dependent and p66Shc-independent effects in its ability to alter anchorage properties.
A third characteristic of circulating lymphocytes assumed by metastatic solid tumors is their ability to home back to specific vascular and lymphatic beds once in the circulation. In this regard, it is noteworthy that Aiolos causes a marked induction of CXCR4, a homing chemokine receptor involved in directing hematopoietic cells to bone marrow, lymph nodes, lung, and other vascular beds commonly targeted by metastatic solid tumors. While the full significance of this effect has yet to be explored, CXCR4 expression has been shown to correlate strongly with both metastatic activity and mortality in breast, lung, and other carcinomas (Hermann et al., 2007; Jung et al., 2013; Muller et al., 2001).
In summary, this study reveals a central mechanism by which epithelial carcinomas acquire certain hematopoietic characteristics to gain metastatic ability, through abnormal expression of a key epigenetic modifier required for lymphocyte development and function. Expression of Aiolos leads to a molecular form of hematopoietic “identity theft” which allows solid tumors to mimic cellular behaviors normally involved in lymphocyte trafficking during immune surveillance. The high prevalence of Aiolos expression in human lung cancers and its strong inverse correlation with the metastasis suppressor p66Shc in primary tumors and single cancer cells substantiates the clinical significance and both prognostic and therapeutic implications of this mechanism.
EXERIMENTAL PROCEDURES
Immunohistochemical Analysis
Serial tissue sections were prepared from archived formalin-fixed paraffin-embedded human lung cancer specimens and quantitative scoring of tissue sections was performed in a blinded fashion. Percentage of positive cells and staining intensity were scored separately and contributed to the aggregate score. The use of all human lung cancer tissues and clinical data was approved by the Institutional Review Board of Tianjin Medical University. Informed consent was obtained from all patients, and samples were deindentified prior to analysis. Kaplan-Meier survival curves were analyzed using Mantel-Cox log-rank tests, and hazard ratios calculated using Mantel-Haenszel tests (GraphPad Prism 6.02). Further details and references are provided in Supplemental Information.
Expression Profiling
The expression and functional profiles of genes were compared between A549 cells expressing empty vector and Aiolos/IKZF3 using Agilent SurePrint G3 Human Gene Expression 8x60K v2 Microarray (Agilent Technologies, Palo Alto, CA, USA). Analysis of functional profiling of Aiolos-regulated genes was performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID) based on the biological pathways from KEGG (Kyoto Encyclopedia of genes and genomes) database. Expression profiling for human lung cancer cell lines was performed using the Illumina Human WG-6 V3 chip on 119 NSCLC, 29 SCLC, 30 HBEC, and 29 HSAEC cell lines. Arrays were background-corrected, quantile-normalized and log-transformed. Further details and references are provided in Supplemental Information.
Anchorage-independent Cell Behavior
Anoikis was assessed by measuring cell death (DNA fragmentation, Roche) after 16 h of culture on cell culture-treated or low attachment plates (Corning). For anchorage-independent growth, 104 A549 cells were suspended in 0.35% agarose the poured over 0.6% agar in 6-well plates. Colonies were counted after 4 weeks. To generate acini, MCF10A cells were grown in reconstituted basement membrane (Matrigel, BD). All animal procedures were approved by Animal Care and Use Committee at Tianjin Medical University and conform to the legal mandates and national guidelines for the care and maintenance of laboratory animals. Further details and references are provided in Supplemental Information.
Single Cell RT-PCR
Fresh human lung tumor surgical samples were trimmed free of adventitial tissue, minced and digested with collagenase to release cancer cells. Single cancer cells were isolated and transferred into freshly prepared cell lysis buffer for RT-PCR. Amplification of SHC1(p66), IKZF3, GAPDH and KRT19 was performed in a single tube, and products quantified by capillary electrophoresis. Details of the protocol and sequences of primers used are provided in Supplemental Information.
Chromosome Conformation Capture
Cells (1 × 106) were cross-linked, lysed, and nuclei were digested with Dpn II. After ligation and subsequent DNA purification, the cross-linking frequencies between the anchor and test fragments were estimated by PCR reactions relative to standards. A PCR product from 6kb upstream of the p66Shc promoter was ligated at high concentration following Dpn II digestion was used to generate standards. The cross-linking and ligation efficiencies between different samples and different experiments were normalized by setting the highest cross-linking frequency for each experimental series to 1.0. Error bars represent the SDs from the means from three to five independent experiments as indicated in the figure legends. Experimental details and primer sequences used in this study are provided in Supplemental Information.
Supplementary Material
HIGHLIGHTS.
Aiolos downregulates multiple cell-cell and cell-matrix adhesion genes
Aiolos epigenetically silences p66Shcand promotes metastatic behavior
p66Shccorrelates inversely with Aiolos in human lung tumors
Expression of Aiolos worsens clinical outcome in subjects with lung cancer
SIGNIFICANCE.
Epigenetic dysregulation is now believed to drive all stages of tumorigenesis in association with key genetic mutations. However, specific epigenetic factors which broadly reprogram cells to allow anchorage independence and metastatic spread—cellular behaviors which are chiefly responsible for cancer mortality—are poorly studied. In this report, we find that carcinoma cells co-opt a hematopoietic factor required for lymphopoiesis to survive anchorage deprivation through repression of SHC1 and other genes. These findings have broad implications for the ability of epithelial tumors to acquire certain hematopoietic characteristics.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (grant 2014CB910100 to Z.L. and 2009CB918903 to Z.Y.), the National Natural Science Foundation of China (grants 31371295, 91019012, 31071128 to Z.L., 81372307, 81071730 to Z.M., 81101546 to X.L.), the Tianjin Municipal Science and Technology Commission (grants 11JCZDJC19000 to Z.L., 11JCZDJC18700 to Z.M. and 13JCQNJC10300 to X.L.), specialized Fund for the Doctoral Program of Higher Education (grant 20121202110001 to Z.L.), the China Postdoctoral Science Foundation (grant 20110490792 to X.L.), the James M. Collins for Biomedical Research and the National Institutes of Health (grants R01-HL067256 and R01-HL061897 to L.S.T.).
Footnotes
Accession Numbers
A549 cell expression profile data have been entered on the GEO website under accession number GSE50812. Human lung cancer cell line expression profiles have been entered under the accession number GSE32036.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Antica M, Cicin-Sain L, Kapitanovic S, Matulic M, Dzebro S, Dominis M. Aberrant Ikaros, Aiolos, and Helios expression in Hodgkin and non-Hodgkin lymphoma. Blood. 2008;111:3296–3297. doi: 10.1182/blood-2007-12-125682. [DOI] [PubMed] [Google Scholar]
- Billot K, Parizot C, Arrouss I, Mazier D, Debre P, Rogner UC, Rebollo A. Differential aiolos expression in human hematopoietic subpopulations. Leukemia Res. 2010;34:289–293. doi: 10.1016/j.leukres.2009.05.016. [DOI] [PubMed] [Google Scholar]
- Billot K, Soeur J, Chereau F, Arrouss I, Merle-Beral H, Huang ME, Mazier D, Baud V, Rebollo A. Deregulation of Aiolos expression in chronic lymphocytic leukemia is associated with epigenetic modifications. Blood. 2011;117:1917–1927. doi: 10.1182/blood-2010-09-307140. [DOI] [PubMed] [Google Scholar]
- Caballero R, Setien F, Lopez-Serra L, Boix-Chornet M, Fraga MF, Ropero S, Megias D, Alaminos M, Sanchez-Tapia EM, Montoya MC, et al. Combinatorial effects of splice variants modulate function of Aiolos. J Cell Sci. 2007;120:2619–2630. doi: 10.1242/jcs.007344. [DOI] [PubMed] [Google Scholar]
- Debnath J, Mills KR, Collins NL, Reginato MJ, Muthuswamy SK, Brugge JS. The role of apoptosis in creating and maintaining luminal space within normal and oncogene-expressing mammary acini. Cell. 2002;111:29–40. doi: 10.1016/s0092-8674(02)01001-2. [DOI] [PubMed] [Google Scholar]
- Derksen PW, Liu X, Saridin F, van der Gulden H, Zevenhoven J, Evers B, van Beijnum JR, Griffioen AW, Vink J, Krimpenfort P, et al. Somatic inactivation of E-cadherin and p53 in mice leads to metastatic lobular mammary carcinoma through induction of anoikis resistance and angiogenesis. Cancer Cell. 2006;10:437–449. doi: 10.1016/j.ccr.2006.09.013. [DOI] [PubMed] [Google Scholar]
- Duhamel M, Arrouss I, Merle-Beral H, Rebollo A. The Aiolos transcription factor is up-regulated in chronic lymphocytic leukemia. Blood. 2008;111:3225–3228. doi: 10.1182/blood-2007-09-113191. [DOI] [PubMed] [Google Scholar]
- Edgren H, Murumagi A, Kangaspeska S, Nicorici D, Hongisto V, Kleivi K, Rye IH, Nyberg S, Wolf M, Borresen-Dale AL, Kallioniemi O. Identification of fusion genes in breast cancer by paired-end RNA-sequencing. Genome Biol. 2011;12:R6. doi: 10.1186/gb-2011-12-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer B, Arcaro A. Current status of clinical trials for small cell lung cancer. Reviews on recent clinical trials. 2008;3:40–61. doi: 10.2174/157488708783330503. [DOI] [PubMed] [Google Scholar]
- Frisch SM, Francis H. Disruption of epithelial cell-matrix interactions induces apoptosis. J Cell Biol. 1994;124:619–626. doi: 10.1083/jcb.124.4.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch SM, Schaller M, Cieply B. Mechanisms that link the oncogenic epithelial-mesenchymal transition to suppression of anoikis. J Cell Sci. 2013;126:21–29. doi: 10.1242/jcs.120907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio M, Migliaccio E, Orsini F, Paolucci D, Moroni M, Contursi C, Pelliccia G, Luzi L, Minucci S, Marcaccio M, et al. Electron transfer between cytochrome c and p66Shc generates reactive oxygen species that trigger mitochondrial apoptosis. Cell. 2005;122:221–233. doi: 10.1016/j.cell.2005.05.011. [DOI] [PubMed] [Google Scholar]
- Glodek AM, Honczarenko M, Le Y, Campbell JJ, Silberstein LE. Sustained activation of cell adhesion is a differentially regulated process in B lymphopoiesis. J Exp Med. 2003;197:461–473. doi: 10.1084/jem.20021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han HJ, Russo J, Kohwi Y, Kohwi-Shigematsu T. SATB1 reprogrammes gene expression to promote breast tumour growth and metastasis. Nature. 2008;452:187–193. doi: 10.1038/nature06781. [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, Bruns CJ, Heeschen C. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007;1:313–323. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, et al. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013;45:242–252. doi: 10.1038/ng.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hystad ME, Myklebust JH, Bo TH, Sivertsen EA, Rian E, Forfang L, Munthe E, Rosenwald A, Chiorazzi M, Jonassen I, et al. Characterization of early stages of human B cell development by gene expression profiling. J Immunol. 2007;179:3662–3671. doi: 10.4049/jimmunol.179.6.3662. [DOI] [PubMed] [Google Scholar]
- Jackman DM, Johnson BE. Small-cell lung cancer. Lancet. 2005;366:1385–1396. doi: 10.1016/S0140-6736(05)67569-1. [DOI] [PubMed] [Google Scholar]
- Jung MJ, Rho JK, Kim YM, Jung JE, Jin YB, Ko YG, Lee JS, Lee SJ, Lee JC, Park MJ. Upregulation of CXCR4 is functionally crucial for maintenance of stemness in drug-resistant non-small cell lung cancer cells. Oncogene. 2013;32:209–221. doi: 10.1038/onc.2012.37. [DOI] [PubMed] [Google Scholar]
- Kilpinen S, Autio R, Ojala K, Iljin K, Bucher E, Sara H, Pisto T, Saarela M, Skotheim RI, Bjorkman M, et al. Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 2008;9:R139. doi: 10.1186/gb-2008-9-9-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Park SH, Cieply B, Schupp J, Killiam E, Zhang F, Rimm DL, Frisch SM. A pathway for the control of anoikis sensitivity by E-cadherin and epithelial-to-mesenchymal transition. Mol Cell Biol. 2011;31:4036–4051. doi: 10.1128/MCB.01342-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Pathak S, Trinh L, Lu R. Interferon regulatory factors 4 and 8 induce the expression of Ikaros and Aiolos to down-regulate pre-B-cell receptor and promote cell-cycle withdrawal in pre-B-cell development. Blood. 2008;111:1396–1403. doi: 10.1182/blood-2007-08-110106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Liu Z, Wu RF, Terada LS. p66Shc restrains Ras hyperactivation and suppresses metastatic behavior. Oncogene. 2010;29:5559–5567. doi: 10.1038/onc.2010.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Myers DP, Wu RF, Nwariaku FE, Terada LS. p66Shc mediates anoikis through RhoA. J Cell Biol. 2007;179:23–31. doi: 10.1083/jcb.200706097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mainiero F, Murgia C, Wary KK, Curatola AM, Pepe A, Blumemberg M, Westwick JK, Der CJ, Giancotti FG. The coupling of alpha6beta4 integrin to Ras-MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J. 1997;16:2365–2375. doi: 10.1093/emboj/16.9.2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Powers SE, Ochiai K, Georgopoulos K, Kee BL, Singh H, Clark MR. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat Immunol. 2009;10:1110–1117. doi: 10.1038/ni.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlen P, Puisieux A. Metastasis: a question of life or death. Nat Rev Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013. doi: 10.1093/emboj/16.8.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410:50–56. doi: 10.1038/35065016. [DOI] [PubMed] [Google Scholar]
- Niebuhr B, Kriebitzsch N, Fischer M, Behrens K, Gunther T, Alawi M, Bergholz U, Muller U, Roscher S, Ziegler M, et al. Runx1 is essential at two stages of early murine B-cell development. Blood. 2013;122:413–423. doi: 10.1182/blood-2013-01-480244. [DOI] [PubMed] [Google Scholar]
- Onder TT, Gupta PB, Mani SA, Yang J, Lander ES, Weinberg RA. Loss of E-cadherin promotes metastasis via multiple downstream transcriptional pathways. Cancer Res. 2008;68:3645–3654. doi: 10.1158/0008-5472.CAN-07-2938. [DOI] [PubMed] [Google Scholar]
- Pelicci G, Lanfrancone L, Grignani F, McGlade J, Cavallo F, Forni G, Nicoletti I, Pawson T, Pelicci PG. A novel transforming protein (SHC) with an SH2 domain is implicated in mitogenic signal transduction. Cell. 1992;70:93–104. doi: 10.1016/0092-8674(92)90536-l. [DOI] [PubMed] [Google Scholar]
- Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D, Rangachari M, Zhu C, Xiao S, Seavitt J, et al. Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol. 2012;13:770–777. doi: 10.1038/ni.2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2011;470:279–283. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynaud D, Demarco IA, Reddy KL, Schjerven H, Bertolino E, Chen Z, Smale ST, Winandy S, Singh H. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9:927–936. doi: 10.1038/ni.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero F, Martinez AC, Camonis J, Rebollo A. Aiolos transcription factor controls cell death in T cells by regulating Bcl-2 expression and its cellular localization. EMBO J. 1999;18:3419–3430. doi: 10.1093/emboj/18.12.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung YM, Xu X, Sun J, Mueller D, Sentissi K, Johnson P, Urbach E, Seillier-Moiseiwitsch F, Johnson MD, Mueller SC. Tumor suppressor function of Syk in human MCF10A in vitro and normal mouse mammary epithelium in vivo. PLoS ONE. 2009;4:e7445. doi: 10.1371/journal.pone.0007445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson EC, Cobb BS, Sabbattini P, Meixlsperger S, Parelho V, Liberg D, Taylor B, Dillon N, Georgopoulos K, Jumaa H, et al. Ikaros DNA-binding proteins as integral components of B cell developmental-stage-specific regulatory circuits. Immunity. 2007;26:335–344. doi: 10.1016/j.immuni.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Timp W, Feinberg AP. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat Rev Cancer. 2013;13:497–510. doi: 10.1038/nrc3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura A, Luzi L, Pacini S, Baldari CT, Pelicci PG. The p66Shc longevity gene is silenced through epigenetic modifications of an alternative promoter. J Biol Chem. 2002;277:22370–22376. doi: 10.1074/jbc.M200280200. [DOI] [PubMed] [Google Scholar]
- Wang JH, Avitahl N, Cariappa A, Friedrich C, Ikeda T, Renold A, Andrikopoulos K, Liang L, Pillai S, Morgan BA, Georgopoulos K. Aiolos regulates B cell activation and maturation to effector state. Immunity. 1998;9:543–553. doi: 10.1016/s1074-7613(00)80637-8. [DOI] [PubMed] [Google Scholar]
- Wary KK, Mainiero F, Isakoff SJ, Marcantonio EE, Giancotti FG. The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell. 1996;87:733–743. doi: 10.1016/s0092-8674(00)81392-6. [DOI] [PubMed] [Google Scholar]
- Xi G, Shen X, Clemmons DR. p66shc inhibits insulin-like growth factor-I signaling via direct binding to Src through its polyproline and Src homology 2 domains, resulting in impairment of Src kinase activation. J Biol Chem. 2010;285:6937–6951. doi: 10.1074/jbc.M109.069872. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.