Abstract
Chloroplast formation is associated with embryo development and seedling growth. However, the relationship between chloroplast differentiation and embryo development remains unclear. Five FtsHi genes that encode proteins with high similarity to FtsH proteins, but lack Zn2+-binding motifs, are present in the Arabidopsis genome. In this study, we showed that T-DNA insertion mutations in the Arabidopsis FtsHi4 gene resulted in embryo arrest at the globular-to-heart–shaped transition stage. Transmission electron microscopic analyses revealed abnormal plastid differentiation with a severe defect in thylakoid formation in the mutant embryos. Immunocytological studies demonstrated that FtsHi4 localized in chloroplasts as a thylakoid membrane-associated protein, supporting its essential role in thylakoid membrane formation. We further showed that FtsHi4 forms protein complexes, and that there was a significant reduction in the accumulation of D2 and PsbO (two photosystem II proteins) in mutant ovules. The role of FtsHi4 in chloroplast development was confirmed using an RNA-interfering approach. Additionally, mutations in other FtsHi genes including FtsHi1, FtsHi2, and FtsHi5 caused phenotypic abnormalities similar to ftshi4 with respect to plastid differentiation during embryogenesis. Taken together, our data suggest that FtsHi4, together with FtsHi1, FtsHi2, and FtsHi5 are essential for chloroplast development in Arabidopsis.
Introduction
Angiosperm embryo and endosperm initiate from double fertilization, where one of the two sperm cells fuse with an egg cell and the other fertilizes with the central cell, respectively [1]. After fertilization, the elongated zygote undergoes an asymmetric cell division to produce a smaller apical and larger basal cell [2]. The apical cell undergoes one-cell, two-cell, four-cell, eight-cell, globular, heart-shaped, torpedo, and bent cotyledon-shaped embryo stages to refine embryonic patterns [3].
The chloroplast, which is an organelle derived from cyanobacteria through endosymbiosis, is a specific type of plastid. Chloroplasts are propagated from a pre-existing plastid through divisions, and formation is initiated from an undifferentiated plastid type; the proplastids. After responding to light, proplastids develop grana, which are stacks of thylakoid membranes for light harvesting, electron transfer, and ATP synthesis. The chloroplast is responsible not only for photosynthesis, but for synthesis and storage of metabolic products (e.g., fatty acids, amino acids, starch) [2], [4].
Chloroplast formation is associated with embryo development and seedling growth. The early differentiation of chloroplasts occurs at the globular-to-heart transition stage during embryogenesis [2], and embryos begin accumulating chlorophyll during the heart-shaped stage. Many developmental and metabolic events occur at the globular-to-heart transition stage. For example, the embryo establishes bilateral symmetry with the emergence of cotyledons [5]. Approximately 40% of embryo-defective mutants are arrested at the globular-heart transition stage [6]. Null mutation of TIC110, which functions in the formation of the chloroplast inner envelope translocation channel, leads to embryo lethality [7]. Two ankyrin repeat-containing proteins, EMB506 and AKRP, which are essential for plastid differentiation, affect embryo transition from the globular to heart-shaped stage [8]. Disrupted plastid Nap7, which likely affects Fe-S biogenesis, leads to embryo arrest at the late globular stage [9]. The type I MADS-box gene AGL23 and the pentatricopeptide repeat protein DELAYED GREENING1 (DG1) are also involved in chloroplast biogenesis during embryogenesis [10], [11]. EMB1303, a chloroplast-localized protein, is essential for chloroplast development. Mutants of emb1303 show delayed embryo development and severe dwarf and albino seedlings with arrested plastid development at the early stage [12]. In addition, EMB1211, a plastid MORN-containing protein, is essential for the transition from the globular to the heart-shaped stage during embryo development [13]. These observations indicate that normal chloroplast development is required for nourishment, and is an important biological process for normal embryogenesis. It has also been proposed that developing chloroplasts release a signal required for regulating nuclear gene expression, which consequentially affects embryo development [14]–[16].
In contrast, mutations in photosynthesis-related genes do not necessarily cause embryo lethality, and often produce homozygous albino seeds that are morphologically normal. Such seeds can typically germinate and grow to various extents on sugar-rich medium, resulting in albino, de-pigmented, pale green to yellow seedlings or variegated seedlings [6], [17], [18]. The majority of plastidic proteins essential for the globular-heart transition are involved in the transcriptional and translational machineries of the plastids [19]. Interestingly, some chloroplast-encoded genes are essential for cell viability [20]. Disruption of the housekeeping chloroplast function often results in embryo lethality, yet rarely in gametophyte lethality [21], [22].
Proteases play crucial roles in the biogenesis and maintenance of chloroplasts. To date, four protease families have been identified in chloroplasts: Clp, FtsH, Lon, and Deg. However, only one of the ClpPR protease complexes, ClpP5, is known to be essential for the transition from the globular to the heart-shaped stage during embryo development [23]. Filamentation temperature-sensitive H (FtsH) is an ATP-dependent metalloprotease that controls plastid protein quality. There are 12 nuclear-encoded FtsH genes in the Arabidopsis genome [24] and four potential FtsH proteases in Synechocystis [25]. Nine of the Arabidopsis FtsHs are targeted to chloroplasts and three are targeted to mitochondria [26]. In Synechocystis, a hetero-oligomeric complex composed of FtsH2 and FtsH3 functions in removing damaged D1 protein [27]. In Arabidopsis, the thylakoid FtsH protease possesses proteolytic activity and is involved in the turnover of the D1 protein in the photosystem II (PSII) reaction center within the context of repair from photoinhibition [28], [29] and degradation of unassembled proteins [30]. Chloroplast-targeted FtsH2 and FtsH8, FtsH1 and FtsH5, and FtsH7 and FtsH9 are closely related pairs. Inactivation of FtsH1 and FtsH5 or FtsH2 and FtsH8 results in a heterotrophic albino phenotype [31].
There are five nuclear-encoded FtsHi genes (FtsHi1 to FtsHi5, and the “i” indicates proteolytic inactivation [32]) in the Arabidopsis genome that display a high degree of similarity to FtsHs at the protein level. However, they lack a Zn-binding site required for proteolytic activity [32], [33]. FtsHi1 is required for chloroplast development [34]. However, how other FtsHi proteins affect embryo development remains unknown.
In this study, we used a reverse-genetics approach to explore the function of the FtsHi genes using Arabidopsis T-DNA insertion mutants. Mutations of the FtsHi4 gene (At5g64580) led to embryo lethality and failed thylakoid formation, similar to other ftshi mutants including mutants of FtsHi1 (At4g23940), FtsHi2 (At3g16290), and FtsHi5 (At3g04340). FtsHi4 was localized in chloroplasts as a thylakoid membrane-associated protein. A significant decrease in D2 and PsbO protein accumulation occurred in the homozygous ftshi4 mutant embryos. Moreover, we demonstrated that knock-down of FtsHi4 expression using an RNA interfering approach resulted in defects in PSII functioning. These results indicated that FtsHi4 is required for PSII formation during embryogenesis. Taken together, our data suggest that FtsHi4, together with other FtsHi proteins, are essential for plastid development during embryogenesis in Arabidopsis.
Materials and Methods
Plant material and growth conditions
The Arabidopsis thaliana ecotype Columbia-0 was used as the wild-type. The ftshi4-1 mutant allele was isolated from a population of transgenic plants generated in our laboratory that displayed white ovules. Mutant seeds obtained from the Arabidopsis Biological Resource Center (ABRC; The Ohio State University) were as follows: ftshi4-2/emb3144 (Salk_113657), ftshi3-1 (GK_723C06), ftshi3-2 (GK_555D09), ftshi1-1/emb2458 (CS16181), ftshi2-1/emb2083-4 (CS16209), ftshi2-2/emb2083-3 (CS16208), ftshi2-3/emb2083-2 (CS16167), and ftshi5 (SAIL_262_D04). Seeds were sterilized in 70% ethanol (with 0.05% Tween20) for 10 min and then washed twice in 95% ethanol and 100% ethanol. After the ethanol evaporated, the seeds were placed on 1/2 MS media (1/2 MS) agar plates supplemented with 50 µg ml−1 kanamycin. Plates were then cold-treated at 4°C for 48-h and allowed to germinate. Plants were grown at 22°C in a greenhouse with a light intensity of 300 µmol m−2s−1 under a 16-h-light/8-h-dark cycle. Measurement of leaf area was performed using standard protocols by LI-3000, A (LI-Cor Inc., Nebraska, USA).
Isolation of ftshi mutants and segregation analysis
The T-DNA flanking sequence was amplified as described previously [35]. Heterozygous ftshi4-1, ftshi4-2, ftshi1, ftshi2-1, ftshi2-2, ftshi2-3, and ftshi5 mutant plants were either self- or reciprocally crossed with wild-type. In both cases, the seeds produced were sown on soil, and plants were genotyped by PCR and phenotyped by analyzing seed development. The following primers were used for genotyping:
LB: 5′-GCTTCCTATTATATCTTCCCAAATTACCAATACA-3′ (for ftshi1, ftshi2, and ftshi5)
FtsHi1LP: 5′-GCCACAACCTCGTTGATTTCTTCT-3′
FtsHi1RP: 5′-CATACTGTTGGCTCAACATGGT-3′
FtsHi2-1RP: 5′-CCCTAACTCATCCCATTGTTCCCA-3′
FtsHi2-1LP: 5′-CCTCTGCATTTTCGTCACCT-3′
FtsHi2-2RP: 5′-CGTGCTGCTGAGTTGAAGAGA-3′
FtsHi2-2LP: 5′-GTACCTTCCTCAACCTCCTCATC-3′
FtsHi2-3RP: 5′-TCTTCTCTATCTCTGCTTCTC-3′
FtsHi2-3LP: 5′-CTGCTCCAACCATGCCATCTG-3′
FtsHi5LP: 5′-GGAAAGCTTTATGTTCCGGAG-3′
FtsHi2RP: 5′-TGCAGCAATAACTGCTGTCAC-3′
LB2: 5′-GATCGACCGGCATGCAAG-3′ (for ftshi4-1)
FtsHi4-1RP: 5′-CAATGTTCTACTCCAATCTGATGCC-3′
FtsHi4-1LP: 5′-TCTCTGTCTAGTTTCGTCCGTCG-3′
LB3: 5′-ATTTGCCGATTTCGGAAC-3′ (for ftshi4-2)
FtsHi4-2RP: 5′-GAGCAGAACTGCAAAACGTTC-3′
FtsHi4-2LP: 5′-CATTCCCGTCTGAAGAATCAG-3′
For phenotyping, siliques were dissected to count the ratio of white seeds to green seeds at 8 days after pollination (DAP). Plants producing 25% white seeds were scored as heterozygous and plants producing 100% green seeds were scored as wild type.
Whole-mount preparations
Ovules of heterozygous ftshi mutants were cleared, as described by Liang et al. [13]. Briefly, siliques from the heterozygous ftshi mutants ranging from 2 to 12 DAP were dissected with hypodermic needles, and the ovules were immersed in HCG solution (80-g chloral hydrate, 10-ml glycerol, 30-ml H2O), mounted on microscopy slides after being cleared, and then observed under a Leica 5500 microscope equipped with DIC optics.
TEM analysis of plastid development
Green (wild type) and white (ftshi mutants) ovules were isolated from the same heterozygous plant siliques when wild-type ovules developed to the torpedo stage. Ovules were punctured with a needle to ensure efficient fixative penetration. The ovules were fixed for 8 h in 200 mM phosphate buffer (pH 7.2) containing 4% glutaraldehyde at room temperature, followed by an overnight incubation in 2% OsO4. The samples were dehydrated in an ethanol series before being embedded in Spur. Ultrathin sections (100 nm) were stained for 15 min in uranyl acetate solution followed by 5 min in lead citrate. Ultrasections were observed under a transmission electron microscope (TEM) (model RILI H-7500) at 80 kV.
Construction of the FtsHi4 hairpin RNA expression vector
A 265-bp DNA fragment of FtsHi4 gene was amplified by PCR with the following primer pairs: FF, 5′-CAGACGGGAATGACTGCT-3′ and FR, 5′-CGACAGGTCTCACGGGTT-3′. This fragment is upstream of the stop codon of the FtsHi4 gene. The PCR products were cloned into the pENTR1A vector at the KpnI and NotI sites, and then recombined into the destination vector pK7GWIWG2 with Gateway LR Clonase II Enzyme Mix (Invitrogen) to generate a hairpin RNA expression vector. This construct was introduced into Arabidopsis via Agrobacterium-mediated floral dipping [36].
Genetic complementation
For the molecular complementation experiment, a 4941-bp genomic region containing the full-length FtsHi4 gene and 866 bp upstream of the FtsHi4 ATG start codon was amplified by PCR using the forward primer gFtsHi4F: 5′-GCTCTAGGTACCTTGTGTTGCTTTTTATGGGTCTC-3′ and the reverse primer gFtshi4R: 5′-GCTCTAGTCGACTTAAAAGAAATGGGTTGCCATCAAA-3′. The PCR product was cloned into the binary vector pCAMBIA1300 at the KpnI and SalI sites in the correct orientation. Constructs were verified by sequencing and used to transform Arabidopsis plants heterozygous for the mutant allele using floral dipping, as described previously [36]. The collected seeds were plated on 1/2 MS culture medium supplied with 25 mg l−1 hygromycin B, and the green seedlings were transplanted into soil.
Real-time RT-PCR expression analysis
Total RNA was extracted according to the EASYspin Plant RNA kit manufacturer's instructions (Galen Biopharm, Beijing, China) with a DNase I (TaKaRa Bio, Ohtsu, Japan) treatment step. Total RNA (1 µg) was subjected to synthesis of single-stranded cDNA using the First Strand DNA Synthesis kit (Toyobo, Osaka, Japan). ACTIN primers were used to detect genomic DNA contamination. Relative quantification values for each target gene were calculated using the 2-⊿⊿CT method [37] using ACTIN as an internal reference gene to compare data from different PCR runs or cDNA samples (qRT-PCR). qRT-PCR analysis provided relative changes in gene expression, with the root normalized to a value of 1. Data were analyzed statistically using Student's t-test. The results shown are representative of two independent experiments, and within each experiment treatments were replicated three times unless otherwise stated. The primers used for the real-time RT-PCR analysis of FtsHi4 gene expression were as follows: FtsHi4F, 5′-GGCTTACCGAGAAGCAGCTGTTG-3′; FtsHi4R, 5′-CTACTCTAGGAGCACATGCACGG-3′. The Actin2 gene was used as a loading control. The primers were as follows: ACTIN2F, 5′-ATGTCTCTTACAATTTCCCG-3′. ACTIN2R, 5′-CCAACAGAGAGAAGATGACT-3′.
Construction of the FtsHi4-GFP fusion and GFP transient assay
The N-terminal 300 bp of the FtsHi4 coding region were amplified by PCR and fused in frame into the expression vector (pTH2) p35S-sGFP using SalI and NcoI restriction endonucleases. The primers used were: sFtsHi4F (5′-GGACCTGGTCGACATGACGTTTTATATCTCGAGCTCG-3′) and sFtsHi4R (5′-GGACCTGCCATGGTAAGTCGTTCTCTTTCAGTTTCT-3′). We used Arabidopsis mesophyll protoplasts for the GFP transient expression assay. Briefly, well-expanded leaves from 3–4-week-old Arabidopsis plants were cut into 0.5–1-mm strips with fresh razor blades and digested in 5–10-ml cellulase/macerozyme solution (1–1.5% cellulase R10, 0.2–0.4% macerozyme R10 (Yakult Honsha, Tokyo, Japan), 0.4 M mannitol, 20 mM KCl, 20 mM MES, 10 mM CaCl2, and 0.1% bovine serum albumin (BSA) (Sigma A-6793; St. Louis, MO, USA), pH 5.7) for 3 h at 23°C. The enzyme solution containing protoplasts was filtered with a 35–75-µm nylon mesh and spun at 80×g for 2 min. The protoplasts were washed once in cold W5 solution (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, 2 mM MES, pH 5.7), resuspended in cold W5 solution at 1–2×105/ml, and maintained on ice for 30 min. The protoplasts were centrifuged at 80×g for 2 min and resuspended in MMg solution (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7) at 1–2×105/ml. A total of 10 µl of DNA (10–20 µg) aliquots and 100 µl of protoplasts were added to a 2-ml microfuge tube and mixed. PEG/Ca solution (110 µl of 40% PEG4000 (Fluka, #81240), 0.2 M mannitol, 1 mM CaCl2) was added, mixed, incubated at 23°C for 10 min, diluted with 0.44-ml W5 solution, and mixed gently. The mixture was centrifuged at 80×g for 1 min to remove the PEG, and the protoplasts were resuspended gently and diluted in 100 µl, followed by the addition of 1 ml of W5 to six-well plates. Protoplasts were then cultured for 6–16 h at 23°C. Cells exhibiting GFP signals were visualized using a confocal laser scanning microscope (LSM510; Carl Zeiss, Jena, Germany).
Thylakoid membrane preparation
Thylakoid membranes were prepared as described previously [38]. Briefly, homogenized leaves in an ice-cold extraction buffer (400 mM sucrose, 50 mM HEPES-KOH, pH 7.8, 10 mM NaCl, and 2 mM MgCl2) were filtered through two layers of cheesecloth. The filtrate was centrifuged at 5000×g for 10 min. Following a wash with extraction buffer, the thylakoid pellets were re-centrifuged and finally suspended in extraction buffer. The chlorophyll contents were determined spectrophotometrically, as described by Porra et al. [39]. The isolated thylakoid membranes were used immediately.
Antibodies and immunolocalization
The polyclonal antibody against FtsHi4 was raised in rabbits using the synthetic peptide sequence SETSGRVFARKSDY. Intracellular localization of FtsHi4 was determined as described previously [40]. Arabidopsis thylakoid membranes were suspended to a final concentration of 100 mg chlorophyll/ml. After 30-min incubation in 10 mM HEPES-KOH, pH 8.0, 10 mM MgCl2, 0.33 M sorbitol, and 1 mM PMSF, the thylakoid membranes were sonicated three times for 15 s on ice. After salt treatment (250 mM NaCl, 200 mM Na2CO3, 1 M CaCl2, or 6 M urea), the membranes were centrifuged at 100,000×g at 4°C for 2 h, washed twice with suspension buffer, and then subjected to immunoblot analyses.
BN-PAGE, SDS-PAGE, and protein identification
BN-PAGE was performed according to Schägger et al. [41]. Following a wash with 0.33 M sorbitol and 50 mM Tris-HCl, pH 7.0, the thylakoid membranes were solubilized in 1% (w/v) DM (20% glycerol, 25 mM Tris-HCl, pH 7.0) at 0.5 mg chlorophyll/mL. After a 10-min incubation at 4°C and centrifugation at 12,000×g for 10 min, the supernatant was combined with 1/10 volume of 5% Serva blue G in 100 mM Tris-HCl, pH 7.0, 0.5 M 6-amino-η-caproic acid, and 30% (w/v) glycerol, and loaded on 6–12% acrylamide gradient gels with a thickness of 0.75 mm. For two-dimensional analysis, BN-PAGE lanes were excised and then soaked for 60 min in SDS sample buffer containing 5% β-mercaptoethanol and layered onto 15% SDS-PAGE gels containing 6 M urea with a thickness of 1 mm. After electrophoresis, proteins were exposed to antibodies, transferred to nitrocellulose membranes, and visualized using the enhanced chemiluminescence method [42].
Measurements of chlorophyll contents
After being cleaned and weighed, the Arabidopsis plant cotyledons and leaves cultured for 3 weeks were ground into homogenate, along with a small amount of calcium carbonate, quartz sand and 1 ml of 95% ethanol. The extract was filtered to 10 ml in a brown volumetric flask with filter paper. The mortar, pestle and residue were washed several times with a small amount of 95% ethanol until no green was visible, after which ethanol was added to a final volume of 10 ml. The extract absorbance was measured with 95% ethanol as a control at 665 and 649 nm, respectively. Chlorophyll contents were calculated as follows based on the extract volume and material weight: Ca = 13.95A665–6.88A649, Cb = 24.96A649–7.32A665.
Measurements of chlorophyll fluorescence
Fluorescence measurements were performed as described previously [43] using a portable fluorometer (PAM-2000, Walz, Fffeltrich, Germany). Before measurements, leaves were dark-adapted for 30 min. The minimum fluorescence yield (F0) was measured under measuring light (650 nm) with low intensity (0.8 µmol m−2 s−1). To estimate the maximum fluorescence yield (Fm), a saturating pulse of white light (3000 µmol m−2 s−1 for 1 s) was applied. The maximum photochemical efficiency of PSII was determined from the ratio of variable (Fv) to maximum (Fm) fluorescence (Fv/Fm = (Fm-Fo)/Fm) [44]. All the above measurements were performed in a dark room under stable ambient conditions.
Yeast two-hybrid assay
The coding sequences of FtsHi1, FtsHi2, FtsHi4 and FtsHi5 were amplified using the gene-specific primers indicated below, and the PCR products were cloned into pGADT7 and pGBKT7 vector, respectively. The yeast two-hybrid assay was performed following the manufacturer's instruction (Clontech, California, USA).
FtsHi1F: GTCGGATCCATGGCGTCTATAGACAATGTTTTCTC
FtsHi1R: GTCCTCGAGTCATACTTGAGCATTGACGTC
FtsHi2F: GTCCATATGATGGCTTGTCGTTTCCCTCTG
FtsHi2R: GTCCCCGGGTTATGAACTGTTTCTTGCGGTAG
FtsHi4F: GTCCCCGGGATGACGTTTTATATCTCGAGCTC
FtsHi4R: GTCCCCGGGTTAAAAGAAATGGGTTGCCATC
FtsHi5F: CTGGAATTCATGATACTGCCCAATGTTTTGGAA
FtsHi5R: CTGGGATCCTTAGGTTGGTGCACTAAGAAGTGC
Results
Isolation and characterization of the ftshi4 mutant
To investigate chloroplast biosynthesis during embryogenesis in Arabidopsis, we generated an Arabidopsis T-DNA insertion population [35] and screened for mutant plants whose siliques contained roughly 25% albino embryos arrested at early embryogenesis; other seeds appeared green due to chlorophyll accumulation. As a result, a mutant plant that produced 24.3% albino seeds (117 of 481, 10 siliques) was identified (Fig. 1C; Table 1). When the mature seeds harvested from this plant were plated on 1/2 Murashige and Skoog (MS) medium containing 50 mg/L kanamycin (Km), the ratio of kanamycin-sensitive versus kanamycin-insensitive seeds was 1∶2 (231∶476, p>0.05), which is consistent with a single insertion line of embryo-lethal defects. We reciprocally crossed wild-type and mutant plants, and no white ovules were observed in the crossed siliques, suggesting that no parental effects were involved. Moreover, each of the Km-resistant plants produced approximately 25% white ovules, as observed in the mutant parents (n = 127). These results indicated that this mutation was recessive and monogenic, and that it affected embryo development but did not cause gametophytic defects.
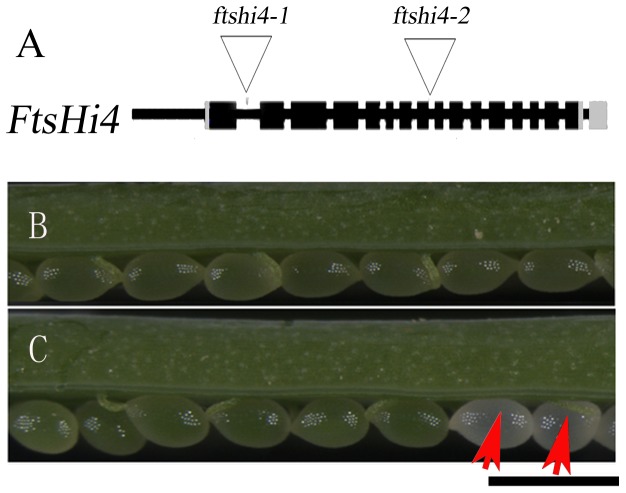
Figure 1. Isolation and characterization of Arabidopsis ftshi4 mutant.
A, Diagram of the T-DNA insertion position in the FtsHi4 gene. Black boxes represent exons. The 5′ untranslated region (UTR) and 3′ UTR are shown in grey boxes. B, 9-DAP dissected wild-type silique. C, 9-DAP dissected ftshi4 (+/−) mutant silique. Bar = 1 mm.
Table 1. Segregation analysis of green and white seeds in developing siliques of heterozygous </emph>ftshi mutants and heterozygous ftshi4 mutants containing FtsHi4 genomic DNA.
| Parental genotype | Green Seeds | White seeds | Number examined | % of white seeds | χ2 |
| ftshi4+/+ | 138 | 1 | 139 | 0.7 | |
| ftshi4-1+/− | 364 | 117 | 481 | 24.3 | 0.008 |
| tshi4-2+/− | 218 | 72 | 290 | 24.8 | 0 |
| gftshi4-1 gftshi4-8 gftshi4-12 gftshi4-22 | 27 19 14 22 | 394 271 213 322 | 421 290 227 344 | 6.41 6.55 6.17 6.40 | 0.0005 0.008 0.007 0 |
| ftshi5+/+ | 231 | 0 | 231 | 0 | |
| ftshi5+/− | 384 | 138 | 522 | 26.4 | 0.02 |
| ftshi2+/− | 180 | 2 | 182 | 1.1 | |
| ftshi2-1+/− | 496 | 150 | 646 | 23.2 | 1.00 |
| ftshi2-2+/− | 418 | 134 | 552 | 24.3 | 0.12 |
| ftshi2-2+/− | 344 | 116 | 460 | 25.2 | 0.001 |
| ftshi1+/+ | 211 | 0 | 211 | 0 | |
| ftshi1+/− | 417 | 141 | 558 | 25.3 | 0.02 |
gftshi4 indicates heterozygous ftshi4 mutant containing FtsHi4 genomic DNA.
To identify the T-DNA insertion site, we performed polymerase chain reaction (PCR)-based genomic walking in the transgenic line using HpaI-digested genomic DNA as template (see Materials and Methods). One PCR product was obtained and sequenced, revealing the presence of a T-DNA insert 367 bp downstream of the ATG start codon of the At5g64580 gene locus. A BLAST search against NCBI indicated that this locus encodes an FstH-like protein named FtsHi4, which is composed of 17 exons and 16 introns, and that the T-DNA was inserted into the first intron (Fig. 1A). The T-DNA insertion position was also confirmed by amplification of the flanking region using primers specific to the T-DNA left border and FtsHi4. Thus, the mutant was designated ftshi4-1.
To further confirm the gene mutation responsible for the observed phenotypes, an independent T-DNA insertion mutant was obtained from the Arabidopsis Biological Resource Center (ABRC) (Salk_113657). The T-DNA was located in the eighth intron, as confirmed by PCR amplification and sequencing of the T-DNA flanking region (Fig. 1A). The Salk_113657 line showed the same phenotypes as ftshi4-1, which also produced 25% white ovules in the heterozygous line (Table 1). F1 plants from the cross between ftshi4-1 and Salk_113657 produced 25% white seeds (Table 2), suggesting that these two mutants were allelic. Accordingly, Salk_113657 was named ftshi4-2.
Table 2. Analysis of reciprocal crosses between wild type and ftshi mutants.
| Maternal | Paternal | Green Seeds | White Seeds | Number Examined | % of white seeds | χ2 |
| ftshi4+/+ | ftshi4-1+/− | 203 | 1 | 204 | 0.49 | |
| ftshi4-1+/− | ftshi4+/+ | 306 | 0 | 306 | 0 | |
| ftshi4+/+ | ftshi4-2+/− | 225 | 0 | 225 | 0 | |
| ftshi4-2+/− | ftshi4+/+ | 244 | 0 | 244 | 0 | |
| ftshi4-1+/− | ftshi4-2+/− | 325 | 107 | 432 | 24.8 | 0.003 |
| ftshi4-2+/− | ftshi4-1+/− | 230 | 73 | 303 | 24.1 | 0.089 |
| ftshi5+/+ | ftshi5+/− | 172 | 0 | 172 | 0 | |
| ftshi5+/− | ftshi5+/+ | 231 | 0 | 231 | 0 | |
| ftshi2+/+ | ftshi2-1+/− | 193 | 1 | 194 | 0.52 | |
| ftshi2-1+/− | ftshi2+/+ | 213 | 0 | 213 | 0 | |
| ftshi3+/+ | ftshi3-2+/− | 160 | 0 | 160 | 0 | |
| ftshi2-2+/− | ftshi2+/+ | 186 | 0 | 186 | 0 | |
| ftshi2+/+ | ftshi2-3+/− | 176 | 1 | 177 | 0.56 | |
| ftshi2-3+/− | ftshi2+/+ | 178 | 0 | 178 | 0 | |
| ftshi2-1+/− | ftshi2-2+/− | 318 | 102 | 420 | 24.3 | 0.079 |
| ftshi2-2+/− | ftshi2-1+/− | 202 | 68 | 270 | 25.2 | 0 |
| ftshi2-1+/− | ftshi2-3+/− | 431 | 140 | 571 | 24.5 | 0.047 |
| ftshi2-3+/− | ftshi2-1+/− | 263 | 85 | 348 | 24.4 | 0.034 |
| ftshi2-2+/− | ftshi2-3+/− | 142 | 49 | 191 | 25.7 | 0.016 |
| ftshi2-3+/− | ftshi2-2+/− | 243 | 76 | 319 | 23.8 | 0.176 |
| ftshi1+/+ | ftshi1+/− | 240 | 1 | 241 | 0.4 | |
| ftshi1+/− | ftshi1+/+ | 158 | 0 | 158 | 0 |
To further verify whether the T-DNA insertion in At5g64580 was responsible for the seed abortion observed, a complementation vector containing a 4941-bp genomic DNA fragment spanning the FtsHi4 gene was constructed and introduced into the heterozygous mutant plants by floral dipping. A total of 6.25% seed abortion was expected if a single copy transgene was introduced into the heterozygous plants and was located on a different chromosome than FtsHi4. Four independent hygromycin-resistant T1 transgenic plants were identified, all of which produced approximately 6.25% white seeds (Table 1). These results indicated that the introduced genomic DNA fragment completely rescued the mutant phenotype.
ftshi4 mutations lead to embryo arrest at the globular to heart-shaped transition stage
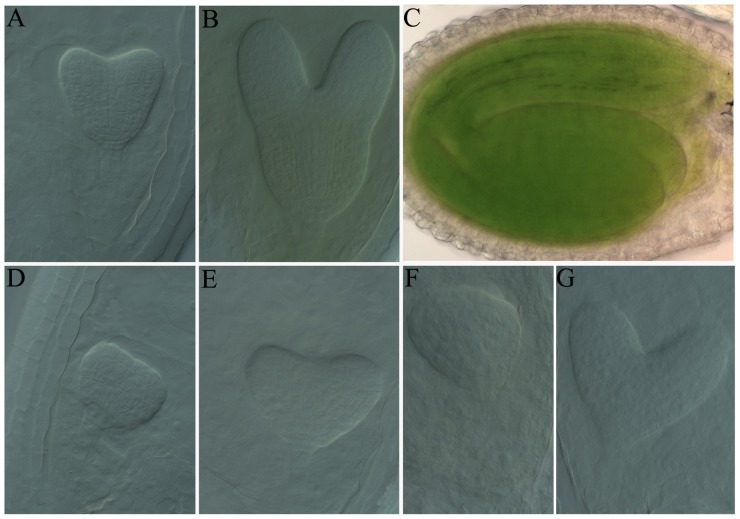
The observation that no homozygous ftshi4 plants were recovered suggested that homozygous mutants are completely embryonic lethal. To investigate embryo development, we cleared the seeds from ftshi4 heterozygous plants and observed them under a Nomarski microscope. At the globular embryo stage, which is the onset of chlorophyll accumulation [2], no obvious defects in embryo development were observed (n = 526). At the heart-shaped stage, the mutant embryos in the same siliques were mostly at the globular stage (Fig. 2A, D). When wild-type embryos reached the mature stage, about 80% (n≥60) of mutant embryos were arrested at the globular stage, whereas the remaining 20% were able to reach the heart-shaped stage, although an obvious abnormal division pattern was observed (Fig. 2C, F, G).
Figure 2. Development of ftshi4 mutant embryos.
A to C, Wild-type embryos from heterozygous ftshi plants undergoing normal development. A, Heart-shaped stage. B, Early torpedo stage. C, Mature embryo. D to G, Mutant embryos from heterozygous ftshi4-1 plants were retarded and morphologically abnormal compared to wild-type embryos from the same silique. D, Mutant embryo development was retarded when wild-type embryo developed to the heart-shaped stage in the same silique. E, Mutant embryo showing abnormalities in the regions that develop an embryo axis and radicle compared with wild-type embryos that developed to the early torpedo stage in the same silique. F, Mutant globular embryo was morphologically abnormal when wild-type embryo reached maturity in the same silique. G, Mutant embryo was arrested at the heart-stage when wild-type embryo reached maturity in the same silique.
Mutations in FtsHi4 affect plastid biogenesis and thylakoid formation
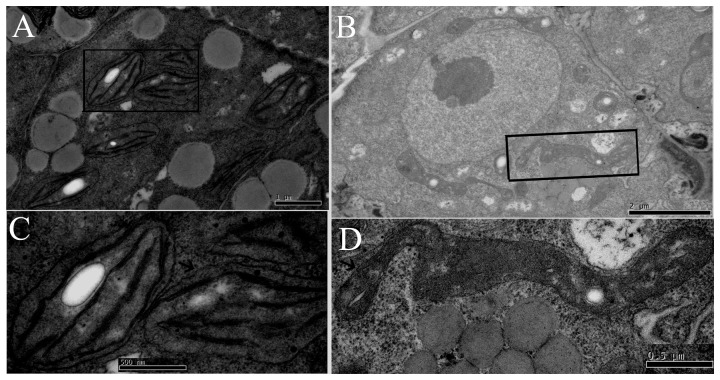
Chloroplast differentiation occurs at the globular-to-heart transition stage during embryogenesis [2] when the embryos started to accumulate chlorophylls. Since the homozygous ftshi4 mutants produced white ovules (Fig. 1C), we explored whether plastid development was impaired in the mutant during embryogenesis. We compared the plastid ultrastructure of the wild-type and the ftshi4-1 mutant embryos in the same siliques of the heterozygous mutant plants at the torpedo stage, coincident with the stage when embryos turn green. In wild-type embryos, thylakoid membranes developed normally and began to stack into grana, an important indicator of plastids that have started to differentiate into spindle-shaped chloroplasts (Fig. 3A, F). In contrast, in ftshi4-1 embryos, plastids were morphologically irregular, polymorphic, and poorly differentiated with very few internal membranes (Fig. 3B, G). No normally developed thylakoids were observed in mutant plastids. These observations indicated that plastid biogenesis and thylakoid differentiation were largely impaired in ftshi4 mutant embryos.
Figure 3. Transmission electron microscopy analyses of chloroplast biogenesis in the ftshi4 mutant embryos.
A, Wild-type torpedo embryo from a heterozygous ftshi4-1 plant with well-developed chloroplasts showing thylakoid membranes beginning to stack into grana. B, Mutant embryo from the same heterozygous ftshi4-1 silique with development-disrupted “plastids”. C, Enlargement of an above-described chloroplast indicated by an arrow.D, Enlargement of an above-described “plastid” indicated by an arrow.
FtsHi4 is an integral thylakoid membrane protein
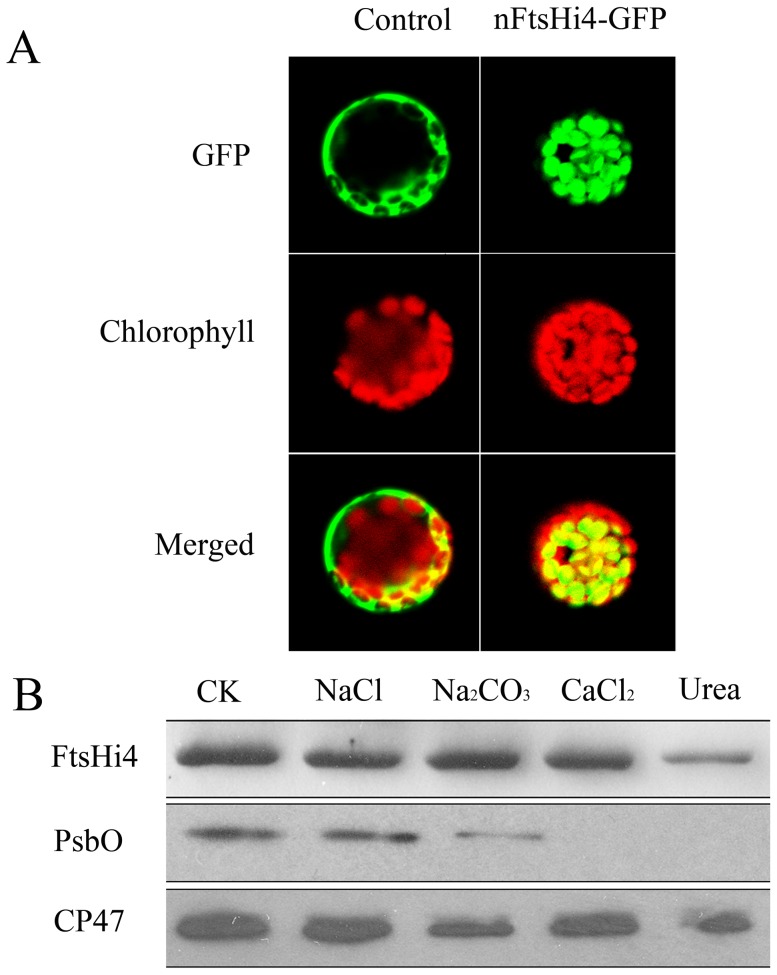
Since plastid biogenesis was impaired in ftshi4 mutants, as revealed by transmission electron microscopy (TEM), we speculated that FtsHi4 may be a chloroplast-localized protein. Analysis using the ChloroP 1.1 Server (http://www.cbs.dtu.dk/services/ChloroP/) software showed that FtsHi4 contained a targeting signal peptide for chloroplasts. To confirm localization, the N-terminal polypeptide of FtsHi4 containing the targeting signal peptide was fused to the N-terminus of green fluorescent protein (GFP) under control of the constitutive CaMV 35S promoter. The resulting construct was introduced into Arabidopsis leaf protoplasts. Green fluorescent signals were co-localized with the chlorophyll autofluorescence in the transformed protoplasts, whereas control GFP lacking the FtsHi4 signal peptide was retained in the cytosol (Fig. 4A). These results indicated that FtsHi4 was targeted to chloroplasts.
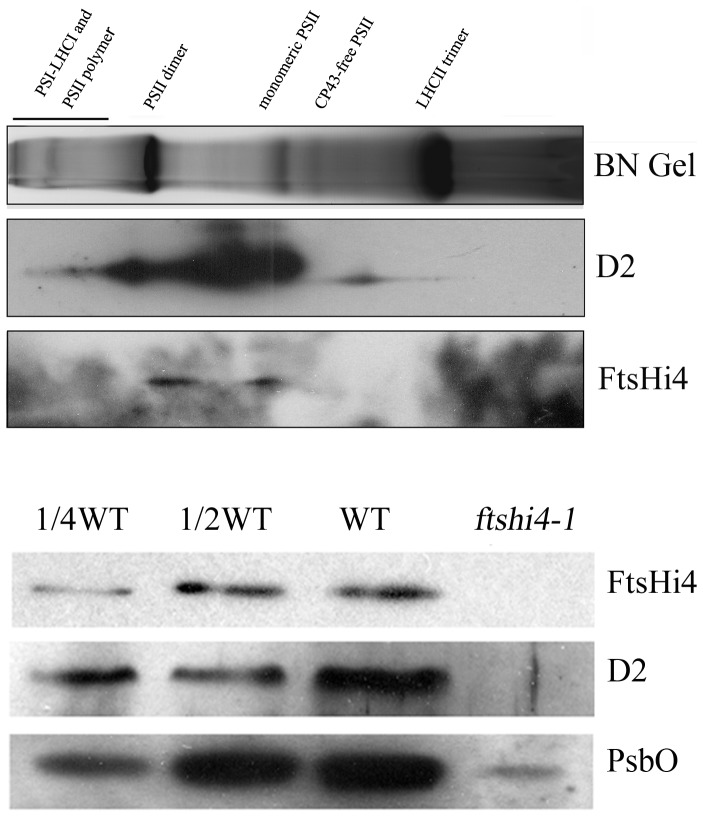
Figure 4. Subcellular and suborganellar localization of FtsHi4.
A, In vivo targeting of green fluorescent protein (GFP) mediated by the FtsHi4 signal peptide in protoplasts. Arabidopsis protoplasts transformed by fusion between the FtsHi4 signal peptide and GFP. GFP fluorescence, chlorophyll autofluorescence, and merged images are shown. Free GFP was used as the control. B, Suborganellar localization of FtsHi4 protein. Thylakoid membranes were prepared from wild-type plants, fractionated by SDS-PAGE, transferred to polyvinylidene difluoride membranes, and visualized using antibodies raised against the FtsHi4 segment, PsbO (a 33-kDa luminal protein of PSII), or CP47 (a core protein of PSII). Membranes that were not subjected to salt treatment (CK) were used as controls.
To further investigate sub-organelle localization of the FtsHi4 protein, the thylakoid membrane fractions were isolated from wild-type plants and treated with alkaline and chaotropic salts to release membrane-associated proteins [40]. In this assay, PsbO (a 33-kDa luminal protein of PSII) and CP47 (a core protein of PSII) were used as markers to distinguish peripheral and integral membrane proteins, respectively. We found that FtsHi4 was retained in the membrane fraction, behaving similar to the integral protein CP47 (Fig. 4B). These results indicated that FtsHi4 was localized to the thylakoid as an integral membrane protein, even though it contains no predicted transmembrane domains.
PSII protein accumulation is defective in the ftshi4 mutant
Chloroplast thylakoid membranes contain a large number of proteins and protein complexes, such as photosystem I (PSI) and PSII, which play various roles in photosynthesis. FtsH proteins were previously shown to be localized near PSII at grana, and are responsible for turnover of the PSII D1 protein [28], [45]. Additionally, FtsHi4 shows a high degree of similarity to FtsH proteins [33]. Thus, we investigated whether FtsHi4 was also localized to PSII-enriched thylakoid membranes. To accomplish this, the thylakoid membrane protein complexes isolated from 4-week-old leaves were separated on blue native (BN)-PAGE gel, and the separated protein complexes corresponding to PSII supercomplexes (band I), monomeric PSI and dimeric PSII (band II), monomeric PSII (band III), dimeric cytochrome b6/f dimer (band IV), trimeric LHCII (band V), and monomeric LHCII (band VI) were fractionated by SDS-PAGE followed by immunoblotting using anti-FtsHi4 and -D2 antibodies. D2 protein is reported to assemble into the PSII complex [46]. Figure 5 showed that the D2 protein was mainly found at band II and III, and the FtsHi4 protein migrated in two protein complexes of ∼460 and ∼240 kDa, respectively, which also corresponded to the PSII dimer and monomer (Fig. 5A). Although some of the D2 complexes migrated at sizes similar to FtsHi4, the overall pattern differed between complexes. These results suggested that FtsHi4 forms a complex.
Figure 5. D2 and PsbO protein accumulation is defective in the ftshi4 mutant.
A, Detection of the PSII core subunit D2 and FtsHi4 by immunoblotting on two-dimensional gel electrophoresis. Membrane samples were solubilized with DM and separated in the first dimension on a blue native gel followed by SDS-PAGE in the second dimension. D2 and FtsHi4 proteins were detected using appropriate antibodies. The separated complexes are designated as: I, PSII supercomplexes; II, monomeric PSI and dimeric PSII; III, monomeric PSII; IV, dimeric cytochrome b6/f dimmer; V, trimeric LHCII; and VI, monomeric LHCII. B, Immunoblot analyses for the accumulation of D2, PsbO and FtsHi4 PSII proteins in wild-type and mutant ovules from heterozygous ftshi4-1 mutant plants. The thylakoid membrane proteins were fractionated by SDS-urea-PAGE, and the blots were probed using antibodies raised against D2, PsbO, or FtsHi4.
To increase our understanding of how defects in thylakoid membrane biogenesis caused by the loss-of-function of FtsHi4 affected PSII protein accumulation in the ftshi4 mutant, immunoblot analyses were conducted using the thylakoid proteins extracted from green and white ovules isolated from the heterozygous ftshi4-1 mutant plants. Accumulation of the FtsHi4 protein was non-detectable in the white ovules, and the two PSII proteins D2 and PsbO were completely lost or decreased significantly, respectively (Fig. 5B). These results indicated that the ftshi4 mutation caused a defect in PSII protein complex formation.
Knock-down of FtsHi4
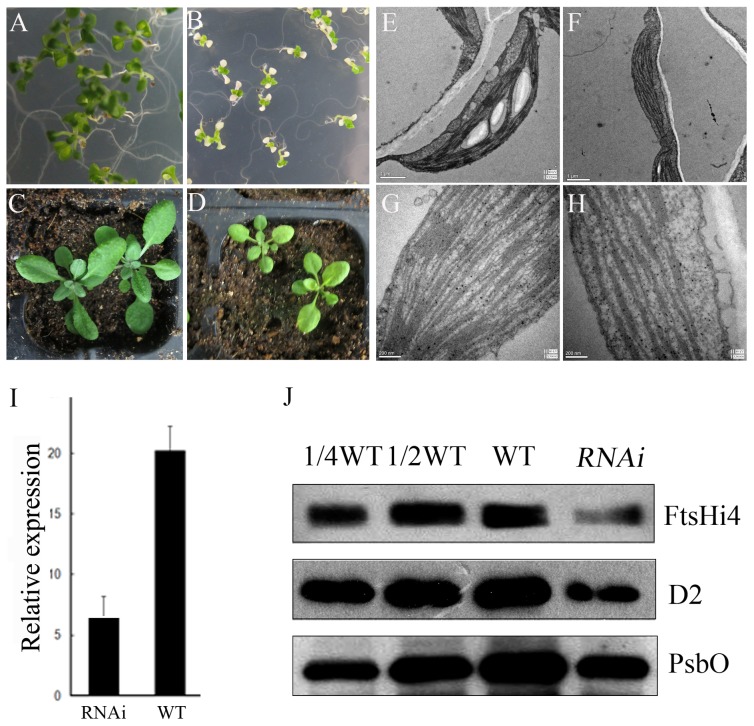
To further confirm the function of FtsHi4, the gene was silenced by double-stranded RNAi using a 265-bp cDNA fragment with 100% specificity to FtsHi4. The expression level of FtsHi4 was examined in transgenic plants using qRT-PCR, which revealed a significant down-regulation of the FtsHi4 gene in the RNAi line compared with WT plants (Fig. 6I). Further immunoblot analysis using an anti-FtsHi4 antibody confirmed the lower signal intensity in total protein preparations (Fig. 6J). Interestingly, the RNAi-FtsHi4 mutant showed a strong phenotype in leaf color. Unlike the wild type, which had green cotyledons and mature leaves, the mutant plants had white cotyledons and yellowish leaves (Fig. 6A, B). In addition, the mutant plants were smaller than WT plants throughout their life cycle (Fig. 6C, D). Under normal conditions, growth was significantly reduced in RNAi plants. As shown in Figure S1, the leaf area of the RNAi plants was ∼40% smaller than WT plants 26 d after germination.
Figure 6. Down regulation of FtsHi4 by RNAi leads to defects in chloroplast development.
A, 2-week-old seedlings of WT grown on half MS medium. B, 2-week-old seedlings of RNAi-FtsHi4 line grown on half MS medium. C, 5-week-old plants of WT grown in soil. D, 5-week-old plants of RNAi-FtsHi4 line grown in soil. E, Chloroplasts from a 5-week-old leaf of wild-type plant. These chloroplasts are well developed, with redundant grana interconnected by stroma thylakoids. Enlargement of such a chloroplast is shown in (G). F, Plastids from 5-week-old leaf of RNAi-FtsHi4 plants, these plastids had straight thylakoids but lacked granal lamellae. Enlargement of such a plastid is shown in (H). ItoJ, qRT-PCR and immunoblot analysis showing expression levels of FtsHi4 in leaves excised from 2-week seedlings described in A and B. Chloroplast proteins were further detected in 2-week seedlings of the RNAi mutant using a specific antibody.
Ultrastructural changes in the chloroplasts of the RNAi-FtsHi4 plants were also assessed. The 5-week-old leaves from wild type and the RNAi-FtsHi4 mutant were sectioned to examine their chloroplast structure. The mutant plastids had straight thylakoids but lacked granal lamellae (Fig. 6E–H). We also analyzed thylakoid membrane protein accumulation of D2 and PsbO with specific antibodies, and found that protein levels of the RNAi-FtsHi4 mutant decreased significantly compared with wild type (Fig. 6J).
Defects in PSII functioning in RNAi mutant plants
Since transgenic RNAi mutant plants displayed chlorosis in both cotyledons and leaves, we measured the chlorophyll contents in RNAi mutant plants. The results showed that both chlorophyll a and chlorophyll b were significantly reduced in the cotyledons of FtsHi4-RNAi mutant (0.20 mg/g in RNAi plants vs. 0.68 mg/g in WT for chlorophyll a; 0.1 mg/g in RNAi plants vs. 0.65 mg/g in WT for chlorophyll b). Unlike cotyledons, chlorophyll a levels in the RNAi plant leaves decreased to 64% compared to WT (0.58 vs. 0.9 mg/g), but the chlorophyll b content was similar in these two genotypes. Consequently, total chlorophyll was significantly reduced in the RNAi plants (Fig. 7A).
Figure 7. Defects in the PSII complex of the RNAi-Ftshi4 mutant block energy transfer within PSII.
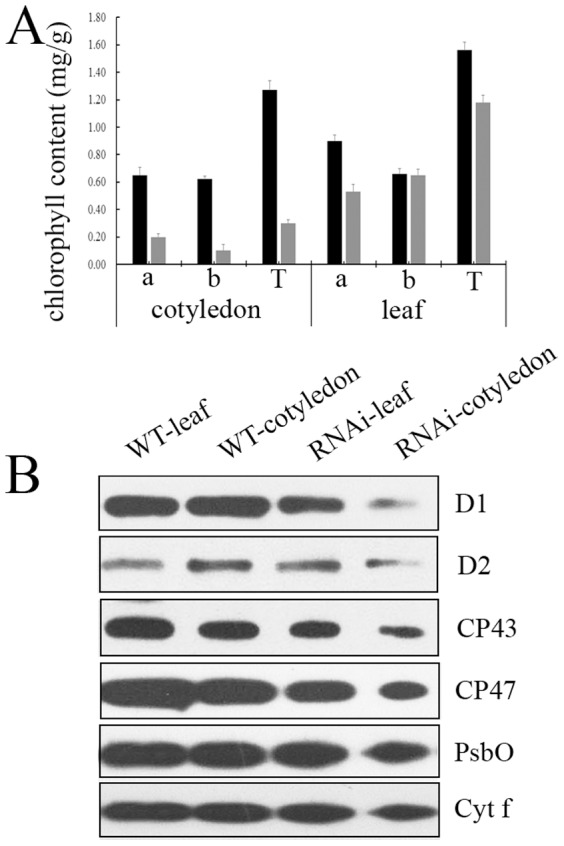
A, The chlorophyll concentrations of wild-type and RNAi-FtsHi4 mutant cotyledons and true leaves. B, Immunoblot analyses for the accumulation of D1, D2, CP43, CP47, PsbO, and Cyt f proteins in wild-type and RNAi-FtsHi4 mutant cotyledons and true leaves. The thylakoid membrane proteins were fractionated by SDS-urea-PAGE, and the blots were probed using antibodies raised against D1, D2, CP43, CP47, PsbO, or Cyt f, respectively.
Fv/Fm (the ratio between variable fluorescence and maximum fluorescence) reflects the maximum potential capacity of the photochemical reactions of PSII [47]. Analyses of the chlorophyll fluorescence Fv/Fm ratio with dark-adapted leaves revealed a significant reduction in RNAi plants compared with WT (0.68 vs. 0.85), suggesting that FtsHi4 knock-down affected energy transfer within PSII.
On the other hand, we analyzed the accumulation of proteins essential for photosynthesis with corresponding antibodies in the transgenic RNAi plants (Fig. 7B). The results demonstrated that levels of D1, D2, CP43 and CP47, the PSII core subunits, Cyt f (the subunit of cytochrome b6f complex) and PsbO (the subunit of the oxygen-evolving complex) were significantly reduced in cotyledons of the RNAi plants compared with WT. Additionally, D1, CP43, and CP47 showed a reduction in leaves, but to a lesser degree than in cotyledons. No significant changes were found for D2, PsbO and Cyt f in leaves (Fig. 7B)
FtsHi4 gene expression pattern
Expression analyses of the FtsHi4 gene in Arabidopsis using qRT-PCR with gene-specific primers showed that FtsHi4 transcripts were ubiquitously present in organs, including roots, stems, cotyledons, young true leaves, rosette leaves, flowers, young siliques and mature siliques (Fig. 8). FtsHi4 transcripts were most abundant in young leaves and present at the lowest levels in roots.
Figure 8. Expression patterns of FtsHi4 gene in Arabidopsis.
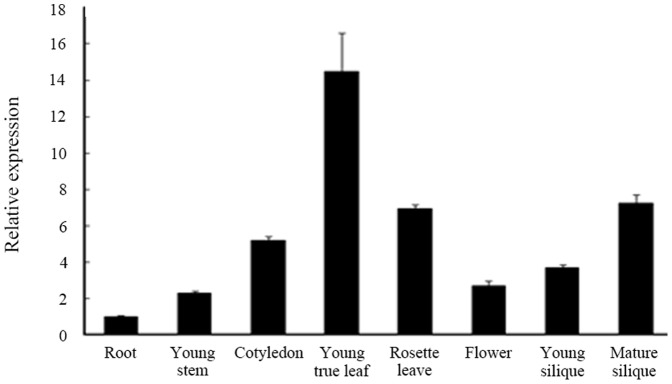
Total RNA was extracted from different tissues of 40-day-old wild-type plants grown in soil. Gene-specific primers were used to detect FtsHi4 transcripts. The ACTIN2 gene was used as a control.
ftshi1, ftshi2 and ftshi5 mutants resemble ftshi4 phenotypes
Since all five FtsHi proteins represent a subclade, we generated T-DNA insertion mutants for the other four genes from ABRC (SAIL_262_D04 for FtsHi1, named ftshi1; emb2083-1, emb2083-2, and emb2083-4 for FtsHi2, named ftshi2-1, ftshi2-2, and ftshi2-3, respectively; GK_723C06 and GK_555D09 for FtsHi3, named ftshi3-1 and ftshi3-2, respectively; and emb2458 for FtsHi5, named ftshi5and) (Fig. S2A). After genotyping by PCR using a combination of gene- and T-DNA left border-specific primers, no homozygous mutants were identified from these lines, suggesting that homozygous mutants may be embryo-lethal. The T-DNA insertion lines for FtsHi3 did not show any visible phenotypes under our conditions (data not shown). Similar to ftshi4, approximately 25% of the ovules in those heterozygous lines of ftshi1, ftshi2, and ftshi5 were albinos with an embryo-lethal phenotype (Fig. S2C, D, E; Table 1). Reciprocal crosses between wild-type and heterozygous mutant plants showed that all of these mutations were recessive without maternal or paternal effects (Table 2).
To investigate embryo development of these mutants, ovules from heterozygous mutant plants were cleared in HCG solution and observed under a differential interference contrast (DIC) microscope. Similar to ftshi4, mutants of ftshi1, ftshi2, and ftshi5 exhibited no evident defects prior to the globular stage, whereas abnormal development was observed when wild-type embryos developed to the heart-shaped stage (Fig. S3). At the mature stage, ∼80% (n≥60) of mutant embryos were arrested at the globular stage, and the remaining 20% reached the heart-shaped stage with an abnormal division pattern (Fig. S3C, D, G, H, K, L).
Since these mutants showed embryo-developmental defects similar to ftshi4, we also examined plastid morphogenesis of the mutants during embryo development. Similar to ftshi4-1, these mutant plastids were morphologically polymorphic and accumulated very few internal membranes (Fig. S4). These observations indicated that FtsHi1, FtsHi2, FtsHi4, and FtsHi5 were required for plastid development during embryogenesis.
FtsHi4 physically interacts with FtsHi2 in yeast cells
In Arabidopsis, two types of FtsH isomers, type A (FtsH5/FtsH1) and type B (FtsH2/FtsH8), form the FtsH heterocomplexes [24], [31], [55]. In this study, each of the ftshi mutants showed very similar phenotype (Fig. 2; Fig.S3) and two protein complexes with different molecular weight were observed in the BN gel analysis when anti-FtsHi4 antibody was used for immunobloting (Fig. 5). Therefore, it's interested to investigate whether the four FtsHis proteins, including FtsHi1, FtsHi2, FtsHi4 and FtsHi5, could interact each other using yeast two-hybrid system. The results showed that there was an interaction between FtsHi2 and FtsHi4. Interestingly, each of these two proteins interacted with itself, respectively (Fig. S5; Table S1).
Discussion
In this report, we described the biological function of FtsHi4, an Arabidopsis FtsH-like gene involved in embryo development by affecting thylakoid biogenesis, using a T-DNA insertion Arabidopsis mutant. FtsHi4 encodes a thylakoid membrane-associated protein, the disruption of which caused a failure in thylakoid formation and led to an embryo-lethal phenotype.
FtsHi4 displays a high degree of similarity to FtsH proteins, a type of Zn2+-metalloprotease that degrades short-lived proteins and misassembled membrane proteins, thus contributing to cellular regulation at the level of protein stability and membrane protein quality control [48]. ftsH was first isolated from E. coli as a temperature-sensitive and cell-division-defective mutant [49]. E. coli FtsH is a membrane-bound protein containing two transmembrane segments at the N-terminus and a cytoplasmic region that includes an ATPase and a protease domain [50]. In E. coli, FtsH functions as both a protease and a chaperone. The lack of a Zn-binding motif in FtsHi suggests that these proteins function as chaperones rather than having protease activity [32].
Among the 12 Arabidopsis FtsHs, 7 have been characterized functionally. FtsH6 contributes to the degradation of Lhcb3 during dark-induced senescence and Lhcb1 and Lhcb3 during high-light acclimation [51]. FtsH1, FtsH5, FtsH2, and FtsH8 form the FtsH complex, which is involved in the repair of PSII (particularly turnover of the D1 protein) [25], [29], [31]. Single FtsH protein mutants are variegated, but not embryo lethal, although the double mutants of atftsh2/atftsh8 and atftsh1/atftsh5 are embryo lethal or seedling lethal, respectively [31]. For example, the ftsh2 mutant and ftsh5 mutant display a variegation phenotype and are sensitive to photoinhibition [52]–[54]. However, in the current study, the ftshi4 mutant was arrested at the globular to heart-shaped transition (Fig. 2D–G). Interestingly, mutants of other FtsHi proteins, including Ftshi1, Ftshi2, and Ftshi5, showed similar phenotypes during embryo development [34] (Figs. 2, 3). The FtsH1, FtsH5, FtsH2, and FtsH8 genes are expressed ubiquitously with similar expression profiles, particularly in green organs; however, expression is low in the roots [24], [55]. At the protein level, FtsH2 is the most abundant FtsH protein in chloroplasts, followed by FtsH5, FtsH8, and FtsH1 [56]. FtsHi4 was expressed in almost all organs examined (Fig. 8). These observations suggest that in contrast to FtsH genes, the expression of FtsHi4 is not confined to green organs. Both the phenotypic difference in embryo development between the ftsh and ftshi mutants and the difference in expression profiling between FtsH and FtsHi genes suggests that these two proteins play different biological roles during chloroplast development. Although functional redundancy between FtsH proteins has been reported, there was no apparent functional redundancy between FtsHi proteins in this study. This is based on the observation that each ftshi mutant showed similar embryo lethality, suggesting that FtsHi forms a homo- or hetero-complex. In agreement with this, we found that FtsHi4 formed two protein complexes of ∼460 and ∼230 kDa, respectively, and the migration pattern of the FtsHi4 complexes differed markedly from that of D2, although some of the D2 complexes migrated to positions similar to the FtsHi4 complexes (Fig. 5). Moreover, it was also found that FtsHi2 and FtsHi4 interacted each other and each of these two proteins interacted with itself in yeast cells (Fig. S5). These results suggest that the FtsHi4 protein is not co-localized with D2, but functions as a complex. However, this requires further investigation using protein-protein interaction experiments.
Characterizing the biochemical function of FtsHi proteins will increase our understanding of how these proteins regulate plastid development. Unfortunately, we did not examine both prokaryotic and eukaryotic expression of FtsHi4 polypeptides in E. coli cells and yeast cells, respectively. Thus, we could not biochemically investigate whether FtsHi4 acts as a molecular chaperone or protease during thylakoid membrane biogenesis. However, there was a complete loss in D2 and a significant reduction in PsbO protein accumulation in the ftshi4 mutant embryos (Fig. 5B). TEM analyses revealed the absence of normally developed thylakoids in mutant embryos (Fig. 3B, G). Thylakoid biogenesis is a complicated process involving physical aspects of formation and differentiation, genetic transmission, signaling cascades, and protein import and sorting. For example, mutations in genes controlling the upstream stages of thylakoid protein assembly cause the loss of thylakoid membrane proteins. Such mutants include csr1 (defective in the targeting pathway to the thylakoids), hcf7 and cps (defective in RNA metabolism), and cpSecY (defective in chloroplast translation) [57]–[59]. It has also been suggested that the nuclear- and plastid-encoded proteins must be targeted to the correct position on the thylakoid with the help of thylakoid-targeting machinery to ensure correct assembly of the thylakoid protein complexes [60]. The phenotypes of these mutants were characterized by abnormal plastid biogenesis and, in most cases, early seedling lethality. The formation of bilayer structures in the thylakoid membrane is an energetically demanding process that depends on the presence of LHC or other membrane proteins, which must be delivered to and combined with the lipid phase [61]. Thus, they likely lack some membrane proteins (e.g., PSII proteins), which may lead to failed thylakoid membrane biogenesis.
The proteins responsible for thylakoid protein complex formation in Arabidopsis play important roles in plastid differentiation. Examples of such proteins include HCF136, which is essential for the stability of PSII in Arabidopsis [62], ALB3, a subunit of the thylakoid Sec protein transport system in Arabidopsis [63], [64], APG2, a component of the Arabidopsis ΔpH-dependent thylakoid protein transport machinery [65], and TerC, a bacterial homologue in Arabidopsis involved in tellurite resistance [60]. Therefore, it is possible that the failed thylakoid membrane biogenesis in the ftshi4 mutant embryos may have been directly caused (at least in part) by an impaired PSII protein complex, which ultimately caused embryo developmental arrest.
The role of plastids during embryogenesis remains unclear, but likely involves the synthesis of metabolites to nourish embryos or the production of signals to regulate nuclear genes. To date, a number of Arabidopsis mutants defective in plastid development have been reported. These mutants are grouped into four classes, one of which includes mutants that are truly embryo-lethal, with embryo development arrested at the globular-to-heart transition stage [23], such as the emb1211 mutant, which is affected in early plastid differentiation [13], the protein import machinery tic110 mutant [7], and the lysophosphatidic acid acyltransferase 1 mutant [22], [66]. Based on analysis of the biological processes of these genes, only those involved in basal cellular functions, such as chloroplast translation machinery and membrane biogenesis, are required for normal embryo development [13]. In this study, we found that mutations in FtsHi proteins (excluding FtsHi3) caused embryo arrest at the transition from the globular to heart-shaped stage, and that these mutants were defective in chloroplast biogenesis and thylakoid formation during embryogenesis (Figs. 2 and 3). These observations indicate that normal functioning of FtsHi proteins is essential for the formation of functional chloroplasts, and these four FtsHi proteins are indispensable for chloroplast biogenesis.
To further explore the possible functions of FtsHi proteins, we examined FtsHi proteins in the ATTEDII co-expression database and found that they are highly associated with each other, but not with other FtsH proteins. Interestingly, some proteins (such as pTAC12/HEMERA) are co-expressed with FtsHi proteins. It was reported that TAC12, together with other TAC members, plays a role in the phytochrome-dependent light signaling pathway to regulate PEP activity and plastid gene expression [67], [68]. In addition, searching of the PPDB proteome database showed that FtsHi1 and FtsHi2 are found in plastids. This result suggests that the FtsHi proteins may be involved in the regulation of plastid gene expression, thereby affecting chloroplast development.
In summary, the impaired accumulation of PSII proteins in the ftshi4 mutant may be caused by failed functional thylakoid membrane formation. Consequently, these defects may have blocked plastid differentiation into functional chloroplasts at the globular to heart-shaped transition stage. However, the role of FtsHi4 and other FtsHi proteins in chloroplast biogenesis in Arabidopsis requires further investigation.
Supporting Information
Growth kinetics of the RNAi- Ftshi4 mutant plants. Values are averages ±S.E. of at least six replicated experiments.
(TIF)
Isolation and characterization of Arabidopsis ftshi1, ftshi2, and ftshi5 mutants. A, Diagram of the T-DNA insertion position in the FtsHi1, FtsHi2, and FtsHi5 genes. Black boxes represent exons. The 5′ untranslated region (UTR) and 3′ UTR are shown in grey boxes. B, 9-DAP dissected wild-type silique. C, 9-DAP dissected ftshi1 (+/−) mutant silique. D, 9-DAP dissected ftshi2 (+/−) mutant silique. E, 9-DAP dissected ftshi5 (+/−) mutant silique. Arrows indicate white ovules with no chlorophyll accumulation. Bar = 1 mm.
(TIF)
Embryo development of ftshi1 , ftshi2 , and ftshi5 mutants. Mutant embryos from heterozygous ftshi1 plants (A–D), heterozygous ftshi2 plants (E–H), and heterozygous ftshi5 plants (I–L) were retarded and morphologically abnormal compared to wild-type embryos from the same silique. A, E, and I, Mutant embryo development was retarded when wild-type embryo developed to the heart-shaped stage in the same silique. B, F, and J, Mutant embryos showed abnormalities in the regions that developed an embryo axis and radicle compared to wild-type embryos that developed to the early torpedo stage in the same silique. C, G, and K, Mutant globular embryos were morphologically abnormal when wild-type embryos reached maturity in the same silique. D, H, and L, Mutant embryos were arrested at the heart-stage when wild-type embryos reached maturity in the same silique. The wild-type embryo development is the same as in Figure 2.
(TIF)
Transmission electron microscopic analysis of plastid development of ftshi1 , ftshi2 , and ftshi5 mutants. A, B, and C, Mutant ftshi1, ftshi2-1, or ftshi5 embryos with development-disrupted “plastids”. Enlargements of the above-described “plastids” are shown in D, E, or F, respectively, and indicated by arrows. Wild-type embryos from each of the same heterozygous ftshi1, ftshi2-1, and ftshi5 siliques are the same as in Figure 3 and therefore not shown.
(TIF)
Yeast two-hybrid assay of FtsHi2 and FtsHi4 protein. Yeast cells transformed with the corresponding vectors grew on the -Trp/-Leu medium (A) and -Ade/-His/-Leu/-Trp medium (B). Numbers of 1-10 represent the yeast hybrid with FtsHi proteins as baits or preys, respectively, which is described as following. 1, bait pGBKT7-53 and prey pGADT7-T. 2, bait pGBKT7-FtsHi2 and prey pGADT7-FtsHi2. 3, bait pGBKT7-FtsHi4 and prey pGADT7-FtsHi4. 4, bait pGBKT7-FtsHi2 and prey pGADT7-FtsHi4. 5, bait pGBKT7-FtsHi4 and prey pGADT7-FtsHi2. 6, bait pGBKT7-FtsHi2 and prey pGADT7. 7, bait pGBKT7-FtsHi4 and prey pGADT7. 8, bait pGBKT7 and prey pGBKT7-FtsHi2. 9, bait pGBKT7 and prey pGBKT7-FtsHi4.10, bait pGBKT7-Lam and prey pGADT7-T.
(TIF)
The interactions between FtsHi proteins in yeast cells.
(DOC)
Acknowledgments
We are grateful to the Flanders Interuniversity Institute for Biotechnology (Belgium) for providing the pHWG plasmid.
Funding Statement
This work was supported by the National Science Foundation of China (grant number 31071077 to C. Z.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, et al. (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699. [DOI] [PubMed] [Google Scholar]
- 2. Mansfield SG, Briarty LG (1991) Early embryogenesis in Arabidopsis thaliana. II. The developing embryo. Canadian Journal of Botany 69: 461–476. [Google Scholar]
- 3.Berleth T, Chatfield B (2002) Embryogenesis: pattern formation from a single cell. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book: American Society of Plant Biologists. [DOI] [PMC free article] [PubMed]
- 4. Ruuska SA, Schwender J, Ohlrogge JB (2004) The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol 136: 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jurgens G (2001) Apical-basal pattern formation in Arabidopsis embryogenesis. EMBO J 20: 3609–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. McElver J, Tzafrir I, Aux G, Rogers R, Ashby C, et al. (2001) Insertional mutagenesis of genes required for seed development in Arabidopsis thaliana. Genetics 159: 1751–1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kovacheva S, Bedard J, Patel R, Dudley P, Twell D, et al. (2005) In vivo studies on the roles of Tic110, Tic40 and Hsp93 during chloroplast protein import. Plant J 41: 412–428. [DOI] [PubMed] [Google Scholar]
- 8. Garcion C, Guilleminot J, Kroj T, Parcy F, Giraudat J, et al. (2006) AKRP and EMB506 are two ankyrin repeat proteins essential for plastid differentiation and plant development in Arabidopsis. Plant J 48: 895–906. [DOI] [PubMed] [Google Scholar]
- 9. Xu XM, Moller SG (2004) AtNAP7 is a plastidic SufC-like ATP-binding cassette/ATPase essential for Arabidopsis embryogenesis. Proc Natl Acad Sci U S A 101: 9143–9148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chi W, Ma J, Zhang D, Guo J, Chen F, et al. (2008) The pentratricopeptide repeat protein DELAYED GREENING1 is involved in the regulation of early chloroplast development and chloroplast gene expression in Arabidopsis. Plant Physiol 147: 573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, et al. (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54: 1037–1048. [DOI] [PubMed] [Google Scholar]
- 12. Huang X, Zhang X, Yang S (2009) A novel chloroplast-localized protein EMB1303 is required for chloroplast development in Arabidopsis. Cell Res 19: 1205–1216. [DOI] [PubMed] [Google Scholar]
- 13. Liang Q, Lu X, Jiang L, Wang C, Fan Y, et al. (2010) EMB1211 is required for normal embryo development and influences chloroplast biogenesis in Arabidopsis. Physiol Plant 140: 380–394. [DOI] [PubMed] [Google Scholar]
- 14. Oelmuller R, Kendrick RE, Briggs WR (1989) Blue-light mediated accumulation of nuclear-encoded transcripts coding for proteins of the thylakoid membrane is absent in the phytochrome-deficient aurea mutant of tomato. Plant Mol Biol 13: 223–232. [DOI] [PubMed] [Google Scholar]
- 15. Taylor WC (1989) Regulatory interactions between nuclear and plastid genomes. Annu Rev Plant Physiol Plant Mol Biol 40: 211–233. [Google Scholar]
- 16. Uwer U, Willmitzer L, Altmann T (1998) Inactivation of a glycyl-tRNA synthetase leads to an arrest in plant embryo development. Plant Cell 10: 1277–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aluru MR, Yu F, Fu A, Rodermel S (2006) Arabidopsis variegation mutants: new insights into chloroplast biogenesis. J Exp Bot 57: 1871–1881. [DOI] [PubMed] [Google Scholar]
- 18. Leister D (2003) Chloroplast research in the genomic age. Trends Genet 19: 47–56. [DOI] [PubMed] [Google Scholar]
- 19. Tzafrir I, Dickerman A, Brazhnik O, Nguyen Q, McElver J, et al. (2003) The Arabidopsis SeedGenes Project. Nucleic Acids Res 31: 90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Drescher A, Ruf S, Calsa T Jr, Carrer H, Bock R (2000) The two largest chloroplast genome-encoded open reading frames of higher plants are essential genes. Plant J 22: 97–104. [DOI] [PubMed] [Google Scholar]
- 21. Hust B, Gutensohn M (2006) Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol (Stuttg) 8: 18–30. [DOI] [PubMed] [Google Scholar]
- 22. Yu B, Wakao S, Fan J, Benning C (2004) Loss of plastidic lysophosphatidic acid acyltransferase causes embryo-lethality in Arabidopsis. Plant Cell Physiol 45: 503–510. [DOI] [PubMed] [Google Scholar]
- 23. Kim J, Rudella A, Ramirez Rodriguez V, Zybailov B, Olinares PD, et al. (2009) Subunits of the plastid ClpPR protease complex have differential contributions to embryogenesis, plastid biogenesis, and plant development in Arabidopsis. Plant Cell 21: 1669–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu F, Park S, Rodermel SR (2004) The Arabidopsis FtsH metalloprotease gene family: interchangeability of subunits in chloroplast oligomeric complexes. Plant J 37: 864–876. [DOI] [PubMed] [Google Scholar]
- 25. Nixon PJ, Barker M, Boehm M, de Vries R, Komenda J (2005) FtsH-mediated repair of the photosystem II complex in response to light stress. J Exp Bot 56: 357–363. [DOI] [PubMed] [Google Scholar]
- 26. Sakamoto W, Zaltsman A, Adam Z, Takahashi Y (2003) Coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15: 2843–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nixon PJ, Michoux F, Yu J, Boehm M, Komenda J (2010) Recent advances in understanding the assembly and repair of photosystem II. Ann Bot 106: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lindahl M, Spetea C, Hundal T, Oppenheim AB, Adam Z, et al. (2000) The thylakoid FtsH protease plays a role in the light-induced turnover of the photosystem II D1 protein. Plant Cell 12: 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bailey S, Thompson E, Nixon PJ, Horton P, Mullineaux CW, et al. (2002) A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J Biol Chem 277: 2006–2011. [DOI] [PubMed] [Google Scholar]
- 30. Ostersetzer O, Adam Z (1997) Light-stimulated degradation of an unassembled Rieske FeS protein by a thylakoid-bound protease: the possible role of the FtsH protease. Plant Cell 9: 957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zaltsman A, Ori N, Adam Z (2005) Two types of FtsH protease subunits are required for chloroplast biogenesis and Photosystem II repair in Arabidopsis. Plant Cell 17: 2782–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sokolenko A, Pojidaeva E, Zinchenko V, Panichkin V, Glaser VM, et al. (2002) The gene complement for proteolysis in the cyanobacterium Synechocystis sp. PCC 6803 and Arabidopsis thaliana chloroplasts. Curr Genet 41: 291–310. [DOI] [PubMed] [Google Scholar]
- 33. Wagner R, Aigner H, Funk C (2012) FtsH proteases located in the plant chloroplast. Physiol Plant 145: 203–214. [DOI] [PubMed] [Google Scholar]
- 34. Kadirjan-Kalbach DK, Yoder DW, Ruckle ME, Larkin RM, Osteryoung KW (2012) FtsHi1/ARC1 is an essential gene in Arabidopsis that links chloroplast biogenesis and division. Plant J 72: 856–867. [DOI] [PubMed] [Google Scholar]
- 35. Lu X, Liu X, An L, Zhang W, Sun J, et al. (2008) The Arabidopsis MutS homolog AtMSH5 is required for normal meiosis. Cell Res 18: 589–599. [DOI] [PubMed] [Google Scholar]
- 36. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 37. Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 38. Zhang L, Paakkarinen V, van Wijk KJ, Aro EM (1999) Co-translational assembly of the D1 protein into photosystem II. J Biol Chem 274: 16062–16067. [DOI] [PubMed] [Google Scholar]
- 39. Porra RJ, Thompson WA, Kriedemann PE (1989) Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochimica et Biophysica Acta (BBA) - Bioenergetics 975: 384–394. [Google Scholar]
- 40. Lennartz K, Plucken H, Seidler A, Westhoff P, Bechtold N, et al. (2001) HCF164 encodes a thioredoxin-like protein involved in the biogenesis of the cytochrome b(6)f complex in Arabidopsis. Plant Cell 13: 2539–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Schagger H, Cramer WA, von Jagow G (1994) Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem 217: 220–230. [DOI] [PubMed] [Google Scholar]
- 42. Constantine NT, Bansal J, Zhang X, Hyams KC, Hayes C (1994) Enhanced chemiluminescence as a means of increasing the sensitivity of western blot assays for HIV antibody. J Virol Methods 47: 153–164. [DOI] [PubMed] [Google Scholar]
- 43. Peng L, Ma J, Chi W, Guo J, Zhu S, et al. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kitajima M, Butler WL (1975) Quenching of chlorophyll fluorescence and primary photochemistry in chloroplasts by dibromothymoquinone. Biochim Biophys Acta 376: 105–115. [DOI] [PubMed] [Google Scholar]
- 45. Yoshioka M, Nakayama Y, Yoshida M, Ohashi K, Morita N, et al. (2010) Quality control of photosystem II: FtsH hexamers are localized near photosystem II at grana for the swift repair of damage. J Biol Chem 285: 41972–41981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nickelsen J, Rengstl B (2013) Photosystem II Assembly: From Cyanobacteria to Plants. Annu Rev Plant Biol 64: 609–635. [DOI] [PubMed] [Google Scholar]
- 47. Krause GH, Weis E (1991) Chlorophyll fluorescence and photosynthesis: The basics. Annu Rev Plant Biol 42: 313–349. [Google Scholar]
- 48. Ito K, Akiyama Y (2005) Cellular functions, mechanism of action, and regulation of FtsH protease. Annu Rev Microbiol 59: 211–231. [DOI] [PubMed] [Google Scholar]
- 49. Santos D, De Almeida DF (1975) Isolation and characterization of a new temperature-sensitive cell division mutant of Escherichia coli K-12. J Bacteriol 124: 1502–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Tomoyasu T, Yamanaka K, Murata K, Suzaki T, Bouloc P, et al. (1993) Topology and subcellular localization of FtsH protein in Escherichia coli. J Bacteriol 175: 1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zelisko A, Garcia-Lorenzo M, Jackowski G, Jansson S, Funk C (2005) AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc Natl Acad Sci U S A 102: 13699–13704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chen M, Choi Y, Voytas DF, Rodermel S (2000) Mutations in the Arabidopsis VAR2 locus cause leaf variegation due to the loss of a chloroplast FtsH protease. Plant J 22: 303–313. [DOI] [PubMed] [Google Scholar]
- 53. Sakamoto W, Tamura T, Hanba-Tomita Y, Murata M (2002) Sodmergen (2002) The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells 7: 769–780. [DOI] [PubMed] [Google Scholar]
- 54. Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41: 1334–1346. [DOI] [PubMed] [Google Scholar]
- 55. Yu F, Park S, Rodermel SR (2005) Functional redundancy of AtFtsH metalloproteases in thylakoid membrane complexes. Plant Physiol 138: 1957–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sinvany-Villalobo G, Davydov O, Ben-Ari G, Zaltsman A, Raskind A, et al. (2004) Expression in multigene families. Analysis of chloroplast and mitochondrial proteases. Plant Physiol 135: 1336–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Asakura Y, Hirohashi T, Kikuchi S, Belcher S, Osborne E, et al. (2004) Maize mutants lacking chloroplast FtsY exhibit pleiotropic defects in the biogenesis of thylakoid membranes. Plant Cell 16: 201–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Barkan A (1993) Nuclear Mutants of Maize with Defects in Chloroplast Polysome Assembly Have Altered Chloroplast RNA Metabolism. Plant Cell 5: 389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roy LM, Barkan A (1998) A SecY homologue is required for the elaboration of the chloroplast thylakoid membrane and for normal chloroplast gene expression. J Cell Biol 141: 385–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kwon KC, Cho MH (2008) Deletion of the chloroplast-localized AtTerC gene product in Arabidopsis thaliana leads to loss of the thylakoid membrane and to seedling lethality. Plant J 55: 428–442. [DOI] [PubMed] [Google Scholar]
- 61. Adam Z, Charuvi D, Tsabari O, Knopf R, Reich Z (2011) Biogenesis of thylakoid networks in angiosperms: knowns and unknowns. Plant Molecular Biology 76: 221–234. [DOI] [PubMed] [Google Scholar]
- 62. Meurer J, Plucken H, Kowallik KV, Westhoff P (1998) A nuclear-encoded protein of prokaryotic origin is essential for the stability of photosystem II in Arabidopsis thaliana. EMBO J 17: 5286–5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Klostermann E, Droste Gen Helling I, Carde JP, Schunemann D (2002) The thylakoid membrane protein ALB3 associates with the cpSecY-translocase in Arabidopsis thaliana. Biochem J 368: 777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, et al. (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Motohashi K, Kondoh A, Stumpp MT, Hisabori T (2001) Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proc Natl Acad Sci U S A 98: 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kim HU, Huang AH (2004) Plastid lysophosphatidyl acyltransferase is essential for embryo development in Arabidopsis. Plant Physiol 134: 1206–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Chen M, Galvao RM, Li M, Burger B, Bugea J, et al. (2010) Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141: 1230–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gao ZP, Yu QB, Zhao TT, Ma Q, Chen GX, et al. (2011) A functional component of the transcriptionally active chromosome complex, Arabidopsis pTAC14, interacts with pTAC12/HEMERA and regulates plastid gene expression. Plant Physiol 157: 1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth kinetics of the RNAi- Ftshi4 mutant plants. Values are averages ±S.E. of at least six replicated experiments.
(TIF)
Isolation and characterization of Arabidopsis ftshi1, ftshi2, and ftshi5 mutants. A, Diagram of the T-DNA insertion position in the FtsHi1, FtsHi2, and FtsHi5 genes. Black boxes represent exons. The 5′ untranslated region (UTR) and 3′ UTR are shown in grey boxes. B, 9-DAP dissected wild-type silique. C, 9-DAP dissected ftshi1 (+/−) mutant silique. D, 9-DAP dissected ftshi2 (+/−) mutant silique. E, 9-DAP dissected ftshi5 (+/−) mutant silique. Arrows indicate white ovules with no chlorophyll accumulation. Bar = 1 mm.
(TIF)
Embryo development of ftshi1 , ftshi2 , and ftshi5 mutants. Mutant embryos from heterozygous ftshi1 plants (A–D), heterozygous ftshi2 plants (E–H), and heterozygous ftshi5 plants (I–L) were retarded and morphologically abnormal compared to wild-type embryos from the same silique. A, E, and I, Mutant embryo development was retarded when wild-type embryo developed to the heart-shaped stage in the same silique. B, F, and J, Mutant embryos showed abnormalities in the regions that developed an embryo axis and radicle compared to wild-type embryos that developed to the early torpedo stage in the same silique. C, G, and K, Mutant globular embryos were morphologically abnormal when wild-type embryos reached maturity in the same silique. D, H, and L, Mutant embryos were arrested at the heart-stage when wild-type embryos reached maturity in the same silique. The wild-type embryo development is the same as in Figure 2.
(TIF)
Transmission electron microscopic analysis of plastid development of ftshi1 , ftshi2 , and ftshi5 mutants. A, B, and C, Mutant ftshi1, ftshi2-1, or ftshi5 embryos with development-disrupted “plastids”. Enlargements of the above-described “plastids” are shown in D, E, or F, respectively, and indicated by arrows. Wild-type embryos from each of the same heterozygous ftshi1, ftshi2-1, and ftshi5 siliques are the same as in Figure 3 and therefore not shown.
(TIF)
Yeast two-hybrid assay of FtsHi2 and FtsHi4 protein. Yeast cells transformed with the corresponding vectors grew on the -Trp/-Leu medium (A) and -Ade/-His/-Leu/-Trp medium (B). Numbers of 1-10 represent the yeast hybrid with FtsHi proteins as baits or preys, respectively, which is described as following. 1, bait pGBKT7-53 and prey pGADT7-T. 2, bait pGBKT7-FtsHi2 and prey pGADT7-FtsHi2. 3, bait pGBKT7-FtsHi4 and prey pGADT7-FtsHi4. 4, bait pGBKT7-FtsHi2 and prey pGADT7-FtsHi4. 5, bait pGBKT7-FtsHi4 and prey pGADT7-FtsHi2. 6, bait pGBKT7-FtsHi2 and prey pGADT7. 7, bait pGBKT7-FtsHi4 and prey pGADT7. 8, bait pGBKT7 and prey pGBKT7-FtsHi2. 9, bait pGBKT7 and prey pGBKT7-FtsHi4.10, bait pGBKT7-Lam and prey pGADT7-T.
(TIF)
The interactions between FtsHi proteins in yeast cells.
(DOC)