Abstract
Twenty-eight xylose-utilizing yeast strains were isolated by enrichment culture from 11 samples of feces from the rectum of Murrah buffalo and Swamp buffalo in Thailand. On the basis of their morphological and biochemical characteristics, including sequence analysis of the D1/D2 region of the large-subunit ribosomal RNA gene (LSU rDNA), they were identified as Candida tropicalis (designated as Group I, 11 isolates), Candida parasilosis (Group II, 2 isolates), Candida mengyuniae (Group III, 2 isolates), Sporopachydermia lactativora (Group IV, 2 isolates), Geotrichum sp. (Group V, 5 isolates) and Trichosporon asahii (Group VI, 6 isolates). All isolates utilized xylose as the sole carbon source but 27 isolates could ferment xylose to ethanol (0.006–0.602 g L−1) and 21 isolates could ferment xylose to xylitol (0.19–22.84 g L−1). Candida tropicalis isolates produced the highest yield of xylitol (74.80%). Their ability to convert xylose to xylitol and ethanol ranged from 15.06 g L−1 to 22.84 g L−1 xylitol and 0.110 g L−1 to 0.602 g L−1 ethanol, respectively.
Keywords: Xylose-utilizing yeast, Buffalo feces, Candida, Geotrichum, Trichosporon
Lignocellulosic biomass varies among plant species but generally consists of around 25% lignin and around 75% carbohydrate polymers (cellulose and hemicellulose). It is the largest known renewable carbon source (2, 22). D-xylose, a pentose sugar and the main hydrolysis product of lignocellulosic biomass, is the second most abundant fermentable material (16). Xylose fermentation is challenging because only a few microorganisms, such as bacteria and yeasts, can readily ferment xylose. Xylose-fermenting yeasts such as Candida shehatae, Pachysolen tannophilus, Brettanomyces naardenensis, Candida tenuis, Scheffersomyces segobiensis, Candida lyxosophila, Candida intermedia, Candida jeffriesii, Spathaspora passalidarum, Spathaspora arborariae, Candida prachuapepsis and Scheffersomyces stipitis strains are ethanol producers from xylose (5, 15, 30, 31). In addition, xylitol is a polyol (sugar alcohol) obtained from the reduction of xylose by strains of Candia boidinii, Candida guilliermondii, Candida maltosa, Candida parapsilosis, Candida tropicalis, Debaryomyces hansenii, Pachysolen tannophilus, Meyerozyma caribbica, Pichia miso, Issatchenkia sp., and Clavispora sp. (11, 33, 35, 40, 41). Xylitol is an alternative sweetener, as sweet as sucrose, equivalent to 2.4 kcal g−1 and laxative nature (145 J g−1 caloric content) (33). It has recently drawn the attention of food and drink manufacturers due to its low caloric value and thus the possibility of its use to reduce and control weight, leading to applications as a sweetener in many products and in the pharmaceutical industry (11). Currently, xylose-fermenting yeasts have been isolated from soil, the gut of beetles, wood, rooting wood, estuarine water from a mangrove forest, the gut of coleopteran insects, fruits, tree bark, etc. (5, 7, 30, 35, 36, 37). In Thailand, the distribution of xylose-utilizing yeasts in herbivore animal feces, especially in buffalo feces, has not yet been reported. Buffaloes are also called water buffaloes. There are two broad categories of buffaloes, river buffaloes and swamp buffaloes. The Murrah buffalo (river buffalo) is the best buffalo breed for milk production (26). Swamp buffaloes are used for multiple purposes in Thailand. They are fed on foliage, crop residues, agro-industrial by-products and non-conventional feed resources (48). Therefore, this study deals with the first attempt to isolate xylose-utilizing yeasts from feces from the rectum of two types of buffaloes, Murrah and Swamp buffaloes, in Thailand, to identify them at a specific level based on their phenotypic characteristics and sequence analysis of the D1/D2 region of the large-subunit ribosomal RNA gene (LSU rDNA D1/D2) and to determine their xylose-fermentation products.
Materials and Methods
Collection of samples and the isolation and maintenance of the yeast isolates
Buffalo fecal samples were collected at Murrah Farm in December 2009, in Chachoengsao province, Thailand. Eleven fecal samples were taken directly from each rectum of Murrah and Swamp buffaloes of different ages (Table 1). Each 0.5 g sample was enriched in a tube containing 10 mL YX medium (0.67% yeast nitrogen base, 5% D-xylose) supplemented with 200 mg L−1 chloramphenicol and 0.25% sodium propionate. The enrichment samples were incubated at 30°C for 3–10 days and were spread on YX agar for their isolation. The number of detected colonies was less than 49/sample. Representative yeast colonies were selected based on colonial characteristics, purified using a single colony isolation method and maintained on a YM agar slant (0.3% yeast extract, 0.3% malt extract, 0.5% peptone, 1% glucose and 1.5% agar) at 4°C or in freezing tubes containing YM broth supplemented with 10% glycerol at −80°C.
Table 1.
Source, isolation and identification of xylose-utilizing yeasts
| Samples | Isolate no. | Group | %Similarity | Identification | aAccession number |
|---|---|---|---|---|---|
| Murrah buffalo (1 month old) | BUF2-1 | VI | 100 | T. asahii | AB741064 |
| BUF2-2 | VI | 100 | T. asahii | AB741065 | |
| BUF2-4 | VI | 100 | T. asahii | AB741066 | |
| Murrah buffalo (2 months old) | BUF3-10 | VI | 100 | T. asahii | AB741069 |
| BUF3-16 | III | 99.8 | C. mengyuniae | AB741072 | |
| BUF3-18 | II | 100 | C. parapsilosis | AB741061 | |
| BUF3-19 | II | 100 | C. parapsilosis | AB741062 | |
| BUF3-3 | VI | 100 | T. asahii | AB741067 | |
| BUF3-5 | IV | 99.4 | S. lactativora | AB741063 | |
| BUF3-6 | III | 99.8 | C. mengyuniae | AB741071 | |
| BUF3-8 | VI | 100 | T. asahii | AB741068 | |
| Murrah buffalo (2.2 months old) | BUF1-1 | IV | 99.2 | S. lactativora | AB741078 |
| Murrah buffalo (2.52 years old) | BUF9 | V | 99 | Geotrichum sp. | AB741075 |
| BUF9-4 | I | 100 | C. tropicalis | AB741058 | |
| BUF9-2 | I | 100 | C. tropicalis | AB741056 | |
| BUF9-5 | I | 100 | C. tropicalis | AB741057 | |
| Murrah buffalo (2.58 years old) | BUF8 | V | 99 | Geotrichum sp. | AB741074 |
| BUF8-1 | I | 100 | C. tropicalis | AB741054 | |
| BUF8-2 | I | 100 | C. tropicalis | AB741055 | |
| Murrah buffalo (2.65 years old) | BUF10 | V | 99 | Geotrichum sp. | AB741076 |
| Swamp buffalo (2 months old) | BUF7 | V | 99 | Geotrichum sp. | AB741073 |
| BUF7-1 | I | 100 | C. tropicalis | AB741053 | |
| Swamp buffalo (3 months old) | BUF5-3 | I | 100 | C. tropicalis | AB741052 |
| Swamp buffalo (3.8 months old) | BUF4-2 | I | 100 | C. tropicalis | AB741060 |
| BUF4-3 | I | 100 | C. tropicalis | AB741050 | |
| BUF4-4 | I | 100 | C. tropicalis | AB741051 | |
| Swamp buffalo (4.33 years old) | BUF11 | V | 97 | Geotrichum sp. | AB741077 |
| Swamp buffalo (5.1 years old) | BUF12-1 | I | 100 | C. tropicalis | AB741059 |
Accession number: LSU rDNA D1/D2 sequences determined in this study and deposited in the DDBJ gene databank in Japan.
Phenotypic characterization
Morphological characteristics were examined according to Yarrow (49) and Kurtzman et al. (18). Formation of true- and pseudo-hyphae were monitored in cornmeal agar at 25°C until 7 days and ascospore production was examined on cornmeal agar, 5% malt extract and YM agar until 2 months. For physiological characteristics, Yeast identification system ID 32 C (bioMérieux, Marcy l’Etoile, France) was used according to the manufacturer’s instructions. The kit allows the evaluation of the assimilation of 30 carbon sources for clinical isolates of pathogenic yeasts. Test strips were incubated at 30°C for 48 h (24 to 48 h is recommended).
DNA sequence and phylogenetic analysis
A loopful of yeast cells was suspended in 100 μL lysis buffer in a 1.5 mL Eppendorf tube (34) and was boiled in a water bath or metal block bath for 15 min. After boiling, 100 μL of 2.5 M potassium acetate (pH 7.5) was added, placed on ice for 1 h, and centrifuged at 14,000 rpm for 5 min. The supernatant was extracted twice with 100 μL chloroform/isoamyl alcohol (24:1 [v/v]) and DNA in the upper layer was precipitated with ethanol, dried and dissolved in 30 μL MilliQ water. The D1/D2 domain of the large subunit ribosomal RNA gene (LSU rDNA D1/D2) was amplified by polymerase chain reaction (PCR) with primers NL1 (5′-GCATATCAATAAGCGGAGGAAAAG-3′) and NL4 (5′-GGTCCGTGTTTCAAGACGG-3′) (32). The PCR condition was performed according to the methods described for the amplification of 26S rDNA D1/D2 domain (20). The PCR product was checked by agarose gel electrophoresis and purified using a QIAquick purification kit (Qiagen, Tokyo, Japan). The purified PCR product was sequenced using the BigDye Terminator v.3.1 Cycle Sequencing RR-100 kit and an ABI Model 3130xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA) following the manufacturer’s instructions. The sequencing reactions were performed using the external primers NL1 and NL4. Sequence data were aligned using the program Chromas Pro software (Technelysium Pty, South Brisbane, Australia). The sequences were compared with available sequence data using a BLASTN search (1) and were aligned with sequences of related species retrieved from GenBank using the multiple alignment program CLUSTAL_X version 1.8 (45). A phylogenetic tree was constructed from the evolutionary distance data with maximum composite likelihood correction using the neighbor-joining method (38). The topology of the phylogenetic tree was tested by performing bootstrap re-sampling from 1,000 replicates (10). Sequences determined in this study were deposited in the DDBJ gene databank (Shizuoka, Japan) and their accession numbers are shown in Table 1.
Production of ethanol and xylitol from xylose
The fermentation of D-xylose was tested by cultivation in YP medium (1% yeast extract and 2% peptone), adjusted to pH 5.5 with HCl and supplemented with 6% D-xylose on 50 mL YP medium in a 250 mL Erlenmeyer flask, shaken on a rotary shaker at 200 rpm at 30°C until 24 h. S. stipitis JCM 10742T (Japan Collection of Microorganisms, RIKEN BioResource Center, Tsukuba, Japan) was used as a reference strain.
Analysis of substrates and products
Fermentation broth was centrifuged at 8,000 rpm for 10 min and the supernatants were used to determine the ethanol concentration using a gas chromatograph (GC-9A; Shimadzu, Kyoto, Japan), the concentration of xylose and xylitol concentration using HPLC with a Lichrospher100 NH2 (5 μm) column of 250 mm × 4 mm (Merck, Germany) and detected with an evaporative light scattering detector (Alltech, Illinois, USA). Acetonitrile/water (91:1) was used as the mobile phase at a flow rate of 1.5 mL min−1.
Results and Discussion
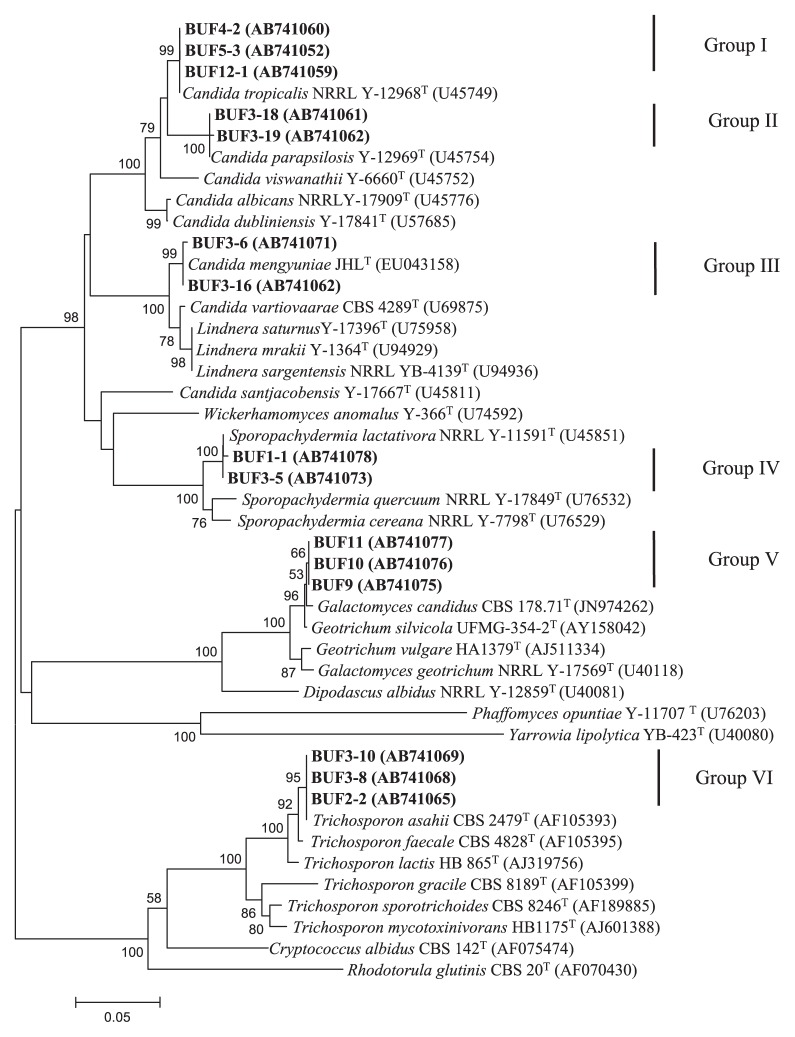
This study presents the first characterization of xylose-utilizing yeast associated with six samples of Murrah buffalo feces and five samples of Swamp buffalo feces that were enriched in YX medium. The cell densities of yeasts in Murrah buffalo feces were higher than in Swamp buffalo feces. However, the xylose-utilizing yeasts obtained from each buffalo fecal sample were less than 100 colony-forming units. The characterization of 28 isolates is summarized in Tables 1 and 2, and Fig. 1.
Table 2.
Differential characteristics of the isolates in Group I, II, III, IV, V, VI and related type strains
| Characteristics | 1 | 2a | 3 | 4a | 5 | 6b | 7 | 8c | 9 | 10d | 11 | 12e |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Assimilation of | ||||||||||||
| l-Arabinose | − | − | + | + | − | − | − | − | − | − | + | + |
| Cellobiose | + | v | − | − | v | − | − | − | − | − | + | + |
| Erythritol | − | − | − | − | − | − | − | − | − | − | + | v |
| Galactose | + | + | + | + | − | + | − | − | + | + | + | + |
| Glycerol | − | v | + | + | + | + | + | + | + | + | − | v |
| Gluconate | + | v | + | v | − | nd | − | − | − | − | + | − |
| Glucosamine | + | v | + | v | − | − | − | − | − | − | + | v |
| Glucose | + | + | + | + | + | + | + | + | + | + | + | + |
| Inositol | − | − | − | − | − | − | − | s | − | − | − | v |
| dl-Lactate | − | v | − | − | + | nd | − | − | l | + | + | v |
| Lactose | − | − | − | − | − | nd | + | v | − | − | + | + |
| Maltose | + | + | + | + | + | − | − | − | − | − | + | + |
| d-Mannitol | + | + | + | + | + | + | − | − | + | + | v | v |
| Melezitose | + | v | + | + | + | − | − | − | − | − | + | v |
| Melibiose | − | − | − | − | − | + | − | − | − | − | − | − |
| α-Methyl-d-glucoside | + | v | + | + | v | + | − | − | − | − | + | + |
| Raffinose | − | − | − | − | + | + | − | − | − | − | − | − |
| l-Rhamnose | − | − | − | − | − | − | − | − | − | − | + | |
| d-Ribose | − | v | − | v | − | − | − | − | − | − | + | + |
| Sucrose | + | v | + | + | + | + | − | − | − | − | + | v |
| Trehalose | + | + | l | + | + | + | − | − | − | − | v | + |
| d-Xylose | + | + | + | + | + | + | + | + | + | + | + | + |
1, Group I (11 isolates); 2, C. tropicalis; 3, Group II (2 isolates); 4, C. parapsilosis; 5, Group III (2 isolates); 6, C. mengyuniae; 7, Group IV (2 isolates); 8, S. lactativora; 9, Group V (5 isolates); 10, G. candidum; 11, Group VI (6 isolates); 12, T. asahii.
+, positive; −, negative; l, latent (longer than 7 days), s, slow; w, weak; v, variable; n, no data.
Data from Lachance et al. (21).
Data from Chen et al. (6).
Data from Rodrigues et al. (37).
Data from de Hoog and Smith (8).
Data from Molnar et al. (28).
Fig. 1.
Phylogenetic tree of xylose assimilation yeasts constructed by the neighbor-joining method based on the D1/D2 domain of LSU rRNA gene sequences. Numbers represent the percentages from 1,000 replicate bootstrap resamplings (frequency <50% is not shown). Bar indicates Knuc.
Identification of isolates
Twenty-eight xylose-utilizing yeast isolates were divided into six groups based on their phenotypic characteristics and the sequences of D1/D2 region analyses (Tables 1 and 2, and Fig. 1), designated as Group I to VI. Five groups were assigned to five ascomycetous yeast species, C. tropicalis (Group I, 11 isolates), C. parapsilosis (Group II, 2 isolates), Candida mengyuniae (Group III, 2 isolates), Sporopachydermia lactativora (Group IV, 2 isolates) and Geotrichum sp. (Group V, 5 isolates) related to Geotrichum candidum. Only one group of basidiomycetous yeast utilizing xylose was assigned to Trichosporon asahii in the genus Trichosporon (Group VI, 6 isolates).
Group I contained 11 isolates. All the isolates assimilated cellobiose, galactose, gluconate and glucosamine, maltose, α-methyl-d-glucoside, mannitol, melezitose, trehalose, sucrose and xylose (Table 2). They had almost the same phenotypic characteristics as C. tropicalis NRRL Y-12968T (21). The isolates had identical sequences in the nucleotide sequence of the D1/D2 domain of the 26S rRNA gene. The representative three isolates were located within the cluster of C. tropicalis in the phylogenetic tree in Fig. 1. Therefore, they were identified as C. tropicalis.
Group II contained 2 isolates, BUF3-18 and BUF3-19. Two isolates assimilated l-arabinose, galactose, gluconate, glucosamine, maltose, mannitol, melezitose, α-methyl-d-glucoside, sucrose, trehalose (latent) and xylose but did not assimilate ribose (Table 2). Their phenotypic characteristics were similar to C. parapsilosis Y-12969T (4). The isolates were closely related to C. parapsilosis Y-12969T based on the identical nucleotide sequence of the D1/D2 domain of the 26S rRNA gene (Table 1, Fig. 1). Therefore, they were identified as C. parapsilosis.
Group III contained 2 isolates, BUF3-6 and BUF3-16. They showed 99.8% similarity of the nucleotide sequence of the D1/D2 domain of the 26S rRNA gene to C. mengyuniae JHLT (Table 1) but some of their phenotypic characteristics were different from this type strain (6). These isolates assimilated dl-lactate, maltose and melezitose but did not assimilate galactose, gluconate, lactose and melibiose (Table 2). The nucleotide sequence of other genes should be confirmed for this group.
Group IV contained 2 isolates, BUF1-1 and BUF3-5, The isolates assimilated glycerol, glucose, lactose and xylose. These isolates could not assimilate inositol but S. lactativora NRRL Y-11591T could (37). The isolates showed 99.2–99.4% similarity of the nucleotide sequence of the D1/D2 domain of the 26S rRNA gene to S. lactativora NRRL Y-11591T (Table 1, Fig. 1), and were identified as S. lactativora.
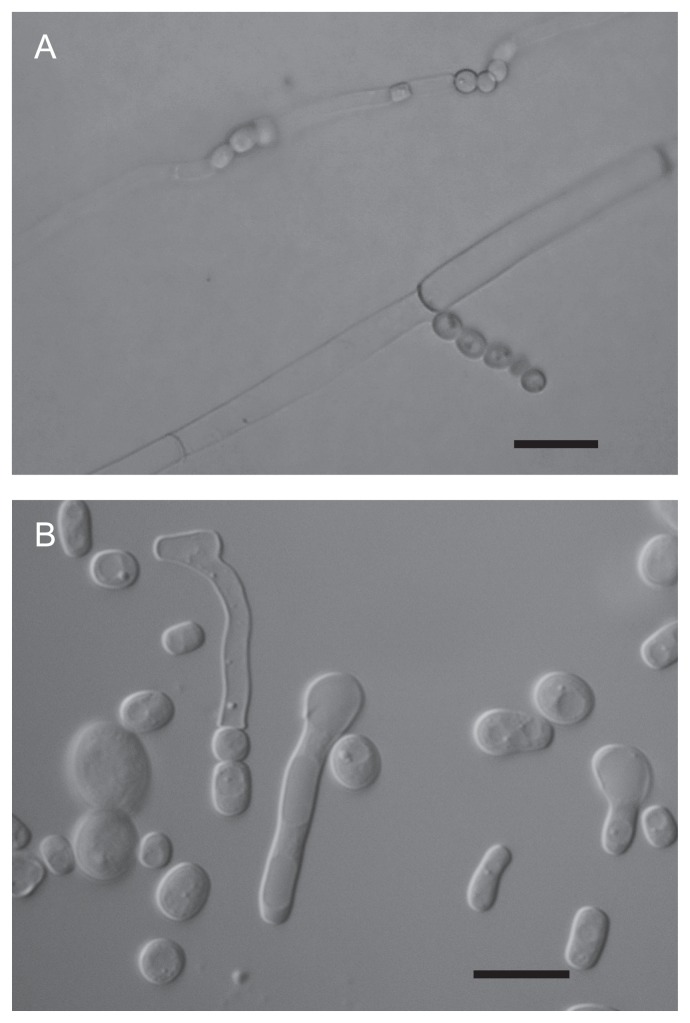
Group V contained 5 isolates, BUF7, BUF8, BUF9, BUF10 and BUF11. The isolates showed 97–99% similarity of the nucleotide sequence of the D1/D2 domain of the 26S rRNA gene to that of Geotrichum candidum or its teleomorph, Galactomyces candidus. In the phylogenetic tree, all these isolates were clustered with their nearest phylogenetic neighbors in the genus Geotrichum and its teleomorphic genus Galactomyces, but were separated from the other Geotrichum species (Fig. 1). From pairwise comparison of these sequences with G. candidum, 4–11 nucleotide (nt) substitutions were found in 537 nucleotides. From these data, this group may include divergent populations within a species or different species from G. candidum. Kurtzman and Robnett (20) compared the divergence among ascomycetous strain pairs with previously determined nuclear DNA reassociation values and it appeared that different species differed by 6 or more nucleotides among the 500–600 nucleotides of the D1/D2 domain. Moreover, the isolates showed 81.4% similarity of the nucleotide sequence of the internal transcribed spacer region (ITS) to that of the type strain of G. candidum (data not shown). The morphological characteristics of these isolates, such as the formation of white, hairy, usually dry colonies, true hyphae, arthroconidia and blastoconidia (Fig. 2), supported that they were members of the genus Geotrichum (8). They assimilated galactose, mannitol, glucose and sorbose and grew on cycloheximide medium at 37°C. However, this data were insufficient for designation to different taxa. Smith et al. (42) assigned strains in the genus Georichum to one taxon on the basis of phenotypic criteria and mating reactions. According to ‘The Yeasts: a taxonomic study, 5th edition (2011)’, many species and genera have been reclassified by molecular taxonomic methodology using such as the sequence divergence of the large subunit (LSU) and small subunit (SSU) rDNA, actin-1 gene, RNA polymerase II (RPB1 and RPB2) gene, translation elongation factor-1α (TEF1α) and mitochondrial regions (19). Consequently, taking these into consideration, we assigned this Group V as Geotrichum sp.
Fig. 2.
Morphology of Geotrichum sp. BUF11 observed by light microscopy. True-hyphae formed on corn meal agar at 25°C for 5 days (A). Vegetative cell grown on 5% malt extract agar at 25°C for 3 days (B). Bar indicates 5 μm.
Group VI contained 6 isolates. The isolates assimilated gluconate, glucosamine, dl-lactate, melezitose, l-rhamnose, and sucrose but did not assimilate glycerol or inositol. The phenotypic characteristics were almost the same as the type strain (28) (Table 2). The isolates had an identical nucleotide sequence of D1/D2 to T. asahii CBS 2479T (Table 1). They were basidiomycetous yeast; therefore, they were identified as T. asahii.
Yeasts associated with buffalo rectum feces
From Table 1, the two types of buffalo feces, Murrah buffalo and Swamp buffalo (Bubalus bulalis), appeared to have different microbiota (or microflora) types of yeast species. In Swamp buffalo feces, only 2 species, C. tropicalis and Geotrichum sp., were found. In each individual feces in young to old animals, C. tropicalis or Geotrichum sp. or both was found. On the other hand, in Murrah buffalo feces, C. tropicalis and Geotrichum sp. were found only in old individuals. In fecal samples from young Murrah buffalos aged 1, 2 and 2.2 months, S. lactativora, T. asahii or both were found. In addition to these two species, C. mengyuniae and Candida parapsilosis were detected in feces from Murrah buffalos aged 2 months. Almost all yeast strains isolated in this study were so-called opportunistic pathogens, such as T. asahii and C. parapsilosis.
Swamp buffalo are very well adapted to hot and humid climates as well as marshy lands (26). Murrah buffalo are highly sensitive to solar radiation, and the poorly evolved body thermoregulation of buffaloes is compensated for by their wallowing activity in fresh water bodies (39). From these reports it was shown that Murrah buffalo may have weaker health than Swamp buffalo. Therefore, the Murrah buffalo in this research might have been infected with yeast pathogens.
Previous studies have concentrated on the search for yeasts in the gastrointestinal tract (GIT) of vertebrates with a focus on farm animals, and researchers have detected various ascomycetous and basidiomycetous yeasts, chiefly representing the genera Candida, Trichosporon, Pichia, Rhodutorula, Debaryomyces, Kluyveromyces and Saccharomyces. (46, 47). C. tropicalis has been isolated from a variety of animal GIT, such as cows, sheep, pigs and beetles (46). Suzuki et al. (44) also reported the occurrence of C. tropicalis in a number of fruits in Thailand. However, this yeast is one of the more frequently encountered clinical yeast species after C. albicans (29). Similarly, C. parapsilosis is considered to be the second most important opportunistic pathogenic yeast (4, 23). T. asahii have a medical association, being commonly found as the causative agent of disseminated mycoses in patients with impaired immunity, and occasionally as the cause of human or animal white piedra (28). S. lactativora strains appear to be cosmopolitan in distribution, such as in soil, fermented food, decaying agriculture residue and bird feces in Antarctica (9, 24, 31), and they could grow at high temperatures of 37°C or more (37).
Species of the genera Geotrichum and Galactomyces, such as Galactomyces geotrichum, Geotrichum silvicola, and Geotrichum sp., have been isolated from animal GIT on farms (8, 46) and are distributed worldwide.
C. mengyuniae, a sulfonylurea herbicide-resistant yeast, was isolated from fields often soaked with discharge from metsulfuron-methyl manufacturing facilities (6). The occurrence of C. mengyuniae in Murrah buffalo of 2 months old may show that this buffalo is eating food contaminated with herbicide.
In addition, xylose-utilizing yeast from gut of passalid beetles, S. stipitis, S. segobiensis, C. shehatae and Candia ergatensis, and yeasts from the digestive tract and feces of animals, C. tropicalis, S. lactativora and T. asahii, have been reported (25, 43, 46). Kazachstania slooffiae, Candida glabrata, I. orientalis (a synonym of Pichia kudriavzevii), Pichia fermentans, T. ashaii and C. tropicalis strains in the gut of piglets kept under experimental conditions, and Kazachstania slooffiae, G. geotrichum, Candida catenulata, Trichosporon coremiiforme and T. asahii in the gut of piglets on a commercial farm were studied (46). In Thailand, xylose-utilizing yeasts, C. tropicalis, C. albicans, Pichia terricola, S. lactativora, Trichosporon mycotoxinivorans and Zygoascus meyerae, have been isolated from herbivore feces, such as from elephants, goats, giraffes, kangaroos, horses, cows and zebras (25). In comparison to this study, we found C. tropicalis, C. parapsilosis, C. mengyuniae, S. lactativora, Geotrichum sp. and T. asahii in buffalo feces.
Ethanol and xylitol fermentation
Twenty-eight isolates in this study utilized xylose as their sole carbon source. All of these isolates could ferment xylose to ethanol (0.006–0.602 g L−1) and 21 isolates could ferment xylose to xylitol (0.19–22.84 g L−1) using YX medium (20% xylose and 0.67% yeast nitrogen base) over 24 h in a 50 mL flask (Table 3). Fermentation results revealed that all yeasts tested were able to consume d-xylose, with consumption rates ranging from 44.60% to 100% in 24 h, except that BUF3-10 consumed 8.63%. This strain had low consumption of xylose but could produce ethanol and xylitol. C. tropicalis strains produced the highest xylitol and ethanol production among the strains (Table 3). Their xylitol production was in the range of 15.06 g L−1 to 22.84 g L−1 and their ethanol production was 0.110 to 0.602 g L−1 in medium containing 60 g L−1 d-xylose at 24 h. Candida tropicalis showed the highest xylitol productivity (Qxy=0.63 g L−1 h−1 to 0.95 g L−1 h−1), with minimum and maximum %Yxy/s values of 29.30% to 74.84%. Strain BUF 9-4 had the highest %Yxy/s value because of its low xylose consumption compared with the same strains and high xylitol productivity (20.02 g L−1). This group prefers to produce xylitol more than ethanol. Candida yeast in particular has been studied with regards to its biotechnological application in the production of xylitol. Barbosa et al. (3) reported that xylitol yield was as high as 0.77 g g−1 or 77% for C. guilliermondii and 0.85 g g−1 or 85% for C. tropicalis. C. parapsilosis showed xylose consumption of around 58% and only produced ethanol of 0.006 to 0.049 g L−1, which was very low. So, yeast strains used xylose as their sole carbon source for growth only. Also, strains assigned as Geotrichum sp. had xylose consumption of 61.20% to 75.30% but very poor ethanol and xylitol production. Strains of C. mengyuniae, S. lactativora and T. asahii produced little ethanol and xylitol production. However, their xylitol production was more than ethanol production (Table 3). S. stipitis JCM 10742T was used as a positive control. This xylose-fermenting yeast produced high ethanol, (%Ye/s=28.5) but showed low xylitol production (%Yxy/s=1) (data not shown).
Table 3.
Xylose fermentation parameters of yeast cultivated in d-xylose culture medium assays under aerobic conditions for 24 h
| Yeast species | Yeast strain | D-Xylose consumption (%)a | Ethanol concentration (g L−1) | %Ye/sb | Xylitol concentration (g L−1) | Qxy (g L−1 h−1)c | %Yxy/sd |
|---|---|---|---|---|---|---|---|
| C. tropicalis | BUF5-3 | 97.80 | 0.329 | 0.34 | 22.34 | 0.93 | 38.10 |
| C. tropicalis | BUF4-2 | 98.15 | 0.169 | 0.17 | 22.84 | 0.95 | 38.80 |
| C. tropicalis | BUF4-3 | 65.53 | 0.225 | 0.34 | 21.19 | 0.88 | 53.90 |
| C. tropicalis | BUF4-4 | 98.57 | 0.178 | 0.18 | 20.15 | 0.84 | 34.10 |
| C. tropicalis | BUF7-1 | 98.64 | 0.297 | 0.30 | 21.35 | 0.89 | 36.10 |
| C. tropicalis | BUF8-1 | 53.15 | 0.333 | 0.63 | 15.06 | 0.63 | 47.20 |
| C. tropicalis | BUF8-2 | 96.92 | 0.602 | 0.62 | 17.04 | 0.71 | 29.30 |
| C. tropicalis | BUF9-2 | 95.80 | 0.474 | 0.49 | 19.69 | 0.82 | 34.30 |
| C. tropicalis | BUF9-4 | 44.60 | 0.452 | 1.01 | 20.02 | 0.83 | 74.80 |
| C. tropicalis | BUF9-5 | 99.72 | 0.133 | 0.13 | 20.12 | 0.84 | 33.60 |
| C. tropicalis | BUF12-1 | 100.0 | 0.110 | 0.11 | 21.62 | 0.90 | 36.00 |
| C. parapsilosis | BUF3-18 | 58.02 | 0.006 | 0.01 | 0.00 | 0.00 | 0.00 |
| C. parapsilosis | BUF3-19 | 58.00 | 0.049 | 0.09 | 0.00 | 0.00 | 0.00 |
| C. mengyuniae | BUF3-6 | 68.59 | 0.060 | 0.09 | 2.59 | 0.11 | 6.30 |
| C. mengyuniae | BUF3-16 | 61.60 | 0.160 | 0.26 | 0.9 | 0.04 | 2.40 |
| S. lactativora | BUF1-1 | 51.57 | 0.071 | 0.14 | 0.00 | 0.00 | 0.00 |
| S. lactativora | BUF3-5 | 80.22 | 0.000 | 0.00 | 3.68 | 0.15 | 7.70 |
| Geotrichum sp. | BUF7 | 75.30 | 0.048 | 0.06 | 0.19 | 0.01 | 0.40 |
| Geotrichum sp. | BUF8 | 65.71 | 0.074 | 0.11 | 0.4 | 0.02 | 1.00 |
| Geotrichum sp. | BUF9 | 68.04 | 0.060 | 0.09 | 0.00 | 0.00 | 0.00 |
| Geotrichum sp. | BUF10 | 61.20 | 0.113 | 0.18 | 0.00 | 0.00 | 0.00 |
| Geotrichum sp. | BUF11 | 70.85 | 0.039 | 0.06 | 0.00 | 0.00 | 0.00 |
| T, asahii | BUF2-1 | 67.29 | 0.042 | 0.06 | 0.00 | 0.00 | 0.00 |
| T. asahii | BUF2-2 | 80.88 | 0.130 | 1.16 | 0.81 | 0.03 | 1.70 |
| T. asahii | BUF2-4 | 65.78 | 0.029 | 0.04 | 1.12 | 0.05 | 2.80 |
| T. asahii | BUF3-10 | 8.63 | 0.028 | 0.32 | 0.34 | 0.01 | 6.50 |
| T. asahii | BUF3-8 | 89.19 | 0.006 | 0.01 | 0.88 | 0.04 | 0.02 |
| T. asahii | BUF3-3 | 58.90 | 0.040 | 0.07 | 0.26 | 0.01 | 0.70 |
d-Xylose consumption (%)—percentage of initial d-xylose consumed;
Ye/s-ethanol yield (g ethanol g−1 d-xylose consumed);
Qxy-volumetric xylitol production rate (g L−1 h−1);
Yxy/l-xylitol yield (g xylitol g−1 d-xylose consumed).
Recently, there have been many attempts to isolate xylose-utilizing yeast that produces high ethanol or xylitol. S. arborariae strain from rotting wood in Brazil was reported to produce ethanol (0.50 g g−1 xylose) (5) and Candida saraburiensis from decaying agriculture residue produced ethanol (3.1–3.6 g L−1 at 72 h) (31). Spathaspora passalidrum from the gut of passalid beetles could produce ethanol (0.4 g g−1 xylose) (13), and C. guilliermondii Xu280, C. maltosa Xu316 and strain YS54 could produce xylitol 0.73 g g−1, 0.70 g g−1 and 0.58 g g−1, respectively (12, 35). Our present study was the first screening of xylose-utilizing yeasts from buffalo feces; therefore, further study is required on xylose fermentation based on several factors that would affect the biosynthesis of xylitol and ethanol production, such as initial xylose concentration, pH, inoculums, culture media, aeration rate and redox imbalance (14, 17, 27, 33).
Conclusion
In this study, xylose-utilizing yeasts distributed in feces from the rectum of buffalo were investigated for the first time. The results showed that they contained members of the genera Candida, Sporopachydermia, Trichosporon and Geotrichum. Only two yeast species, Candida tropicalis and Geotrichum sp., were isolated from young and old Swamp buffalo feces as well as from old Murrah buffalo feces. Moreover, T. asahii and C. parapsilosis isolates were found in young Murrah buffalo feces. In addition, C. mengyuniae, known as a sulfonylurea herbicide-resistant yeast, was isolated from the same young Murrah buffalo feces. All strains of xylose-fermenting yeasts utilized xylose as their sole carbon source. Some strains could ferment xylose to ethanol and xylitol. In particular, C. tropicalis showed the highest yield of xylitol production.
Acknowledgements
This study was supported by the Thailand Research Fund and Chulalongkorn University for a 2008 Royal Golden Jubilee Ph.D. Program as a scholarship to W.L. and the Higher Education Research Promotion and National Research University Project of Thailand, Office of the Higher Education Commission (ENB 275).
References
- 1.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aristdou A, Penttilä M. Metabolic engineering application to renewable resource utilization. Curr Opin Biotechnol. 2000;11:187–198. doi: 10.1016/s0958-1669(00)00085-9. [DOI] [PubMed] [Google Scholar]
- 3.Barbosa MFS, de Medeiros MB, de Mancilha IM, Schneider H, Lee H. Screening of yeasts for production of xylitol from d-xylose and some factors which affect xylitol yield in Candida guilliermondii. J Ind Microbiol. 1988;3:241–251. [Google Scholar]
- 4.Bourcier T, Touzeau O, Thomas F, Chaumeil C, Baudrimont M, Borderie V, Laroche L. Candida parapsilosis keratitis. Cornea. 2003;22:51–55. doi: 10.1097/00003226-200301000-00012. [DOI] [PubMed] [Google Scholar]
- 5.Cadete RM, Santos RO, Melo MA, Mouro A, Gonçaleves DL, Stambuk BU, Gomes FCO, Lachance MA, Rosa CA. Spathaspora arborariae sp. nov., a d-xylose-fermenting yeast species isolated from rotting wood in Brizil. FEMS Yeast Res. 2009;9:1338–1342. doi: 10.1111/j.1567-1364.2009.00582.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen B, Huang X, Zheng JW, Li SP, He J. Candida mengyuniae sp. nov., a metsufuron-methyl resistant yeast. Int J Syst Evol Microbiol. 2009;59:1237–1241. doi: 10.1099/ijs.0.004614-0. [DOI] [PubMed] [Google Scholar]
- 7.Choi JH, Moon KH, Ryu YW, Seo JH. Production of xylitol in cell recycle fermentations of Candida tropicalis. Biotech Lett. 2000;22:1625–1628. [Google Scholar]
- 8.de Hoog GS, Smith M. Galactomyces Redhead & Malloch 1977. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Vol. 2. Elsevier; Amsterdam: 2011. pp. 413–420. [Google Scholar]
- 9.Fell JW, Phaff HJ. Three new yeasts: Cryptococcus dimennae, Cryptococcus kuetzingii and Cryptococcus lactativorus spp. nov. Antonie van Leeuwenhoek. 1967;33:464–472. [Google Scholar]
- 10.Felsenstein J. Phylogenies from molecular sequences: inference and reliability. Annu Rev Genet. 1988;22:521–565. doi: 10.1146/annurev.ge.22.120188.002513. [DOI] [PubMed] [Google Scholar]
- 11.Granström T, Aristidou AA, Leisola M. Metabolic flux analysis of Candida tropicalis growing on xylose in an oxygen-limited chemostat. Metab Eng. 2002;4:248–256. doi: 10.1006/mben.2002.0230. [DOI] [PubMed] [Google Scholar]
- 12.Guo C, Zhao C, He P, Lu D, Shen A, Jiang N. Screening and characterization of yeasts for xylitol production. J Appl Microbiol. 2006;101:1096–1104. doi: 10.1111/j.1365-2672.2006.02994.x. [DOI] [PubMed] [Google Scholar]
- 13.Hou X. Anaerobic xylose fermentation by Spathaspora passalidarum. Appl Microbiol Biotechnol. 2012;94:205–214. doi: 10.1007/s00253-011-3694-4. [DOI] [PubMed] [Google Scholar]
- 14.Jeffries TW. Engineering yeasts for xylose metabolism. Curr Opin Biotechnol. 2006;17:320–326. doi: 10.1016/j.copbio.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 15.Jeffries TW, Kurtzman CP. Strain selection, taxonomy, and genetics of xylose-fermenting yeasts. Enzyme Microb Technol. 1994;16:922–932. [Google Scholar]
- 16.Jeffries TW, Alexander MA. Production of ethanol from xylose by Candida shehatae grown under continuous or fed-batch conditions. In: Kent Kirk T, Chang HM, editors. Biotechnology in pulp and paper manufacture. Butterworth-Heineman; Boston: 1990. pp. 311–321. [Google Scholar]
- 17.Jeon YJ, Shin HS, Rogers PL. Xylitol production from a mutant strain of Candida tropicalis. Lett Appl Microbiol. 2011;53:106–113. doi: 10.1111/j.1472-765X.2011.03078.x. [DOI] [PubMed] [Google Scholar]
- 18.Kurtzman CP, Fell JW, Boekhout T, Robert V. Methods for isolation, phenotypic characterization and maintenance of yeasts. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Vol. 1. Elsevier; Amsterdam: 2011a. pp. 87–110. [Google Scholar]
- 19.Kurtzman CP, Fell JW, Boekhout T, Robert V. Gene sequence analyses and other DNA-based methods for yeast species recognition. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th ed. Vol. 1. Elsevier; Amsterdam: 2011b. pp. 137–144. [Google Scholar]
- 20.Kutzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie van Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 21.Lachance MA, Boekhout T, Scorzetti G, Fell JW, Kurtzman CP. Candida Berkhout 1923. In: Kurtzman CP, Fell JW, Boekhout T, editors. The Yeasts, a Taxonomic Study. 5th. Elsevier; Amsterdam: 2011. pp. 987–1278. [Google Scholar]
- 22.Lee J. Biological conversion of lignocellulosic biomass to ethanol. J Biotechnol. 1997;56:1–24. doi: 10.1016/s0168-1656(97)00073-4. [DOI] [PubMed] [Google Scholar]
- 23.Lin D, Wu LC, Rinaldi MG, Lehmann PF. Three distinct genotypes within Candida parapsilosis from clinical sources. J Clin Microbiol. 1995;33:1815–1821. doi: 10.1128/jcm.33.7.1815-1821.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim SL, Tay ST. Diversity and killer activity of yeasts in Malaysian fermented food samples. Trop Biomed. 2011;28:438–443. [PubMed] [Google Scholar]
- 25.Lorliam W, Akaracharanya A, Jindamorakot S, Suwannarangsee S, Tanasupawat S. Characterization of xylose-utilizing yeasts isolated from herbivore faeces in Thailand. Sci Asia. 2013;39:26–35. [Google Scholar]
- 26.Mahadevan P. Distribution, ecology and adaptation of river buffaloes. In: Tulooh MH, Holmes JHG, editors. Buffalo Production, Production-system Approach. World Animal Science, Elsevier; Amsterdam: 1992. pp. 1–58. [Google Scholar]
- 27.McMillan JD. Xylose Fermentation to Ethanol: A review. NREL/TP 421-4944. National Renewable Energy Laboratory; Colorado: 1993. [Google Scholar]
- 28.Molnar O, Schatzmayr G, Fuchs E, Prillinger H. Trichosporon mycotoxinivorans sp. nov., a new yeast species useful in biological detoxification of various mycotoxins. Syst Appl Microbiol. 2004;27:661–671. doi: 10.1078/0723202042369947. [DOI] [PubMed] [Google Scholar]
- 29.Moran GP, Derek JS, Coleman DC. Emergence of Non-Candida albicans Candida species as pathogen. In: Calderone RA, editor. Candida and Candidiasis. ASM Press; Washington DC: 2002. p. 37. [Google Scholar]
- 30.Nguyen NH, Suh SO, Marshall CJ, Blackwell M. Morphological and ecological similarities: wood-boring beetles associated with novel xylose-fermenting yeasts, Spathaspora passalidarum gen. sp. nov. and Candida jeffriesii sp. nov. Mycol Res. 2006;110:1232–1241. doi: 10.1016/j.mycres.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 31.Nitiyon S, Boonmak C, Am-In S, Jindamorakot S, Kawasaki H, Yongmanitchai W, Limtong S. Candida saraburiensis sp. nov. and Candida prachuapensis sp. nov., xylose-utilizing yeast species isolated in Thailand. Int J Syst Evol Microbiol. 2011;61:462–468. doi: 10.1099/ijs.0.023317-0. [DOI] [PubMed] [Google Scholar]
- 32.O’Donnell K. Fusarium and its near relatives. In: Reynolds DR, Taylor JW, editors. The Fungal Holomorph—Mitotic, Meiotic and Pleomorphic Speciation in Fungal Systematics. Wallingford, UK: 1993. pp. 225–233. [Google Scholar]
- 33.Prakasham RS, Rao RS, Hobbs PJ. Current trends in biotechnological production of xylitol and future prospects. Curr Trends Biotechnol Pharm. 2009;3:8–36. [Google Scholar]
- 34.Raeder U, Broda P. Rapid preparation of DNA from filamentous fungi. Lett Appl Microbiol. 1985;1:17–20. [Google Scholar]
- 35.Rao RS, Bhadra B, Shivaji S. Isolation and characterization of xylitol-producing yeasts from the gut of colleopteran insects. Curr Microbiol. 2007;55:441–446. doi: 10.1007/s00284-007-9005-8. [DOI] [PubMed] [Google Scholar]
- 36.Rao RS, Bhadra B, Shivaji S. Isolation and characterization of ethanol-producing yeasts from fruits and tree barks. Lett Appl Microbiol. 2008;47:19–24. doi: 10.1111/j.1472-765X.2008.02380.x. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues de Miranda L. Sporopachydermia. In: Rodrigues de Miranda L, Kreger-van Rij NJW, editors. The Yeasts, A Taxonomic Study. 3rd. Elsevier; Amsterdam: 1984. pp. 427–430. [Google Scholar]
- 38.Saitou N, Nei M. The neighbor-joining method: a new method for resconstructing phylogenetics trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 39.Sharma RL, Godara R, Thilagar MB. Epizootiology, pathogenesis and immunoprophylactic trends to control tropical bubaline fasciolosis: an overview. J Parasit Dis. 2011;35:1–9. doi: 10.1007/s12639-011-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Silva CJSM, Mussatto SI, Roberto IC. Study of xylitol production by Candida guilliermondii on a bench bioreactor. J Food Eng. 2006;75:115–119. [Google Scholar]
- 41.Sirisansaneeyakul S, Staniszewski M, Rizzi M. Screening of yeasts for production of xylitol from d-xylose. J Ferm Bioeng. 1995;80:565–570. [Google Scholar]
- 42.Smith MT, Poot GA, de Cock AWAM. Reexamination of some species of the genus Geotrichum Link: Fr. Antonie van Leeuwenhoek. 2000;77:71–81. doi: 10.1023/a:1002030518989. [DOI] [PubMed] [Google Scholar]
- 43.Suh SO, Marshall CJ, Mchugh JV, Blackwell M. Wood ingestion by passalid beetles in the presence of xylose-fermenting gut yeasts. Mol Ecol. 2003;12:3137–3145. doi: 10.1046/j.1365-294x.2003.01973.x. [DOI] [PubMed] [Google Scholar]
- 44.Suzuki M, Nakase T, Daengsubha W, Chaowsangket W, Suyanandana P, Komagata K. Identification of yeasts isolated from fermented foods and related materials in Thailand. J Gen Appl Microbiol. 1987;33:205–220. [Google Scholar]
- 45.Thompson JD, Gibson TJ, Pleniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Urubschurov V, Janczyk P, Pieper R, Souffrant WB. Biological diversity of yeasts in the gastrointestinal tract of weaned piglets kept under different farm conditions. FEMS Yeast Res. 2008;8:1349–1356. doi: 10.1111/j.1567-1364.2008.00444.x. [DOI] [PubMed] [Google Scholar]
- 47.Urubschurov V, Janczyk P, Souffrant WB, Freyer G, Zeyner A. Establishment of intestinal microbiota with focus on yeasts of unweaned and weaned piglets kept under different farm conditions. FEMS Microbiol Ecol. 2011;77:493–502. doi: 10.1111/j.1574-6941.2011.01129.x. [DOI] [PubMed] [Google Scholar]
- 48.Wanapat M, Rowlinson P. Nutrition and feeding of swamp buffalo: feed resources and rumen approach. Ital J Anim Sci. 2007;6:67–73. [Google Scholar]
- 49.Yarrow D. Methods for the isolation, maintenance and identification of yeasts. In: Kurtzman CP, Fell JW, editors. The Yeasts, a Taxonomic Study. 4th. Elsevier; Amsterdam: 1998. pp. 77–100. [Google Scholar]