Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) glycosylation, the covalent attachment of N-acetylglucosamine to serine and threonine residues of proteins, is a post-translational modification that shares many features with protein phosphorylation. O-GlcNAc is essential for cell survival and plays important role in many biological processes (e.g. transcription, translation, cell division) and human diseases (e.g. diabetes, Alzheimer's disease, cancer). However, detection of O-GlcNAc is challenging. Here, a method for O-GlcNAc detection using in vitro sulfation with two N-acetylglucosamine (GlcNAc)-specific sulfotransferases, carbohydrate sulfotransferase 2 and carbohydrate sulfotransferase 4, and the radioisotope 35S is described. Sulfation on free GlcNAc is first demonstrated, and then on O-GlcNAc residues of peptides as well as nuclear and cytoplasmic proteins. It is also demonstrated that the sulfation on O-GlcNAc is sensitive to OGT and O-β-N-acetylglucosaminidase treatment. The labeled samples are separated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis and visualized by autoradiography. Overall, the method is sensitive, specific and convenient.
Keywords: CHST, glycosylation, O-GlcNAc, OGA, OGT
Introduction
O-GlcNAc post-translational modification occurs by the addition of a single N-acetylglucosamine residue to serine/threonine residues of cellular proteins. Originally thought to exist solely on nuclear and cytosolic proteins (Comer and Hart 2001), it has recently been discovered on extracellular proteins (Love and Hanover 2005). Unlike other types of glycosylation, the sugar residue is not elongated into complex oligosaccharides. In fact, O-GlcNAc shares many features with protein phosphorylation, a fundamental mechanism for intracellular communication. It is postulated that O-GlcNAc has a mutually exclusive relationship with phosphorylation (Comer and Hart 2001; Love and Hanover 2005; Hart et al. 2011). While a variety of kinases are involved in phosphorylation, intracellular O-GlcNAc is introduced by a single O-GlcNAc transferase (OGT) in the nucleus and cytoplasm (Haltiwanger et al. 1990) and extracellular O-GlcNAc is introduced by a single extracellular O-GlcNAc transferase (EOGT) in the secretary pathway (Sakaidani et al. 2012; Tashima and Stanley 2014). In addition, all O-GlcNAc residues are removed by a single O-β-N-acetylglucosaminidase (OGA) (Gao et al. 2001). O-GlcNAc is involved in many cellular processes, including stress responses, transcription, translation, cell signaling and cell cycle regulation (Hart et al. 2007). O-GlcNAc is also involved in many human diseases including diabetes, Alzheimer's disease and cancer (Slawson et al. 2006; Ma and Hart 2013; Ma and Vosseller 2013; Pearson et al. 2013).
Despite its importance, detection of O-GlcNAc is a challenging task. O-GlcNAc has no effect on protein mobility during electrophoresis because of its small size and neutral charge. Traditionally, the uridine diphosphate (UDP)-[3H]Gal-based galactosyltransferase labeling approach has been the gold standard for O-GlcNAc detection (Torres and Hart 1984; Holt and Hart 1986). Although antibodies such as CTD110.6 (Comer et al. 2001), RL-2 (Snow et al. 1987), O-GlcNAc specific IgG monoclonal antibodies (Teo et al. 2010) and lectins (Roquemore et al. 1994) have been used to detect O-GlcNAc, these reagents suffer from weak binding affinity and limited specificity for O-GlcNAc (Isono 2011). Recently, methods for labeling O-GlcNAc with chemically modified galactose using recombinant galactosyltransferases have been reported. For example, Keto-Gal or GalNAz has been covalently linked to O-GlcNAc using galactosyltransferases or mutant galactosyltransferase Gal-T1 (Y289L) (Ramakrishnan and Qasba 2002; Khidekel et al. 2003; Clark et al. 2008).
Previously, we reported a method for detecting specific glycosaminoglycans and glycan epitopes with radioisotope 35S (Wu et al. 2011) by taking advantage of the observation that in vivo sulfation is usually incomplete (Wu and Lech 2005). Here we extend the method to detect O-GlcNAc using the N-acetylglucosamine (GlcNAc)-specific sulfotransferases, N-acetylglucosamine 6-O-sulfotransferase 1 and 2 (carbohydrate sulfotransferase 2 (CHST2) and carbohydrate sulfotransferase 4 (CHST4), respectively) (Uchimura et al. 2002). Both enzymes introduce sulfate to 6-O of nonreducing N-acetylglucosamine (GlcNAc) residues within various glycan structures (Grunwell et al. 2002). In principle, this 35S labeling approach is similar to UDP-[3H] Gal-based galactosyltransferase labeling, but has the advantage of being much more sensitive.
Results
Free GlcNAc can be sulfated by specific sulfotransferases
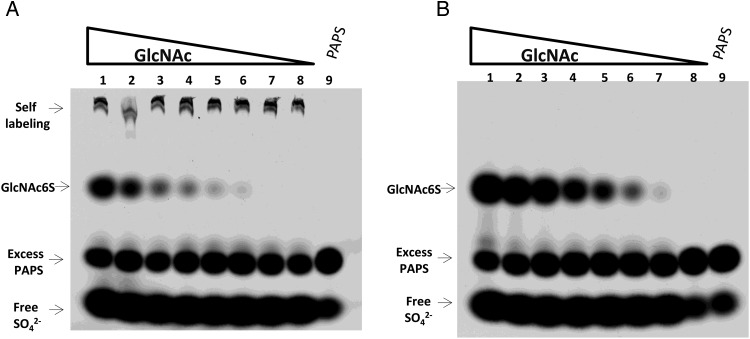
CHST2 and CHST4 are active on oligosaccharides with different reducing end monosaccharides, including GlcNAcβ1-6Man, GlcNAcβ1-2Man and GlcNAcβ1-3Gal (Uchimura et al. 2002). We surmised that these enzymes are active on GlcNAc as well. This hypothesis was tested using an electrophoresis-based sulfotransferase assay (Wu et al. 2010). When CHST2 or CHST4 was incubated with highly radioactive PAP35S and a series of GlcNAc concentrations, a sulfated product formed (Figure 1). Since both CHST2 and CHST4 are GlcNAc 6-O-sulfotranferases, the product was assigned as GlcNAc6S. The Michaelis–Menten constants on GlcNAc of these two enzymes were then determined using a phosphatase-coupled sulfotransferase assay (Prather et al. 2012) (Table I). It should be noted that CHST2 also catalyzed the addition of sulfate to itself, presumably through its N-glycans. Comparing the two enzymes, CHST4 has a lower Km for GlcNAc and a higher Vmax.
Fig. 1.
Sulfation of free GlcNAc by CHST2 and CHST4. Each labeling reaction started with 107 CPM PAP35S, differing concentrations of GlcNAc, and 0.5 µg sulfotransferase. The reactions were incubated at 37°C for 4 h and then resolved with 8% SDS–PAGE. (A) GlcNAc sulfated by CHST2. From lane 1 to 8, GlcNAc decreased from 400 µM to 5.1 pM with a 5-fold serial dilution. The sulfated product GlcNAc6S can be seen in lane 6 with 128 pM GlcNAc. Bands due to CHST2 self-labeling can be seen in all reactions. Lane 9 contained PAPS only. Free sulfate due to PAP35S degradation can be seen in all lanes. (B) The same experiments were repeated for CHST4. GlcNAc6S can be seen in lane 7 with 25.6 pM GlcNAc.
Table I.
Kinetics of CHST2 and CHST4 on GlcNAc
| Km (mM)a | Vmax (pmol/min/µg)a | |
|---|---|---|
| CHST2 | 3.1 | 59.9 |
| CHST4 | 1.2 | 315 |
aThese parameters were measured at constant concentration of PAPS of 0.36 mM at room temperature.
Sulfation of O-GlcNAc peptide
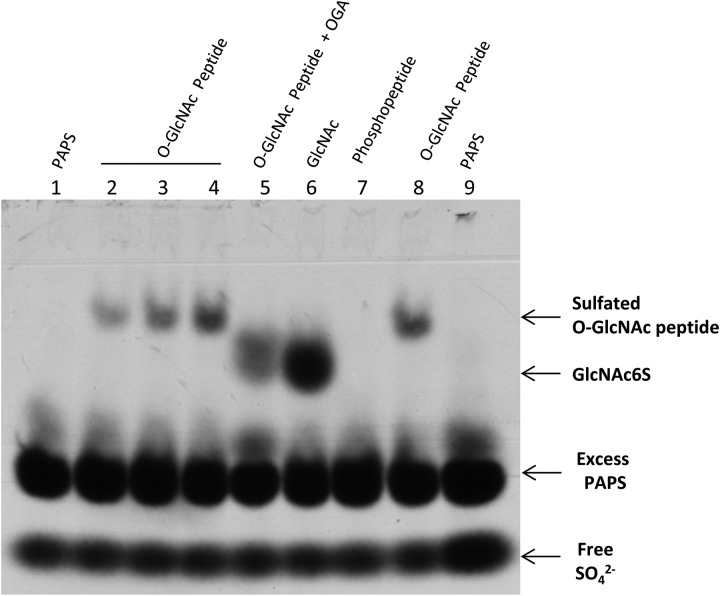
Since CHST2 and CHST4 introduced sulfate to nonreducing end GlcNAc as well as free GlcNAc, it was compelling to test whether sulfation of O-GlcNAc residues on proteins is possible. This hypothesis was first tested on an artificial O-GlcNAc peptide with CHST4 (Figure 2). As expected, CHST4 introduced sulfate to the peptide. Pretreatment of the peptide with OGA, a glycosidase that specifically removes O-GlcNAc, shifted the sulfated band to a position corresponding to GlcNAc6S. In addition, replacement of O-GlcNAc with phosphate completely abolished the labeling, again suggesting that the sulfation was on the O-GlcNAc residue.
Fig. 2.
Sulfation of an O-GlcNAc peptide by CHST4. Lane 2, 3, 4 and 8 contained 0.1, 0.2, 0.4 and 0.4 nmol of an O-GlcNAc peptide, respectively. Lane 5 contained 0.4 nmol of the O-GlcNAc peptide that was pretreated with OGA. Lane 6 contained 2.5 nmol of GlcNAc. Lane 7 contained 0.4 nmol of phosphopeptide. Lane 1 and 9 contained PAP35S only. PAP35S degradation can be seen in all lanes. Samples were separated on 8% SDS–PAGE.
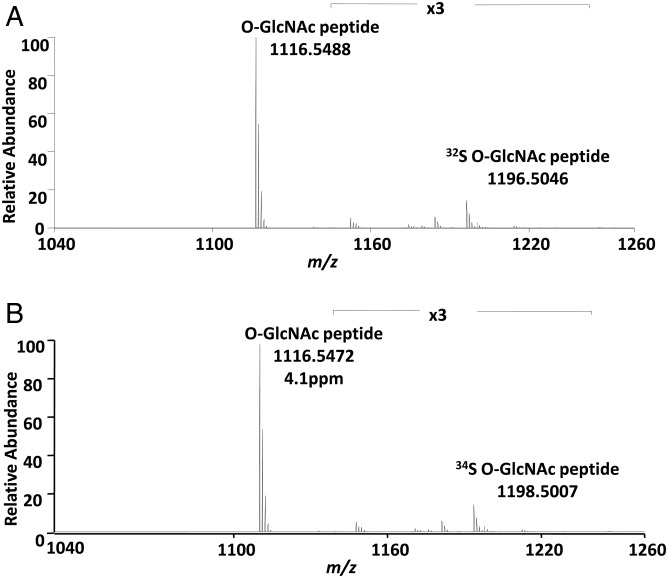
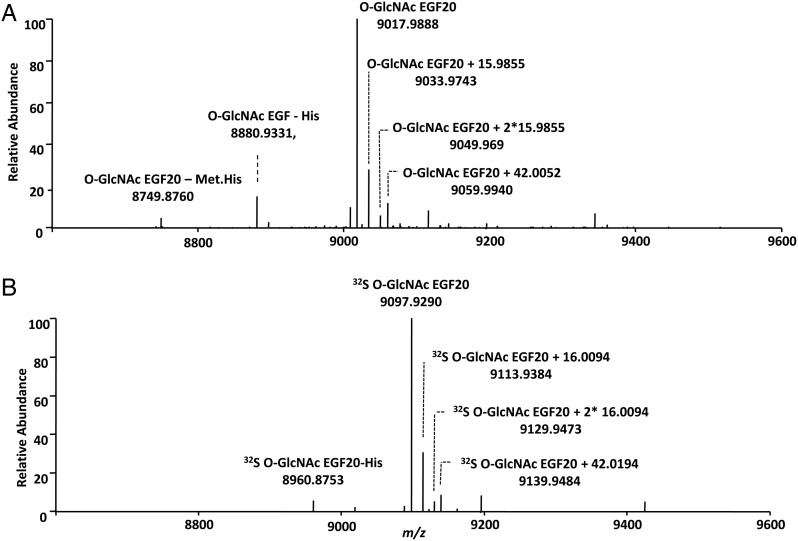
The sulfation of O-GlcNAc peptide was also confirmed by mass spectrometry analysis (Figure 3). The sulfated O-GlcNAcylated peptide was not observed in positive-ion MS mode due to the lability of the sulfate group which resulted in prompt fragmentation and neutral loss of the sulfate moiety. However, high-quality mass spectra were observed in the negative-ion mode. The presence of the ions at m/z 1196.5046 and m/z 1198.5007 proved that the O-GlcNAcylated peptide was sulfated with S-32 and S-34, respectively.
Fig. 3.
Mass spectra of the O-GlcNAc peptide in negative-ion mode. (A) The O-GlcNAc peptide sulfated using PAP32S. (B) The O-GlcNAc peptide sulfated using PAP34S. The ions m/z 1196.5046 and m/z 1198.5007 represent the sulfated peptides with a mass accuracy better than 5 ppm and confirm the addition of sulfate. The peptide sequence is TAPTSTIAPG.
Sulfation of O-GlcNAcylated Notch receptor epidermal growth factor repeat
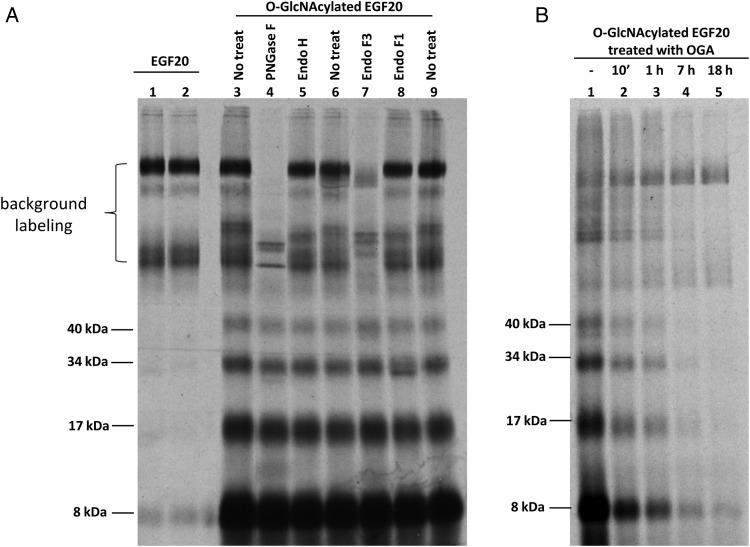
To determine whether CHST2 and CHST4 can sulfate O-GlcNAcylated proteins, a recombinant epidermal growth factor (EGF) repeat EGF20 was first O-GlcNAcylated and then sulfated using CHST2 (Figure 4). When unmodified EGF20 was used as a substrate, bands due to background labeling (likely CHST2 self-labeling) were observed. In contrast, when O-GlcNAcylated EGF20 was used as a substrate, several faster moving bands appeared under nonreducing conditions. When O-GlcNAcylated EGF20 was first sulfated and then deglycosylated with the N-glycan-specific endoglycosidases, peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F (PNGase F) and endo-β-N-acetylglucosaminidase F3 (Endo F3), only the bands due to background labeling were affected. However, the faster moving bands were not affected by the N-deglycosylation, suggesting that the background labeling was on N-glycans and the faster moving bands were due to labeling on O-glycans. This was further confirmed by treatment of O-GlcNAcylated EGF20 with OGA, which reduced the intensities of the fast moving bands, but left those due to background-labeling unchanged. These observations strongly suggested that sulfation occurred on O-GlcNAc residues of O-GlcNAcylated EGF20. Most of the fast moving bands also disappeared under reducing conditions during sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE), suggesting that these bands are likely due to disulfide-linked oligomers of the EGF20 repeat (Supplementary data, Figure S1).
Fig. 4.
O-GlcNAcylated EGF20 labeled by CHST2. Each reaction contained 2 µg of EGF20 or O-GlcNAcylated EGF20 and 0.5 µg CHST2. All samples were resolved on 12% SDS–PAGE under nonreducing conditions. (A) EGF20 (lane 1 and 2) and O-GlcNAcylated EGF20 (lane 3–9) labeled by CHST2. Four bands (molecular weight of 8, 17, 34 and 40 kDa) only with O-GlcNAcylated EGF20 were not susceptible to N-glycosidase treatment (lane 4, 5, 7 and 8). Bands seen in EGF20 labeling were susceptible to PNGase F (lane 4) and Endo F3 (lane 7) treatment. (B) Deglycosylation on O-GlcNAcylated EGF20 abolished the CHST2 labeling. O-GlcNAcylated EGF20 was treated with OGA for the indicated time before CHST2 labeling. The bands at 8, 17, 34 and 40 kDa were abolished by OGA treatment. All samples were treated with Endo F3 after sulfation to partially remove the background labeling.
An intriguing question regarding sulfation is whether it affects the electrophoretic mobility of the target protein during SDS–PAGE due to its negative charge. However, when sulfated O-GlcNAcylated EGF20 was run side by side with the unsulfated version of the repeat, no change on the mobility was observed (Supplementary data, Figure S2), likely due to that sulfated-O-GlcNAc has the same charge as SDS under the conditions for electrophoresis.
To further confirm that the O-GlcNAcylation and sulfation occurred on the EGF20, the modified repeat was also analyzed by mass spectrometry (Figure 5). Extensive dissociation was obtained using ECD which yielded near complete sequence coverage of the EGF20 and maintained the labile modification. The mass spectra indicated that EGF20 was fully O-GlcNAcylated after EOGT treatment (Figure 5A) and was fully sulfated after further CHST2 treatment (Figure 5B). The ECD spectra of the O-GlcNAcylated EGF20 also confirmed that O-GlcNAc occurred on Thr 41 (Supplementary data, Figure S3 and Table S1) (Matsuura et al. 2008). The sulfation site could not be directly identified because the sulfate group did not survive the dissociation using various ionization processes; however, it can be inferred from the precursor ion m/z values to be associated with the O-GlcNAc group.
Fig. 5.
Positive-ion mass spectra of O-GlcNAcylated EGF20 and sulfated O-GlcNAcylated EGF20. Three variants on the EGF20 sequence were observed, corresponding to the full length sequence (HMRSPWPLDD IDECSSNPCQ HGGTCYDKLN AFSCQCMPGY TGQKCETNLE SRGPFEGKPI PNPLLGLDST RTGHHHHHH) with m/z of 9017.9888, the sequence with His truncation with m/z of 8880.9331, and the sequence with His-Met truncation at the N-terminus with m/z of 8749.8760. Mass spectra of (A) O-GlcNAcylated EGF20, (B) O-GlcNAcylated EGF20 sulfated using PAP32S. The mass accuracy was better than 2.7 ppm for the assigned masses. The molecular weight of EGF20 indicated the presence of 3 disulfide bonds, in agreement with the Uniprot database report for EGF (P07207) that indicated linkage between the cysteines C14-C25, C19-C34 and C36-C45.
Sulfation of nuclear and cytoplasmic extracts
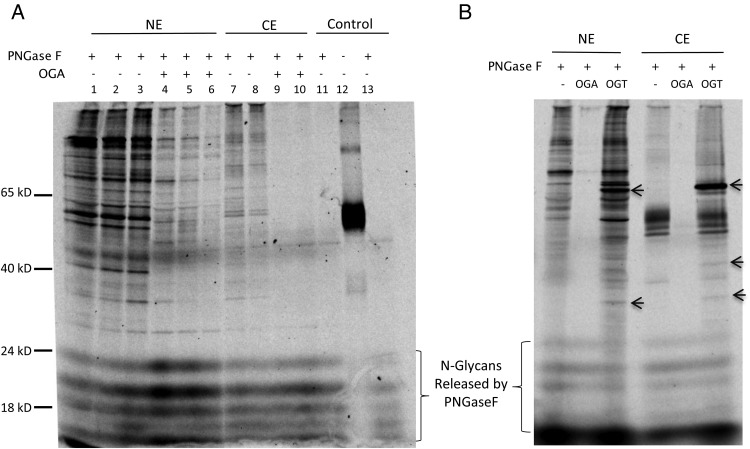
Nuclear and cytoplasmic extracts of HEK293 cells from human embryonic kidney (Graham et al. 1977) were also tested for CHST2 labeling (Figure 6). Both nuclear and cytoplasmic extracts were labeled with CHST2. When the nuclear extract was pretreated with OGA, the labeling was significantly reduced. In contrast, when the samples were pretreated with OGT, some bands on the gel were intensified. More interestingly, OGT treatment also resulted in several novel bands, suggesting that O-GlcNAcylation sites on these proteins were almost completely unoccupied in vivo.
Fig. 6.
Nuclear and cytoplasmic extracts labeling by CHST2 is sensitive to OGA and OGT treatment. All samples were labeled by 0.5 µg recombinant human CHST2 with 107 CPM of S-35 PAPS in 25 µL and incubated at 37°C for 20 min and were resolved on 12% SDS–PAGE under reducing condition. NE, nuclear extract. CE, cytoplasmic extract. (A) CHST2 labeling is sensitive to OGA treatment. Lane 1, 2, 3 contained 8, 4, 2 µg of NE, respectively. Lane 4, 5, 6 contained 8, 4, 2 µg of OGA-treated NE, respectively. Lane 7 and 8 contained 4 and 2 µg of CE, respectively. Lanes 9 and 10 contained 4 and 2 µg of OGA-treated CE. Lane 11, 12 and 13 contained no extracts. Except lane 12, all other reactions were treated with PNGase F to remove the labeling on N-glycans. All pretreatment with OGA lasted 20 min. (B) CHST2 labeling is sensitive to OGT treatment. For comparison, OGA treatment was done simultaneously. All reactions contained 2 µg of sample and were treated with PNGase F after labeling to remove N-glycans. The novel bands appeared in OGT treated samples were indicated with arrows. The samples were pretreated with OGA or OGT for 16 h before labeling.
Difference on CHST2 and CHST4 labeling
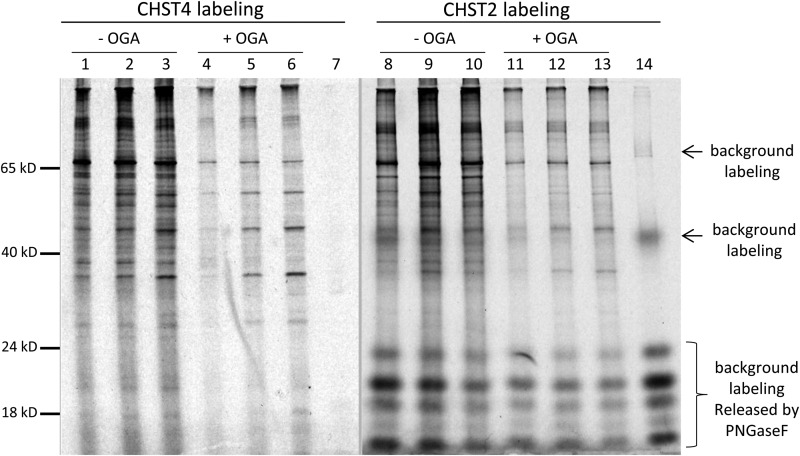
To address whether there is any difference on the preferences of CHST2 and CHST4 labeling, nuclear extract of HEK293 cells was labeled by these two sulfotransferases side by side (Figure 7). While the majority of the labeled bands were similar in both cases, some bands were more significant in one labeling than the other, suggesting that the two enzymes have certain level of difference on their substrate preferences.
Fig. 7.
Comparison of CHST2 and CHST4 labeling on nuclear extract of HEK3 cells. All samples of the nuclear extract of HEK293 cells were labeled with 0.5 µg recombinant human CHST2 or CHST4 with 107 CPM of PAP35S and deglycosylated with PNGase F to remove the background labeling. Amount of nuclear extract applied: 1 µg in lane 1, 4, 8 and 11; 2 µg in lane 2, 5, 9 and 12; and 4 µg in lane 3, 6, 10, 13. The samples in lane 4, 5, 6, 11, 12, and 13 were pretreated with OGA for 10 min before labeling. Lane 7 and 14 contained no nuclear extract to show the background labeling. All samples were resolved on 12% SDS–PAGE under reducing condition.
Discussion
We have presented a method for detecting O-GlcNAc using GlcNAc-specific sulfotransferases (Supplementary data, Figure S4). Evidence of sulfation of O-GlcNAc was shown for peptides, recombinant proteins and cell extracts. The labeling is convenient and specific, and can be accomplished in 20 min. Although the sulfotransferases also sulfate nonreducing end GlcNAc residues of N- and O-glycans, sulfation of O-GlcNAc can be confirmed by OGT, EOGT, OGA and N-specific glycosidase treatment. Sulfation on O-GlcNAc is also supported by mass spectrometry analysis. This method may be used to detect O-GlcNAc on secreted and membrane proteins. It is especially useful for detecting O-GlcNAc on cytoplasmic and nuclear proteins that are usually devoid of other types of glycosylation. This method may also be used to detect arginine GlcNAcylation (Li et al. 2013; Pearson et al. 2013).
The sensitivity of the method is dependent upon several factors, including the kinetics of the sulfotransferase reaction and the specific radioactivity of donor substrate PAP35S. Despite the Michaelis-Menton constants of the sulfotransferases for free GlcNAc in the millimolar range, sulfated GlcNAc can be detected starting from picomolar concentrations of GlcNAc using the described labeling conditions. In theory, the maximum level of sensitivity is reached when all O-GlcNAc residues in a sample are sulfated; therefore, the ultimate sensitivity should be solely dependent on the specific radioactivity applied. Typically, labeled samples are separated on SDS–PAGE to obtain radiograms of the proteins. If necessary, 35S -labeled samples can also be detected by liquid scintillation counting, which is usually far more sensitive. In general, O-GlcNAc on microgram levels of cellular extracts or nanogram levels of pure proteins or peptides can be detected reliably with this method.
All O-GlcNAc samples were labeled so far by CHST2 or CHST4 to certain extent. The different labeling of HEK293 nuclear extracts by CHST2 and CHST4 may reflect the difference in specificity of the two sulfotransferases. Therefore, more information will be obtained by using both enzymes on complex samples. The actual number of O-GlcNAc-modified proteins in a nuclear extract appears be large, judging by the presence of highly smeared bands in the autoradiograms of the labeled extracts of HEK293 cells (Figures 6 and 7) and reported data (Trinidad et al. 2012). While sulfated O-GlcNAcylated EGF20 could be detected using MS without artifactual loss of the sulfate group, the sulfate group dissociated using electron capture dissociation (ECD). It was possible, however, to infer the location of the sulfate group from that of the O-GlcNAc that remained bound to the peptide during dissociation. With these caveats in mind, the use of proteomics methods to identify O-GlcNAcylated proteins in proteomes appears feasible.
When a nuclear extract pretreated with OGT was labeled, several novel bands appeared (Figure 6B), suggesting that these proteins in vivo contained no or very low levels of O-GlcNAcylation, which further suggests that O-GlcNAcylation on different proteins are differently regulated in vivo. The mechanism for achieving different levels of modification on different proteins by a single OGT enzyme is not clear.
Materials and methods
3′-Phosphoadenosine-5′-phosphosulfate (PAPS), PAP34S, Endo F1, Endo H, EndoF3, PNGase F, recombinant B. thetaiotaomicron O-GlcNAcase (OGA), recombinant human CHST2, CHST4, cluster of differentiation 39 like 3 (CD39L3) and recombinant mouse inositol monophosphatase domain containing 1 (IMPAD1) and EOGT were from R&D Systems (Minneapolis, MN). Human OGT from amino acid 382 to 1046 was cloned and expressed in E. coli as previously described with a C-terminal His tag (Gross et al. 2005). The EGF repeat of EGF20 of Drosophila Notch receptor from amino acid 791–829 (Matsuura et al. 2008) was expressed with N-terminal sequence HMRSPWPLD and C-terminal sequence LESRGPFEGKPIPNPLLGLDSTRTGHHHHHH in E. coli. Recombinant OGT and EGF20 were purified using nickel-affinity and gel filtration chromatography. The lysates of OGT and EGF20 expression cells was first loaded onto a 20 mL nickel affinity column with an ÄKTA™ (GE Healthcare) prime and the bound proteins were then eluted with 200 mL each of 0.1 and 0.5 M imidazole solution at pH 6.5. The nickel-purified proteins were further separated on a 450 mL Superdex™-200 gel filtration column with an ÄKTA FPLC system in 25 mM Tris, 150 mM NaCl, pH 7.5. PAP35S was synthesized from carrier-free Na235SO4 (43 Ci/mg, American Radiolabeled Chemicals, Inc.) as previously described (Wu et al. 2002). O-GlcNAc peptide (C33374) and Phosphopeptide (C33373) that had identical amino acid sequences were from Invitrogen. GlcNAc and UDP-GlcNAc were from Sigma Aldrich.
Cell extract preparation
HEK293 cells were grown overnight in a 10 cm dish in Iscove's modified Dulbecco's medium supplemented with 5% fetal bovine serum at 37°C with 5% of CO2. 1 × 107 cells were harvested using 5 mL of fresh cold phosphate-buffered saline and centrifuged for 5 min at 450×g. The cell pellet was gently suspended using 600 µL of isotonic lysis buffer (10 mM Tris–HCl 7.5, 2 mM MgCl2, 3 mM CaCl2, 0.3 M sucrose, 0.2 mM PMSF and 0.5 mM DTT) and incubated for 15 min on ice. After centrifugation for 5 min at 420g, the pellet was suspended with 150 µL of isotonic lysis buffer. The cells were then slowly passed through a syringe with a 27 gauge needle for 6 times. The whole lysate was centrifuged for 20 min at 11,000×g and the supernatant (cytoplasmic fraction) was collected. The pellet was briefly washed with isotonic lysis buffer and then extracted using 150 µL extraction buffer (10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.9, 1.5 mM MgCl2, 0.2 mM ethylenediaminetetraacetic acid, 25% Glycerol, 0.42 M NaCl, 0.2 mM PMSF, and 0.5 mM DTT). The lysate was occasionally vortexed and incubated on ice for 30 min. The supernatant (nuclear fraction) was harvested after centrifugation at 21,000×g for 5 min. Extracts were stored at −80°C.
Preparation of O-GlcNAcylated EGF20
O-GlcNAcylated EGF20 was prepared by mixing 40 µg EGF20, 5 µg EOGT, 1 µg CD39L3, and 1 mM UDP-GlcNAc in 600 µL reaction buffer (25 mM Tris, 0.15 M NaCl, pH 7.5) and incubating the mixture at 37°C overnight. CD39L3 was added to remove the byproduct UDP to increase O-GlcNAcylation. A control reaction containing the same components except EOGT was also run side by side. Both the reaction and control mixtures were then dialyzed in reaction buffer overnight to remove UDP-GlcNAc.
O-GlcNAcylation of cell extracts
A sample of 50 µg cell extract was mixed with 1 µg OGT, 1 µg CD39L3, and 1 mM UDP-GlcNAc in 50 µL reaction buffer (25 mM Tris, 0.15 M NaCl, 10 mM Mn2+, pH 7.5) and was then incubated at room temperature overnight. CD39L3 was introduced to remove the byproduct UDP to increase O-GlcNAcylation. A control reaction containing the same components except OGT was also run side by side.
Removal of O-GlcNAc with OGA treatment
To remove O-GlcNAc, a sample was incubated with 0.2 µg OGA in 50 µL of a buffer containing 50 mM NaAc at pH 5.5 for 10 min to 18 h at 37°C.
In vitro sulfation and N-deglycosylation
For a typical sulfotransferase-labeling reaction, 5 µL sample was mixed with 5 µL PAP35S (>107 CPM, counts per minute) and 0.5 µg CHST2 or CHST4 in a 20 µL buffer containing 100 mM Tris, 150 mM NaCl, pH 7.5. Reactions were incubated at 37°C for at least 20 min and then stopped with 5 µL SDS stop/loading buffer (100 mM Tris, 10% SDS, 30% glycerol, 60 mM β-mercaptoethanol and 0.01% bromophenol blue, pH 8.0). In some cases, higher input of PAP35S and longer reaction time were used to increase the signals for detection. In other cases, 0.5 µg PAP-specific phosphatase IMPAD1 along with 10 mM MgCl2 was added to the reactions to remove the product inhibition caused by PAP (Prather et al. 2012). For N-deglycosylation, a completed labeling reaction was further treated with 0.2 µg of PNGase F or Endo F3 and incubated at 37°C for an additional 20 min.
Electrophoresis
Sulfated GlcNAc and O-GlcNAc peptide were separated on 8% SDS–PAGE in the running buffer of 40 mM Tris, pH 8.0, for 30 min (Wu et al. 2010). Sulfated O-GlcNAcylated EGF20 and cell extract were separated on 12% SDS–PAGE for normal protein separation. After electrophoresis, gels were transferred to cellulose chromatography paper (Fisher Scientific, Cat# 05714-1) and dried with a gel dryer at 80°C under vacuum. The dried gels were then exposed to X-ray films for at least 30 min.
Mass spectrometry analysis
Profile mass spectra and tandem mass spectra (MS/MS) were acquired using an LTQ Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA) coupled with a Triversa Nanomate robot (Advion Biosystems, Inc., Ithaca, NY) which was operated in positive/negative mode. The temperature of the transfer line was set at 100°C. The Triversa Nanomate parameters were as follows: pressure N2 0.3 PSI, voltage ±1.4 kV. All samples were purified by micro-scale C18 purification using Ziptips (Millipore, Billerica, MA) prior to nanospray in 50:50 acetonitrile:water 0.1% formic acid. Profile MS of the O-GlcNAc and sulfated O-GlcNAc peptides and EGF20 were obtained in both the negative-ion and positive-ion modes. The mass spectrometry acquisition parameters were as follows: full scan MS, m/z 200–1000, with a resolving power of 30,000 (O-GlcNAc peptide) and 100,000 (EGF20) at m/z 400 (fwhm) using 1E6 target value, 500 ms maximum injection time and 1–5 microscans. Higher-energy collisional dissociation (HCD) and electron transfer dissociation (ETD) MS/MS data were acquired in positive mode. Tandem MS scans were obtained with a resolving power of 15,000 (O-GlcNAc peptide) or 100,000 (EGF20) (fwhm), with 1E5 target value, 1000 ms max inject time, 5 microscans and an isolation window of 1–5 m/z. The HCD energy used was 15–30%, and the ETD reaction time was set up at 100 ms. MS and MS/MS data were processed with the Xtract module of the Xcalibur™ 2.2 (Thermo Scientific).
ECD MS/MS data were acquired on a 12T-SolariX™ nanoESI-Quadropole-hybrid_FTICR mass spectrometer (Bruker Daltonics, Billerica, MA). The spectra were acquired over a mass range m/z 250–2500. The transient used was 1 M point provided 160,000 resolution at (m/z 400). The ECD pulse length of electron was 10 ms, the ECD bias 1.5 and the cathode heater was 1.5 A. The data were deconvoluted using the Data Analysis software (version 4.2 Bruker).
Supplementary data
Supplementary data for this article is available online at http://glycob.oxfordjournals.org/.
Funding
The work at Boston University was funded by NIH grant P41GM104603.
Abbreviations
CD39L3, cluster of differentiation 39 like 3; CHST2, carbohydrate sulfotransferase 2; CHST4, carbohydrate sulfotransferase 4; CPM, counts per minute; ECD, electron capture dissociation; EGF, epidermal growth factor; Endo F3, endo-β-N-acetylglucosaminidase F3; EOGT, extracellular O-GlcNAc transferase; ETD, electron transfer dissociation; GlcNAc, N-acetylglucosamine; HCD, higher-energy collisional dissociation; IMPAD1, inositol monophosphatase domain containing 1; OGA, O-β-N-acetylglucosaminidase; O-GlcNAc, O-linked β-GlcNAc; OGT, O-β-N-acetylglucosaminyltransferase; PAP, 3′-phosphoadenosine-5′-phosphate; PAPS, 3′-phosphoadenosine-5′-phosphosulfate; PBS, phosphate-buffered saline; PNGase F, peptide-N4-(N-acetyl- β-glucosaminyl)asparagine amidase F; SDS, sodium dodecyl sulfate ; SDS–PAGE, sodium dodecyl sulphate–polyacrylamide gel electrophoresis; UDP, uridine diphosphate.
Supplementary Material
Acknowledgements
We thank Dr. Cindy Zuo and Dr. Wen-Chieh Liao for technique support, Dr. Ruyi Hao for critical reading of the manuscript, and many other coworkers at R&D Systems whose contribution was made through product development.
References
- Clark PM, Dweck JF, Mason DE, Hart CR, Buck SB, Peters EC, Agnew BJ, Hsieh-Wilson LC. Direct iN-gel fluorescence detection and cellular imaging of O-GlcNAc-modified proteins. Journal of the American Chemical Society. 2008;130:11576–11577. doi: 10.1021/ja8030467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Hart GW. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 2001;40:7845–7852. doi: 10.1021/bi0027480. [DOI] [PubMed] [Google Scholar]
- Comer FI, Vosseller K, Wells L, Accavitti MA, Hart GW. Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Analytical Biochemistry. 2001;293:169–177. doi: 10.1006/abio.2001.5132. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. The Journal of Biological Chemistry. 2001;276:9838–9845. doi: 10.1074/jbc.M010420200. [DOI] [PubMed] [Google Scholar]
- Graham FL, Smiley J, Russell WC, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. The Journal of General Virology. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- Gross BJ, Kraybill BC, Walker S. Discovery of O-GlcNAc transferase inhibitors. Journal of the American Chemical Society. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]
- Grunwell JR, Rath VL, Rasmussen J, Cabrilo Z, Bertozzi CR. Characterization and mutagenesis of Gal/GlcNAc-6-O-sulfotransferases. Biochemistry. 2002;41:15590–15600. doi: 10.1021/bi0269557. [DOI] [PubMed] [Google Scholar]
- Haltiwanger RS, Holt GD, Hart GW. Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide beta-N-acetylglucosaminyltransferase. The Journal of Biological Chemistry. 1990;265:2563–2568. [PubMed] [Google Scholar]
- Hart GW, Housley MP, Slawson C. Cycling of O-linked beta-N-acetylglucosamine on nucleocytoplasmic proteins. Nature. 2007;446:1017–1022. doi: 10.1038/nature05815. [DOI] [PubMed] [Google Scholar]
- Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross talk between O-GlcNAcylation and phosphorylation: Roles in signaling, transcription, and chronic disease. Annual Review of Biochemistry. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt GD, Hart GW. The subcellular distribution of terminal N-acetylglucosamine moieties. Localization of a novel protein-saccharide linkage, O-linked GlcNAc. The Journal of Biological Chemistry. 1986;261:8049–8057. [PubMed] [Google Scholar]
- Isono T. O-GlcNAc-specific antibody CTD110.6 cross-reacts with N-GlcNAc2-modified proteins induced under glucose deprivation. PLoS One. 2011;6:e18959. doi: 10.1371/journal.pone.0018959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidekel N, Arndt S, Lamarre-Vincent N, Lippert A, Poulin-Kerstien KG, Ramakrishnan B, Qasba PK, Hsieh-Wilson LC. A chemoenzymatic approach toward the rapid and sensitive detection of O-GlcNAc posttranslational modifications. Journal of the American Chemical Society. 2003;125:16162–16163. doi: 10.1021/ja038545r. [DOI] [PubMed] [Google Scholar]
- Li S, Zhang L, Yao Q, Li L, Dong N, Rong J, Gao W, Ding X, Sun L, Chen X, et al. Pathogen blocks host death receptor signalling by arginine GlcNAcylation of death domains. Nature. 2013;501:242–246. doi: 10.1038/nature12436. [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Science's STKE: Signal Transduction Knowledge Environment. 2005;2005:re13. doi: 10.1126/stke.3122005re13. [DOI] [PubMed] [Google Scholar]
- Ma J, Hart GW. Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Review of Proteomics. 2013;10:365–380. doi: 10.1586/14789450.2013.820536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Z, Vosseller K. O-GlcNAc in cancer biology. Amino Acids. 2013;45:719–733. doi: 10.1007/s00726-013-1543-8. [DOI] [PubMed] [Google Scholar]
- Matsuura A, Ito M, Sakaidani Y, Kondo T, Murakami K, Furukawa K, Nadano D, Matsuda T, Okajima T. O-linked N-acetylglucosamine is present on the extracellular domain of notch receptors. The Journal of Biological Chemistry. 2008;283:35486–35495. doi: 10.1074/jbc.M806202200. [DOI] [PubMed] [Google Scholar]
- Pearson JS, Giogha C, Ong SY, Kennedy CL, Kelly M, Robinson KS, Lung TW, Mansell A, Riedmaier P, Oates CV, et al. A type III effector antagonizes death receptor signalling during bacterial gut infection. Nature. 2013;501:247–251. doi: 10.1038/nature12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather B, Ethen CM, Machacek M, Wu ZL. Golgi-resident PAP-specific 3′-phosphatase-coupled sulfotransferase assays. Analytical Biochemistry. 2012;423:86–92. doi: 10.1016/j.ab.2012.01.003. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan B, Qasba PK. Structure-based design of beta 1,4-galactosyltransferase I (beta 4Gal-T1) with equally efficient N-acetylgalactosaminyltransferase activity: Point mutation broadens beta 4Gal-T1 donor specificity. The Journal of Biological Chemistry. 2002;277:20833–20839. doi: 10.1074/jbc.M111183200. [DOI] [PubMed] [Google Scholar]
- Roquemore EP, Chou TY, Hart GW. Detection of O-linked N-acetylglucosamine (O-GlcNAc) on cytoplasmic and nuclear proteins. Methods in Enzymology. 1994;230:443–460. doi: 10.1016/0076-6879(94)30028-3. [DOI] [PubMed] [Google Scholar]
- Sakaidani Y, Ichiyanagi N, Saito C, Nomura T, Ito M, Nishio Y, Nadano D, Matsuda T, Furukawa K, Okajima T. O-linked-N-acetylglucosamine modification of mammalian Notch receptors by an atypical O-GlcNAc transferase Eogt1. Biochemical and Biophysical Research Communications. 2012;419:14–19. doi: 10.1016/j.bbrc.2012.01.098. [DOI] [PubMed] [Google Scholar]
- Slawson C, Housley MP, Hart GW. O-GlcNAc cycling: How a single sugar post-translational modification is changing the way we think about signaling networks. Journal of Cellular Biochemistry. 2006;97:71–83. doi: 10.1002/jcb.20676. [DOI] [PubMed] [Google Scholar]
- Snow CM, Senior A, Gerace L. Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. The Journal of Cell Biology. 1987;104:1143–1156. doi: 10.1083/jcb.104.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashima Y, Stanley P. Antibodies that detect O-GlcNAc on the extracellular domain of cell surface glycoproteins. The Journal of Biological Chemistry. 2014;289:11132–11142. doi: 10.1074/jbc.M113.492512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nature Chemical Biology. 2010;6:338–343. doi: 10.1038/nchembio.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CR, Hart GW. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. The Journal of Biological Chemistry. 1984;259:3308–3317. [PubMed] [Google Scholar]
- Trinidad JC, Barkan DT, Gulledge BF, Thalhammer A, Sali A, Schoepfer R, Burlingame AL. Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Molecular & Cellular Proteomics: MCP. 2012;11:215–229. doi: 10.1074/mcp.O112.018366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchimura K, El-Fasakhany FM, Hori M, Hemmerich S, Blink SE, Kansas GS, Kanamori A, Kumamoto K, Kannagi R, Muramatsu T. Specificities of N-acetylglucosamine-6-O-sulfotransferases in relation to L-selectin ligand synthesis and tumor-associated enzyme expression. The Journal of Biological Chemistry. 2002;277:3979–3984. doi: 10.1074/jbc.M106587200. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Ethen CM, Larson S, Prather B, Jiang W. A versatile polyacrylamide gel electrophoresis based sulfotransferase assay. BMC Biotechnology. 2010;10:11. doi: 10.1186/1472-6750-10-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Lech M. Modification degrees at specific sites on heparan sulphate: An approach to measure chemical modifications on biological molecules with stable isotope labelling. The Biochemical Journal. 2005;389:383–388. doi: 10.1042/BJ20041827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ZL, Prather B, Ethen CM, Kalyuzhny A, Jiang W. Detection of specific glycosaminoglycans and glycan epitopes by in vitro sulfation using recombinant sulfotransferases. Glycobiology. 2011;21:625–633. doi: 10.1093/glycob/cwq204. [DOI] [PubMed] [Google Scholar]
- Wu ZL, Zhang L, Beeler DL, Kuberan B, Rosenberg RD. A new strategy for defining critical functional groups on heparan sulfate. FASEB Journal: Official Publication of the Federation of American Societies for Experimental Biology. 2002;16:539–545. doi: 10.1096/fj.01-0807com. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.