Abstract
Background
The polyunsaturated arachidonic and docosahexaenoic acids (AA and DHA) participate in cell membrane synthesis during neurodevelopment, neuroplasticity, and neurotransmission throughout life. Each is metabolized via coupled enzymatic reactions within separate but interacting metabolic cascades.
Hypothesis
AA and DHA pathway genes are coordinately expressed and underlie cascade interactions during human brain development and aging.
Methods
The BrainCloud database for human non-pathological prefrontal cortex gene expression was used to quantify postnatal age changes in mRNA expression of 34 genes involved in AA and DHA metabolism.
Results
Expression patterns were split into Development (0 to 20 years) and Aging (21 to 78 years) intervals. Expression of genes for cytosolic phospholipases A2 (cPLA2), cyclooxygenases (COX)-1 and -2, and other AA cascade enzymes, correlated closely with age during Development, less so during Aging. Expression of DHA cascade enzymes was less inter-correlated in each period, but often changed in the opposite direction to expression of AA cascade genes. Except for the PLA2G4A (cPLA2 IVA) and PTGS2 (COX-2) genes at 1q25, highly inter-correlated genes were at distant chromosomal loci.
Conclusions
Coordinated age-related gene expression during the brain Development and Aging intervals likely underlies coupled changes in enzymes of the AA and DHA cascades and largely occur through distant transcriptional regulation. Healthy brain aging does not show upregulation of PLA2G4 or PTGS2 expression, which was found in Alzheimer's disease.
Introduction
The human brain undergoes marked structural and functional changes after birth, such as synaptic growth followed by synaptic pruning, progressive myelination, neuroplasticity, and changes in energy metabolism, which likely underlie maturation and maintenance of cognitive and behavioral abilities [1]–[4]. Programmed changes are largely completed by 21 years of age, although myelination continues through 40 years in regions such as the prefrontal association neocortex [5]–[7]. After about 21 years, homeostatic mechanisms are important for maintaining brain integrity, but even with optimal health, neuropathological age changes are reported [5]–[9]. Furthermore, aging is a risk factor for Alzheimer's and Parkinson's diseases as well as other neurodegenerative diseases and contributes to worsening symptoms of schizophrenia and bipolar disorder [10], [11].
In a genome-wide aging study of brain gene expression in humans and rhesus macaques, Somel et al found that expression variations of energy metabolism, synaptic plasticity, vesicular transport, and mitochondrial functions in the prefrontal cortex translated to related biological functions of the gene products [12]. DNA damage is increased in promoters of genes whose expression decreases with age, which may reduce the expression of selectively vulnerable genes involved in learning, memory and neuronal survival [13]. Epigenetic modifications also occur, as human brain aging is accompanied by a global promoter hypomethylation and hypermethylation of certain promoters, including those for brain derived neurotrophic factor (BDNF) and synaptophysin [14].
Lipids are constituents of brain cell membranes; their metabolism consumes approximately 25% of the brain's ATP, and contribute to neurotransmission and gene transcription [15]–[18]. Furthermore, neurodevelopmental and neurodegenerative diseases have been associated with disturbances in brain lipid composition and related enzymes [19]–[23]. Therefore, we thought it of interest to examine the expression during brain development and aging of a limited number of genes involved in lipid metabolism. We focused on the pathways of two polyunsaturated fatty acids (PUFAs), arachidonic acid (AA, 20∶6n-6) and docosahexaenoic acid (DHA, 22∶6n-3), within their respective coupled metabolic cascades.
In the brain, AA and DHA are mainly esterified in the stereospecifically numbered (sn)-2 position of phospholipids, and in triacylglycerols and cholesteryl esters to a lesser extent [19], [24]. During neurotransmission, AA and DHA may be hydrolyzed from phospholipids by receptor-mediated activation of specific phospholipases A2 (PLA2). For example, Ca2+-dependent cytosolic cPLA2 and Ca2+-independent iPLA2 selectively release AA and DHA, respectively [25], [26]. These PLA2s belong to large families and are found in the brain within neurons and astrocytes [27]–[29]. At synapses, cPLA2 co-localizes with cyclooxygenase (COX)-2, which converts the AA to eicosanoids including prostaglandin E2 (PGE2) [30]–[32].
Once released by a selective PLA2, unesterified AA and DHA may be recycled into phospholipid by an acyltransferase following its activation by an acyl-CoA synthetase (ACSL) to acyl-CoA (Figure S1) [33]–[35]. ACSLs and acyltransferases also belong to enzyme families with varying specificities to AA compared with DHA. ACSL4 is more selective for AA, while ACSL6 is more selective for DHA [36,37l]. The lysophosphatidylcholine acyltransferase LPCAT3 is more selective for AA, LPCAT4 for DHA [38]. Another fraction of unesterified AA and DHA in brain undergoes enzymatic oxidation within distinct metabolic cascades [25], [39], [40], or non-enzymatic loss to reactive oxygen species and other bioactive products. COXs, lipoxygenases (LOXs), and cytochrome P450 epoxygenases (CYP450s) convert AA to eicosanoids such as prostaglandins or leukotrienes, involved in inflammatory responses, and DHA to neuroprotectins and resolvins, which show neuroprotective properties (Figure S1).
In the present study, we focused on transcriptional regulation of PUFA metabolizing enzymes during human development and aging. We used the BrainCloud database, which contains mRNA expression levels of 30,176 gene expression probes [41]. This database was constructed from brains of 269 subjects without a neuropathological or a neuropsychiatric diagnosis, with ages ranging from the fetal period to 78 years [41].
We examined age-related expression of 34 genes largely involved in deacylation-reacylation and enzymatic oxidation of AA and DHA. Based on the literature, we hypothesized that expression of genes for enzymes involved in direct synthesis of prostaglandins and leukotrienes from AA (e.g. COX, CYP450, PTGES) would increase with aging, while expression of genes involved with neuroprotectin and resolvin synthesis from DHA would decrease with aging. Furthermore, because functional coupling has been reported between some genes in the AA or DHA cascades [33], [35], [42]–[48], we expected that genes within the AA and DHA metabolic cascades would be expressed cooperatively.
Methods
The 34 genes included in this study are listed in Table 1. Expression data for these genes was exported from the BrainCloud database, which can be accessed and downloaded from http://braincloud.jhmi.edu/. The database contains gene expression data from postmortem prefrontal cortex from healthy individuals ranging from fetal ages to 78 years [41]. We studied the postnatal brain in two groups of subjects, a Development group aged 0.00548 to 20.95 years (87 subjects) and an Aging group aged 21.02 to 78.23 years (144 subjects). Fetal data was excluded. The brains were collected from the Office of the Chief Medical Examiner in the District of Columbia and Virginia, Northern District, as well as from the National Institute of Childhood Health and Development Brain and Tissue Bank for Developmental Disorders [41]. Subjects' deaths were classified as natural causes, accident, or homicide [41]. The population of individuals in the Development interval consists of 26 females and 61 males, 32 of whom are African-American, 52 of whom are Caucasian, and 3 of whom are Hispanic [41]. The population of individuals in the Aging interval consists of 47 females and 97 males, 80 of whom are African-American, 4 of whom are Asian, 57 of whom are Caucasian, and 3 of whom are Hispanic [41]. See Supplemental Table 7 of Colantuoni et al for more information about the postmortem interval, pH, and RNA integrity of each sample [41]. The intervals were chosen from evidence that most brain development, including development of the prefrontal cortex, is largely completed by 20 years of age [49], [50]. Henceforth, when referring to the intervals, they will be capitalized (e.g. Development and Aging) to distinguish from the processes (e.g. development and aging).
Table 1. Correlation of mRNA expression with age over Development and Aging intervals and significance of difference between intervals.
| Development | Aging | Development and Aging Difference | ||||||
| Family | Gene | PUFA Preference | Expression pattern | Pearson's r | p-value | Pearson's r | p value | p-value |
| PLA2 | PLA2G4A | AA | 6 | 0.546 | <0.0001 | −0.125 | 0.1342 | 0.3201 |
| PLA2G4B | AA | 3 | −0.480 | <0.0001 | 0.315 | 0.0001 | 0.4026 | |
| PLA2G4C | AA | 2 | 0.678 | <0.0001 | 0.111 | 0.1850 | <0.0001 | |
| PLA2G4F | AA | 3 | −0.248 | 0.0207 | 0.115 | 0.1685 | 0.4291 | |
| PLA2G2D | AA | 9 | 0.109 | 0.3144 | 0.069 | 0.4125 | 0.0597 | |
| PLA2G10 | AA | 1 | 0.056 | 0.6075 | 0.111 | 0.1846 | 0.0071 | |
| PLA2G2F_hi | AA | 9 | −0.125 | 0.2498 | 0.032 | 0.7070 | 0.2576 | |
| PLA2G6_hi | DHA | 3 | 0.078 | 0.4745 | 0.041 | 0.6217 | 0.5958 | |
| PNPLA8 | DHA | 9 | 0.226 | 0.0351 | −0.120 | 0.1532 | 0.0001 | |
| PNPLA7_hi | DHA | 9 | −0.206 | 0.0553 | −0.142 | 0.0897 | 0.0146 | |
| PNPLA6 | DHA | 1 | −0.211 | 0.0501 | 0.233 | 0.0050 | 0.0509 | |
| COX | PTGS1_hi | AA | 6 | 0.497 | <0.0001 | −0.101 | 0.2292 | 0.6592 |
| PTGS2_hi | AA | 6 | 0.177 | 0.1020 | −0.270 | 0.0011 | 0.0029 | |
| PGES | PTGES | AA | 6 | 0.648 | <0.0001 | −0.346 | <0.0001 | 0.0479 |
| PTGES2 | AA | 9 | 0.000 | 0.9990 | −0.014 | 0.8711 | 0.9506 | |
| PTGES3_avg | AA | 6 | 0.455 | <0.0001 | −0.072 | 0.3893 | 0.5366 | |
| LOX | ALOX5 | None | 9 | 0.259 | 0.0155 | −0.091 | 0.2795 | 0.0775 |
| ALOX12B | None | 1 | 0.113 | 0.2979 | 0.265 | 0.0013 | <0.0001 | |
| ALOX15B | None | 4 | −0.280 | 0.0086 | 0.274 | 0.0009 | <0.0001 | |
| Fatty Acid Binding Protein | FABP7 | None | 5 | −0.690 | <0.0001 | −0.149 | 0.0746 | <0.0001 |
| Acyl-CoA Synthetase | ACSL4_hi | AA | 6 | −0.238 | 0.0267 | −0.123 | 0.1433 | 0.0005 |
| ACSL6 | DHA | 7 | −0.039 | 0.7208 | 0.032 | 0.6992 | 0.0235 | |
| ACSL3_hi | None | 9 | 0.192 | 0.0742 | −0.098 | 0.2408 | 0.0015 | |
| Acyltransferase | LPCAT3 | AA | 8 | −0.748 | <0.0001 | −0.075 | 0.3722 | <0.0001 |
| LPCAT4_hi | DHA | 1 | 0.204 | 0.0580 | 0.037 | 0.6590 | 0.0001 | |
| TXS | TBXAS1_avg | None | 1 | 0.220 | 0.0407 | −0.163 | 0.0504 | <0.0001 |
| Cytochrome p450 | CYP4F3 | None | 9 | 0.018 | 0.8704 | 0.269 | 0.0011 | <0.0001 |
| CYP4F11 | None | 9 | −0.195 | 0.0708 | 0.150 | 0.0734 | 0.0803 | |
| CYP4F22 | None | 6 | −0.098 | 0.3653 | 0.059 | 0.4833 | 0.0097 | |
| CYP4F2_hi | None | 1 | 0.173 | 0.1088 | 0.092 | 0.2750 | 0.0604 | |
| CYP2C8 | AA | 9 | 0.054 | 0.6199 | 0.023 | 0.7862 | 0.0407 | |
| CYP2J2 | AA | 9 | 0.190 | 0.0776 | 0.061 | 0.4707 | 0.7175 | |
| Transcription Factor | TFAP2D | AA | 1 | 0.065 | 0.5491 | 0.052 | 0.5394 | 0.6465 |
| NFKB1 | AA | 9 | 0.235 | 0.0286 | 0.172 | 0.0394 | 0.0061 | |
The PUFA preferences indicates the preferred substrate for the enzyme, “none” indicates that the enzyme is a significant part of both metabolic pathways. Gene expression patterns are determined from Figure 1. Negative r-values indicate the gene expression level decreases as age increases. A t-test was used to determine if the expression levels for Development were significantly different from Aging. Significant (p<0.05) p-values are bolded. Development: n = 87, Aging: n = 144.
Some genes involved in AA or DHA metabolism were not found in the database and thus were excluded from the analysis (e.g. ALOX15). Some were detected by more than one probe (e.g. PTGES3). Pearson's correlation coefficients were calculated to compare expression data from the probes. If the Pearson's r-value was ≥0.8, as for TBXAS1 (r = 0.811 p<0.0001 Development; r = 0.834 p<0.0001 Aging) and PTGES3 (r = 0.910 p<0.0001 Development; r = 0.856 p<0.0001 Aging), then the average of the expression data for the probes was used and the gene was identified as GENENAME_avg. If not, the probe with the highest intensity was used and labeled GENENAME_hi. Expression data exported from BrainCloud are already linearly corrected for background and log2 ratios of the sample signal to the reference signal (reference is pooled RNA from all subjects) and normalized using loess correction as described in Colantuoni et al 2011[41], [51].
The resulting expression data in the Development and Aging periods were analyzed with Cluster 3.0 software [52], with no filtering or adjustment. Distance between probes was calculated using the Euclidean distance calculation and clustered using the centroid linkage method. The Euclidean distance calculation takes the difference between two gene expression levels directly while taking into account the magnitude of changes in gene expression [53]. Euclidean distance also eliminates possible errors in distance measurements when using the centroid linkage clustering method [53]. Distance is defined as 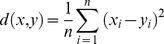 where x and y are each two series of numbers, in this case the age-sorted gene expression values for any two given genes [53]. The output.cdt file was loaded into the TreeView program [54] to generate figures showing correlations between the genes of interest. Pearson's correlations were performed for each gene to determine correlation with age and statistical significance. A t-test was performed using Partek Genomics Suite (Version 6.6 Copyright 2012, Partek Inc., St. Louis, MO, USA) to determine if expression levels in the Development and Aging intervals were significantly different for each gene. A similarity matrix was created for both Development and Aging, comparing expression data between genes using Partek Genomics Suite. This matrix then was clustered using Euclidian distance and centroid linkage clustering to generate a heat map of genes with correlated expression changes for the Development and Aging intervals.
where x and y are each two series of numbers, in this case the age-sorted gene expression values for any two given genes [53]. The output.cdt file was loaded into the TreeView program [54] to generate figures showing correlations between the genes of interest. Pearson's correlations were performed for each gene to determine correlation with age and statistical significance. A t-test was performed using Partek Genomics Suite (Version 6.6 Copyright 2012, Partek Inc., St. Louis, MO, USA) to determine if expression levels in the Development and Aging intervals were significantly different for each gene. A similarity matrix was created for both Development and Aging, comparing expression data between genes using Partek Genomics Suite. This matrix then was clustered using Euclidian distance and centroid linkage clustering to generate a heat map of genes with correlated expression changes for the Development and Aging intervals.
Results
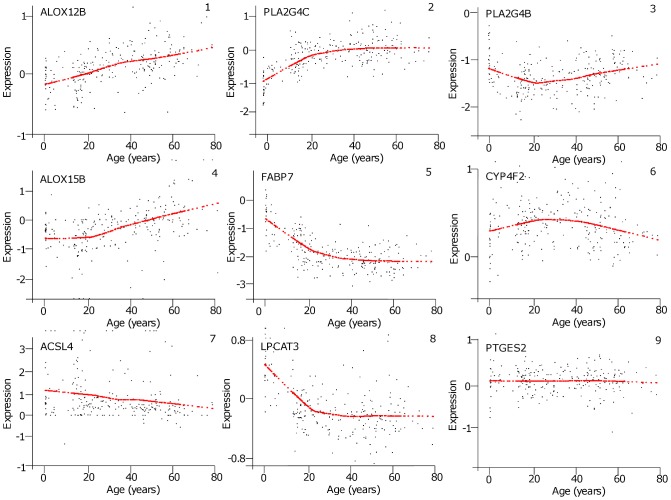
Figure 1 illustrates nine representative graphs of expression data produced by the BrainCloud program, which represent characteristic trends seen in the two age intervals. The values 1 and −1 on the y-axes represent a two-fold change in gene expression in the positive or negative direction, respectively [51], [55]. As noted, some probes change at a fairly steady rate throughout life, either increasing (Fig 1.1, ALOX12B) or decreasing (Fig 1.7, ACSL4_hi) continuously throughout both the Development and Aging periods. Some probes change at different rates, increasing (Fig. 1.2, PLA2G4C) or decreasing (Fig. 1.5, FABP7), but usually changing more quickly during Development than Aging. Others decrease during Development and increase during Aging (Fig. 1.3, PLA2G4B), or increase during Development and decrease during Aging (Fig. 1.6, CYP4F2_hi). Other genes do not have significant changes in expression levels during life (Fig. 1.9, PTGES2). Referring to the patterns in Figure 1, Table 1 lists pattern classification for each gene. There was not a distinct trend of either up- or down-regulation with age for either AA or DHA metabolism genes.
Figure 1. Representative expression patterns.
Graphs were exported directly from BrainCloud. Representative graphs for patterns 1–9 shown here are identified by an example gene. Expression patterns of all genes included in the study are shown in Table 1. The first expression pattern increases at a relatively equal rate throughout life, while the seventh pattern decreases at a relatively equal rate throughout both intervals. Pattern 2 increases sharply during Development then the increase slows during Aging; pattern 5 is the opposite with a sharp decrease during development and a gradual decrease during aging. Pattern 3 decreases during Development, then increases during Aging and pattern 6 increases during Development then decreases during Aging. Pattern 4 remains steady during Development then increases during Aging. Pattern 8 decreases during Development then doesn't change during Aging. Pattern 9 shows minimal changes during both intervals. The units for the y-axes are log2(Sample/Ref). A two-fold change in gene expression occurs at one and negative one on the y-axis. The y-axes are not all identical.
Table 1 also indicates correlations between gene expression and age over the Development and Aging intervals, and whether the correlation with age differed significantly during those two intervals. Gene expression of the AA-selective cPLA2 enzymes (PLA2G4A, PLA2G4B, and PLA2G4C) was correlated with age during Development, whereas only PLA2G4B (cPLA2 IVB) expression correlated with age during the Aging interval (r = 0.577, p<0.001). During Development, expression of PLA2G4A (cPLA2 IVA) and PLA2G4C (cPLA2 IVC) correlated positively with age, while that of PLA2G4B correlated negatively (p<0.001). Only PLA2G4C showed a significant difference in correlation with age between Development and Aging.
PTGS1 (COX-1) and PTGES3 (prostaglandin (PG) E synthase 3, cPGES) correlated positively (p<0.001) with age during Development, whereas PTGS2 (COX-2) (p<0.01) and PTGES3 (p<0.001) correlated negatively with age during the Aging interval. Age-correlations differed significantly for PTGS2, suggesting different roles in neurodevelopment and cell maintenance in the conversion of AA to PGE2.
FABP7 (fatty acid binding protein 7, which has a high affinity for brain DHA [56]) and LPCAT4 correlated significantly with age during the Development but not the Aging interval. Like PLA2G4B, PTGES (PGE synthase 1, mPGES1) and ALOX15B (15-LOX-B), which selectively converts AA to 5S-HETE, were significantly correlated with age during both intervals, but in opposite directions, showing a switch in gene expression pattern. During the Aging interval, PNPLA6 (an iPLA2 selective for DHA), ALOX12B (12-LOX-B), and CYP4F3 (cytochrome p450 family 4, subfamily F, polypeptide 3) expression levels were correlated positively with age, whereas PTGS2_hi (COX-2) had a negative correlation with age. Other genes in the list were not, or were weakly, correlated with age and often displayed variable expression patterns (Figure 1, Table 1).
A t-test was also used to compare gene expression patterns during Development and Aging. More than half of the comparisons were statistically significant, which confirms that these selected intervals are relevant to analyze variation of gene expression throughout the life span (Table 1).
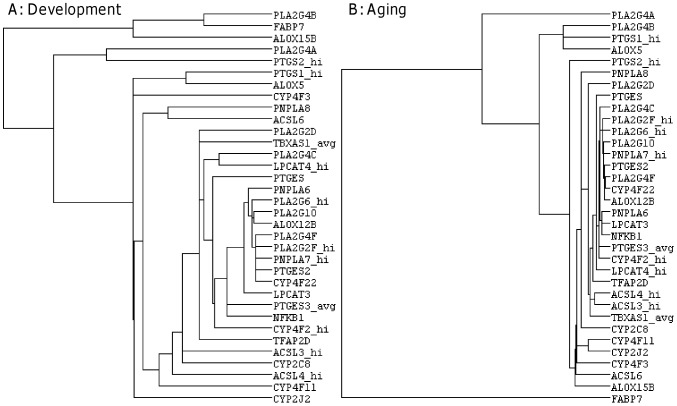
Correlations in expression levels between the genes are illustrated in correlation trees for the two age intervals, Development (Figure 2A) and Aging (Figure 2B). Some gene groups are closely correlated in both of the two intervals, such as PLA2G4F (cPLA2 IVF) and CYP4F22 (cytochrome P450 family 4 subfamily F polypeptide 22), or PTGS1 (COX-1) and ALOX5 (5-LOX). In the Aging tree, FABP7 (fatty acid binding protein) expression was not associated with any other genes because it was very downregulated as compared to the other genes (Figure 2B).
Figure 2. Hierarchal clustering for Development (A) and Aging (B) intervals.
Clustering was performed using Cluster 3.0 and resulting trees obtained using TreeView software. Horizontal length indicates relative relatedness of gene expression levels based on cluster calculations. Genes with short horizontal distances between them have closely correlated expression levels and genes with long branches do not. Development: n = 87, Aging: n = 144.
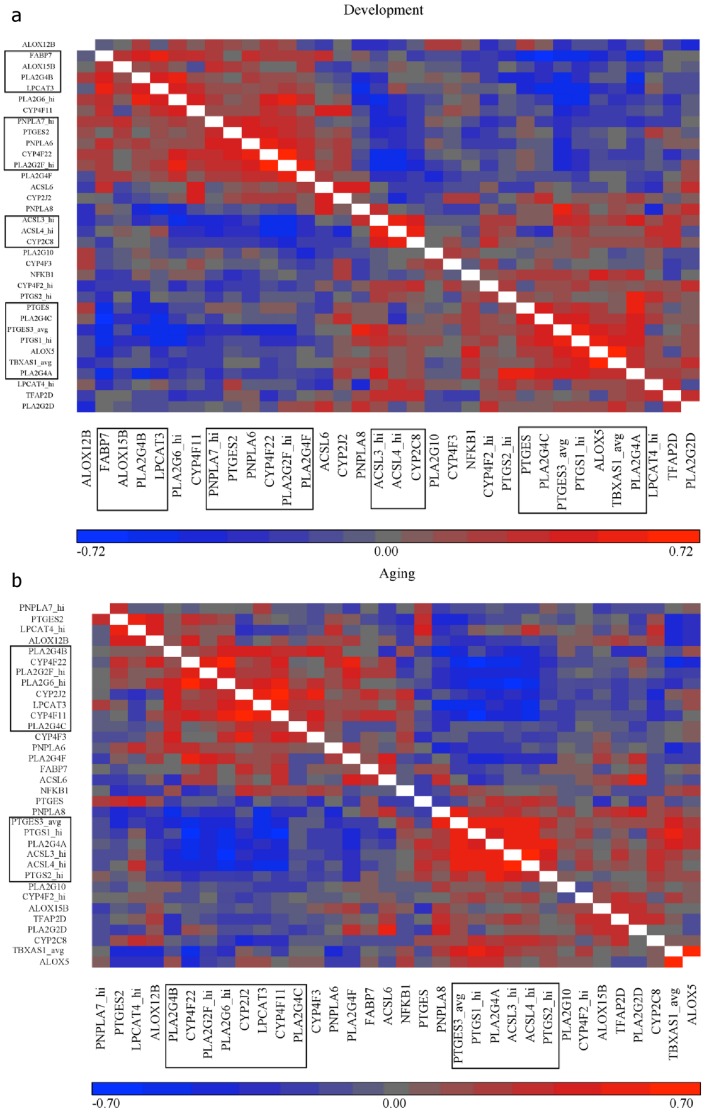
Similarity matrices were calculated showing correlations between each pair of genes for the Development (Figure 3A) and Aging intervals (Figure 3B). From each of these matrices, groups of genes appeared highly correlated, either positively (red), or negatively (blue). During Development, two groups of highly correlated genes were PNPLA7_hi (patatin-like phospholipase domain-containing protein 7, an iPLA2), PTGES2 (mPGES-2), PNPLA6 (neuropathy target esterase, iPLA2 delta), CYP4F22 (CYP450 family 4, subfamily F, polypeptide 22), PLA2G2F_hi (sPLA2 IIF), PLA2G4F (cPLA2 IVF) (Group 1) and PTGES (mPGES1), PLA2G4C (cPLA2 IVC), PTGES3_avg (cPGES), PTGS1_hi (COX-1), ALOX5 (5-LOX), TBXAS1_avg (thromboxane-A synthase 1, TXS), PLA2G4A (cPLA2 IVA) (Group 2). Another group made of ACSL3_hi (ACSL3), ACSL4_hi (ACSL4), and CYP2C8 (CYP450 family 2, subfamily C, polypeptide 8) (Group 3) showed a strong negative correlation with Group 1. Yet another group including FAPB7 (fatty acid binding protein), ALOX15B (15-LOX-B), PLA2G4B (cPLA2 IVB), and LPCAT3 (LPCAT3) (Group 4) correlated negatively with Group 2.
Figure 3. Similarity matrices showing correlations between genes in the Development (A) and Aging (B) intervals.
Calculations and figures were generated using Partek Genomics Suite (Version 6.6 2012). Red indicates positive correlation and blue negative correlation. Scale on the bottom shows the range of colors with values at the extreme of each color; the development interval (A) ranges from -0.72 (bright blue) to 0.72 (bright red) while the aging interval (B) ranges from −0.70 (bright blue) to 0.70 (bright red). The x- and y- axes are the same; thus the two halves of the matrix (split by the white boxes) are simply reflected versions of one another. Black boxes denote the groups of genes that are highly positively or negatively correlated with one another. Development: n = 87, Aging: n = 144.
The Aging matrix (Figure 3B) showed only two groups of high correlation. The first group was PLA2G4B (cPLA2 IVB), CYP4F22 (CYP450 family 4, subfamily F, polypeptide 22), PLA2G2F_hi (sPLA2 IIF), PLA2G6_hi (iPLA2 VI), CYP2J2 (CYP450 family 2, subfamily J, polypeptide 2), LPCAT3 (LPCAT3), CYP4F11 (CYP450 family 4, subfamily F, polypeptide 11), and PLA2G4C (cPLA2 IVC) (Group 1); these genes were highly positively correlated with each other. The second highly positively correlated group was PTGES3_avg (cPGES), PTGS1_hi (COX-1), PLA2G4A (cPLA2 IVA), ACSL3_hi (ACSL3), ACSL4_hi (ACSL4), and PTGS2_hi (COX-2) (Group 2). The blue area on the heat map indicates a strong negative correlation between Group 1 and Group 2.
Pearson's correlation coefficients were calculated for each pair of genes located on the same chromosome and only significantly correlated (p<0.01) gene pairs are presented in Table 2. Among pairs correlated at p<0.0001, PLA2G4A (cPLA2 IVA) and PTGS2 (COX-2) were positively correlated for both the Development and Aging intervals, while PTGES3 (cPGES) and LPCAT3 (LPCAT3) were inversely correlated for both intervals. PLA2G4A and PTGS2 are close to each other on chromosome 1 [31], [32]. However, PTGES3 and LPCAT3 are not located on the same arm of chromosome 12. Furthermore, the loci of many genes on chromosome 19 are very close to each other without being significantly correlated. Thus it appears that proximity in locus is not associated with correlation in expression for the AA and DHA metabolism genes.
Table 2. Correlations between pairs of genes on the same chromosome.
| Chromosome | Gene | Locus | Interval | Gene Pair | Pearson's r | p-value |
| 1 | PLA2G4A | 1q25 | Development | PLA2G4A and PTGS2 | 0.577 | <0.0001 |
| PLA2G2D | 1p36.12 | Aging | PLA2G4A and PTGS2 | 0.541 | <0.0001 | |
| PLA2G2F | 1p35 | Aging | PLA2G4A and PLA2G2F | −0.420 | <0.0001 | |
| PTGS2 | 1q25.2-q25.3 | Aging | PLA2G4A and CYP2J2 | −0.425 | <0.0001 | |
| Aging | PTGS2 and PLA2G2F | −0.266 | 0.0091 | |||
| Aging | PTGS2 and CYP2J2 | −0.227 | 0.0013 | |||
| Aging | PLA2G2F and CYP2J2 | 0.169 | 0.0027 | |||
| 6 | FABP7 | 6q22-q23 | ||||
| TFAP2D | 6p12.3 | |||||
| 7 | PNPLA8 | 7q31 | Aging | PNPLA8 and TBXAS | 0.262 | 0.0015 |
| TBXAS1 | 7q34-q35 | |||||
| 9 | PNPLA7 | 9q34.3 | Development | PNPLA7 and PTGES2 | 0.474 | 0.0004 |
| PTGS1 | 9q32-q33.3 | Development | PTGES and PTGS1 | 0.282 | 0.0031 | |
| PTGES | 9q34.3 | Aging | PTGES and PTGES2 | 0.310 | 0.0002 | |
| PTGES2 | 9q34.12 | Aging | PNPLA7 and PTGES2 | 0.487 | 0.0002 | |
| Aging | PTGES2 and PTGS1 | −0.189 | 0.0032 | |||
| 10 | ALOX5 | 10q11.2 | ||||
| CYP2C8 | 10q24.1 | |||||
| 12 | PTGES3 | 12q13.13 | Development | PTGES3 and LPCAT3 | −0.589 | <0.0001 |
| LPCAT3 | 12p13.31 | Aging | PTGES3 and LPCAT3 | −0.526 | <0.0001 | |
| 15 | PLA2G4B | 15q11.2-q21.3 | ||||
| PLA2G4F | 15q15.1 | |||||
| LPCAT4 | 15q14 | |||||
| 17 | ALOX12B | 17p13.1 | ||||
| ALOX15B | 17p13.1 | |||||
| 19 | PLA2G4C | 19q13.3 | Development | CYP4F22 and PNPLA6 | 0.514 | <0.0001 |
| PNPLA6 | 19p13.2 | Development | CYP4F11 and CYP4F22 | 0.294 | 0.0057 | |
| CYP4F3 | 19p13.12 | Aging | CYP4F3 and CYP4F11 | 0.432 | <0.0001 | |
| CYP4F11 | 19p13.1 | Aging | CYP4F3 and PLA2G4C | 0.333 | <0.0001 | |
| CYP4F22 | 19p13.12 | Aging | CYP4F11 and CYP4F22 | 0.393 | <0.0001 | |
| CYP4F2 | 19p13.12 | Aging | CYP4F22 and PNPLA6 | 0.301 | 0.0002 | |
| Aging | CYP4F11 and PLA2G4C | 0.222 | 0.0074 |
All chromosomes that contain multiple AA or DHA metabolism gene loci are listed. Loci were found using the HUGO Gene Nomenclature Committee database (genenames.org). Correlations between genes are split into Development and Aging intervals. Only correlations where p<0.01 are shown. Development: n = 87, Aging: n = 144.
Pearson's correlation values were also calculated for genes in the same family (e.g. the LOX or PGES family), for genes known to be coupled (e.g. PLA2G4A and PTGS2), or between transcription factors and their associated genes (Table 3). The correlations between PLA2G4A/PTGS2 (cPLA2 IVA/COX-2) and PTGES2/PTGES3 (mPGES-2/cPGES) were the only significant correlations in both the Development and Aging intervals (PLA2G4A/PTGS2 p<0.0001; PTGES2/PTGES3 p = 0.0009 Development, p<0.0001 Aging). Furthermore, the transcription factors NFKB1 (nuclear factor of kappa light polypeptide gene enhancer in B-cells 1, NF-κB) and TFAP2D (transcription factor AP-2 delta, AP-2) were not correlated (r<0.2, p>0.05) with their associated genes, PTGS2 (COX-2) and PLA2G4A (cPLA2 IVA), respectively. Therefore, gene expression within functional families did not follow the same pattern throughout life and there was no correlated expression between transcription factors and the genes they regulate. However, lack of correlation between transcription factors and genes they regulate would be expected since these transcription factors regulate multiple genes, and the genes studied are regulated by multiple transcription factors.
Table 3. Correlations between expression levels of functionally coupled enzymes.
| Genes | Interval | Pearson's r | p-value | |
| cPLA2 and COX-2 | PLA2G4A and PTGS2 | Development | 0.577 | <0.0001 |
| Aging | 0.541 | <0.0001 | ||
| LOX | ALOX12B and ALOX15B | Development | −0.006 | 0.9581 |
| Aging | 0.156 | 0.0621 | ||
| ALOX12B and ALOX5 | Development | −0.125 | 0.2502 | |
| Aging | −0.327 | <0.0001 | ||
| ALOX15B and ALOX5 | Development | −0.233 | 0.0302 | |
| Aging | −0.013 | 0.8799 | ||
| PGES | PTGES and PTGES2 | Development | 0.070 | 0.5217 |
| Aging | 0.310 | 0.0002 | ||
| PTGES and PTGES3 | Development | 0.200 | 0.0633 | |
| Aging | −0.130 | 0.1209 | ||
| PTGES2 and PTGES3 | Development | −0.349 | 0.0009 | |
| Aging | −0.448 | <0.0001 | ||
| Transcription factor and gene it regulates | PLA2G4A and TFAP2D | Development | 0.191 | 0.0764 |
| Aging | 0.121 | 0.1496 | ||
| PTGS2 and NFKB1 | Development | 0.140 | 0.1965 | |
| Aging | 0.072 | 0.3898 |
Gene pairs that are in the same family, function in sequential steps of a part of the pathways, or are transcription factors/target gene pairs. Significant (p<0.05) p-values are bolded. Development: n = 87, Aging: n = 144.
We performed Pearson's r correlations for functionally similar pairs of genes that are selective for one PUFA over the other: PLA2G4A/PLA2G6 (cPLA2 IVA/iPLA2 VI), ACLS4/ACLS6 (ACSL4/ACSL6), and LPCAT3/LPCAT4 (LPCAT3/LPCAT4) (Table 4). Within the PLA2 family, there was a significant negative correlation during Development (r = −0.274, p = 0.0102) and Aging (r = −0.476, p<0.0001) between the AA-selective PLA2G4A (cPLA2 IVA) and the DHA-selective PLA2G6 (iPLA2 VI). During Aging, there was no significant correlation between the acyl-CoA synthetase genes, but the acyltransferase genes were negatively correlated (r = −0.240, p = 0.003).
Table 4. Correlation between AA and DHA metabolism.
| Interval | Pearson's r | p-value | ||
| cPLA2 IVA and iPLA2 VI | PLA2G4A and PLA2G6 | Development | −0.274 | 0.0102 |
| Aging | −0.476 | <0.0001 | ||
| ACSL | ACLS4 and ACSL6 | Development | −0.255 | 0.017 |
| Aging | −0.089 | 0.287 | ||
| LPCAT | LPCAT3 and LPCAT4 | Development | 0.055 | 0.6146 |
| Aging | −0.240 | 0.0038 | ||
Functionally similar genes with specificity for either AA or DHA are correlated to show how the two pathways are associated during Development and Aging. Significant (p<0.05) p-values are bolded. Development: n = 87, Aging: n = 144.
Discussion
We examined age variations throughout life span in human brain expression levels of a limited set of genes involved in PUFA metabolism. We chose AA and DHA metabolism because these PUFAs and their metabolites influence multiple brain processes, including neurotransmission, synaptic growth, gene transcription, membrane fluidity, and the pathological processes of apoptosis, neuroinflammation and excitotoxicity [57]–[61].
We analyzed two postnatal age intervals, Development (0–20 years), and Aging (21 years and older), chosen on the basis of known functional and structural brain changes [1]–[3], [12]. Confirming these intervals as separate time periods involving distinct aspects of brain function and structure, we showed that expression patterns of most genes were statistically different between Development and Aging. Correlations between gene expression level and age were generally lower in the Aging interval than the Development interval, suggesting that with aging, gene expression regulation is less connected to programmed brain changes. Thus as an individual ages, gene expression likely depends more on individual factors, such as health status, environmental stress, nutrition, and other factors influencing lipid metabolism [4], [18], [62]–[64].
Generally, significant correlations between genes were not related to chromosomal location. However, we did find strong positive correlations between expression of PLA2G4A (cPLA2 IVA, locus 1q25) and PTGS2 (COX-2, locus 1q25.2-q25.3) in both the Development and Aging intervals. The coding regions for PLA2G4A and PTGS2 are separated by only about 149 kb of DNA along the long arm of chromosome 1 (1q) [65]. cPLA2 IVA (PLA2G4A) selectively releases AA from the sn-2 position of phospholipids, while COX-2 (PTGS2) catalyzes the rate-limiting step of released AA's conversion to PGE2 [36], [65]. Their highly correlated expression supports the functional coupling between these two AA-selective enzymes that has been reported in cell cultures and in the brain in vivo [25], [43], [44]. Functionally, inducible COX-2 can only convert AA-released by cPLA2 and is not active on exogenous AA [47], [66]. The co-localization and high correlation of expression levels of PLA2G4A (cPLA2 IVA) and PTGS2 (COX-2) in Development and Aging also indicate tight transcriptional co-regulation and co-evolution.
We also identified a larger group of genes whose expression was inter-correlated during the Aging period. This group includes PTGS1_hi (COX-1), PTGS2_hi (COX-2), PLA2G4A (cPLA2 IVA), ACSL3_hi (ACSL3), ACSL4_hi (ACSL4), and PTGES3_avg (cPGES), all of which had a high positive correlation with each other. These genes operate together in a multi-enzymatic cascade catalyzing the conversion of AA to specific eicosanoids [25], [44], [67], and their high positive correlations indicate cooperative regulation during Aging.
mRNA and protein levels of cPLA2 IVA (PLA2G4A), sPLA2 IIA (PLA2G2A), COX-1 and -2 (PTGS1, PTGS2), mPGES1 (PTGES1), and LOX-12 and -15 (ALOX12B, ALOX15B), are increased in Alzheimer's disease in the frontal cortex [68], hippocampus [69]–[72], and cerebellum [69]. In contrast to these reported increases, both PLA2G4A (cPLA2 IVA) and PTGS2 (COX-2) belong to expression pattern groups that decrease during healthy Aging and both genes correlate negatively with age. Genes whose mRNA levels decline with age have significantly greater promoter DNA damage [13], so some mechanism may prevent normal downregulation of PLA2G4A and PTGS2 in Alzheimer's disease. Furthermore, the expression of DHA selective iPLA2 VIA (PLA2G6) is reduced in Alzheimer's disease [68], but we found that its expression is increased in Aging, which shows another disconnection between healthy and pathological aging.
There is some evidence that the brain DHA concentration in brain is reduced with age, especially in patients who develop neurodegenerative disease [19], [22]. DHA regulates membrane fluidity, gene transcription, and can be metabolized to anti-inflammatory neuroprotectins and resolvins [73]–[76]. However, unlike AA selective enzymes, expression of DHA selective enzymes (PLA2G6, ACSL6, LPCAT4) was not specifically correlated during Aging (Figure 3B). Although BrainCloud is a powerful database, there are some limitations to the program. First, its expression data are obtained only from postmortem prefrontal cortex gray matter [41]. This brain region has comparatively prolonged myelination and is reported to show disproportionate degeneration with aging as compared to other neocortical regions [2], [6], [49]. Expression patterns would be expected to differ between regions and many age related changes in brain occur in white matter, which is not analyzed in the BrainCloud project [49]. Finally, BrainCloud does not distinguish between cell types. The Allen Brain Atlases found that astrocytes, oligodendrocytes, and neurons exhibit different age-related changes in gene expression [77]–[79]. On the other hand, to-date BrainCloud has the largest number of samples of gene expression data in the prefrontal cortex. The Allen Human Brain Atlas contains data from only 3 individuals, all male, while the Loerch study contains data from 28 human samples [41], [77], [78]. As such, BrainCloud is an extremely powerful tool for studying age-related gene expression changes in a diverse sample population (including both sexes and four races).
In the future, it would be of interest to investigate possible mechanisms of the age-related changes in mRNA levels. Methylation of gene promoters, histone acetylation and methylation state, transcription factors, miRNAs, DNA sequences of cis-elements (transcription factor binding sites), and feedback regulation by AA and DHA and their metabolites likely play a role in changing mRNA expression levels [80]. Generally, gene groups whose expression decreases with age appear to have higher promoter GC content than other genes [12], suggesting differences in methylation state, and human brain aging is associated with a global hypomethylation [14]. Gene-specific promoter methylation can now be analyzed in BrainCloudMethyl, a database similar to BrainCloud that contains CpG methylation data [81]. Histone acetylation and methylation state have been shown to affect aging of cells [82], [83] and miRNAs have been shown to influence aging in stem cells [84] and the brain [85]. Cis-element sequences affect binding of transcription factors and thus could affect levels of transcripts. Of the two transcription factors included in this study, TFAP2D (AP-2) and NFKB1 (NF-κB), only one (NFKB1) correlated significantly with age during both Development and Aging and had significantly different expression patterns between the two intervals. Transcription factors not included in this study also likely differ in expression during these intervals. These multiple factors likely cooperate to regulate gene expression during Development and Aging and contribute not only to the pattern switch from Development to Aging, but also to the switch from healthy to pathological aging. Their role in the regulation of the genes examined here warrants further investigation.
While there does not appear to be a trend of either upregulation or downregulation with age for AA or DHA related genes, the genes involved in the AA pathway were highly positively correlated with each other, indicating cooperative regulation during aging, which was not the case for the genes involved in DHA metabolism. Further, AA and DHA genes were negatively correlated with each other, indicating independent or inverse regulation, and competition between these two major pathways. These findings are consistent with previous studies in which reduced dietary n-3 PUFA content led to downregulation of DHA and upregulation of brain AA metabolic cascade enzymes in the rat brain, whereas the reverse was true when reducing dietary n-6 PUFA content [86]. Furthermore, mood stabilizers that are used to treat bipolar disorder, lithium, carbamazepine, and valproate, downregulate the AA but not the DHA cascade in rat brain [87].
In summary, we have demonstrated coupled and distinct patterns of changes in mRNA expression of two metabolic brain cascades, that not only suggest different roles of the individual cascades in healthy human brain Development and Aging, but also in underlying processes such as brain growth and neurotransmission during these life periods. The known inverse coupling of the AA and DHA cascades is underscored by profound and coordinated regulation of gene expression of their enzymes. The interaction between transcriptional and phenotypic mechanisms in the normal as well as pathological brain deserves further exploration.
Supporting Information
The brain arachidonic and docosahexaenoic acid cascades. After AA within phospholipid is released by cPLA2 or sPLA2, a portion is converted to prostaglandin H2 (PGH2) by COX-1 or COX-2, to hydroxyeicosatetraenoic acid (20-HETE), to hydroperoxyeicosatetraenoic acids (HPETES) by lipoxygenase (LOX) subtypes 5, 12 or 15. PGH2 is converted to prostaglandin E2 (PGE2) by membrane prostaglandin synthase-1 and 2 (mPGES-1, 2) or cytosolic prostaglandin synthase (cPGES). PGH2 also can be converted to thromboxane A2 (TXA2) by thromboxane synthase (TXS). In brain, the COX-1 is constitutively expressed, whereas COX-2 is inducible. cPGES uses PGH2 produced by COX-1, whereas mPGES-1 uses COX-2-derived endoperoxide. Unconverted AA has a Co-A group added by ACSL4 and is re-incorporated into the membrane by LPCAT3. After DHA within the phospholipids is released by iPLA or sPLA, some is metabolized to neuroprotectins (NPD1) and resolvins (RvD1). DHA can also be activated to DHA-CoA by ACSL6 and be re-esterified into membrane phospholipids by LPCAT4. Modified from Kim et al [88].
(TIF)
Acknowledgments
We thank Drs. Barbara K. Lipska and Chuck C. Chen for their comments during manuscript preparation.
Funding Statement
This work was supported by the Intramural Research Programs of the National Institute on Aging at the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Huttenlocher PR (1990) Morphometric study of human cerebral cortex development. Neuropsychologia 28: 517–527. [DOI] [PubMed] [Google Scholar]
- 2.Yakovlev PI, Lecours AR (1967) The myelogenetic cycles of regional maturation of the brain. In: Minkowski A, editor. Regional Development of the Brain in Early Life. Philadelphia: F. A. Davis. pp. 3–70. [Google Scholar]
- 3. Shaw P, Eckstrand K, Sharp W, Blumenthal J, Lerch JP, et al. (2007) Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci U S A 104: 19649–19654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chugani HT, Phelps ME, Mazziotta JC (1987) Positron emission tomography study of human brain functional development. Ann Neurol 22: 487–497. [DOI] [PubMed] [Google Scholar]
- 5. Koss E, Haxby JV, DeCarli CS, Schapiro MB, Friedland RP, et al. (1991) Patterns of performance preservation and loss in healthy aging. Dev Neuropsychol 7: 99–113. [Google Scholar]
- 6. Morrison JH, Baxter MG (2012) The ageing cortical synapse: hallmarks and implications for cognitive decline. Nature reviews Neuroscience 13: 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Azari NP, Rapoport SI, Grady CL, Schapiro MB, Salerno JA, et al. (1992) Interregional correlations of resting cerebral glucose metabolism in old and young women. Brain Res 589: 279–290. [DOI] [PubMed] [Google Scholar]
- 8. Hatanpaa K, Isaacs KR, Shirao T, Brady DR, Rapoport SI (1999) Loss of proteins regulating synaptic plasticity in normal aging of the human brain and in Alzheimer disease. J Neuropathol Exp Neurol 58: 637–643. [DOI] [PubMed] [Google Scholar]
- 9. Freo U, Ricciardi E, Pietrini P, Schapiro MB, Rapoport SI, et al. (2005) Pharmacological modulation of prefrontal cortical activity during a working memory task in young and older humans: A PET study with physostigmine. Am J Psychiatry 162: 2061–2770. [DOI] [PubMed] [Google Scholar]
- 10. Gogtay N, Greenstein D, Lenane M, Clasen L, Sharp W, et al. (2007) Cortical brain development in nonpsychotic siblings of patients with childhood-onset schizophrenia. Arch Gen Psychiatry 64: 772–780. [DOI] [PubMed] [Google Scholar]
- 11. Gogtay N, Ordonez A, Herman DH, Hayashi KM, Greenstein D, et al. (2007) Dynamic mapping of cortical development before and after the onset of pediatric bipolar illness. J Child Psychol Psychiatry 48: 852–862. [DOI] [PubMed] [Google Scholar]
- 12. Somel M, Guo S, Fu N, Yan Z, Hu HY, et al. (2010) MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome research 20: 1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lu T, Pan Y, Kao SY, Li C, Kohane I, et al. (2004) Gene regulation and DNA damage in the ageing human brain. Nature 429: 883–891. [DOI] [PubMed] [Google Scholar]
- 14. Keleshian VL, Modi HR, Rapoport SI, Rao JS (2013) Aging is associated with altered inflammatory, arachidonic acid cascade, and synaptic markers, influenced by epigenetic modifications, in the human frontal cortex. Journal of neurochemistry 125: 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 15. Belkind-Gerson J, Carreon-Rodriguez A, Contreras-Ochoa CO, Estrada-Mondaca S, Parra-Cabrera MS (2008) Fatty acids and neurodevelopment. Journal of pediatric gastroenterology and nutrition 47 Suppl 1: S7–9. [DOI] [PubMed] [Google Scholar]
- 16. Dobbing J, Sands J (1979) Comparative aspects of the brain growth spurt. Early Hum Dev 3: 79–83. [DOI] [PubMed] [Google Scholar]
- 17. Purdon AD, Rapoport SI (1998) Energy requirements for two aspects of phospholipid metabolism in mammalian brain. Biochem J 335: 313–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Purdon AD, Rapoport SI (2007) Energy consumption by phospholipid metabolism in mammalian brain. In: Gibson G, Dienel G, editors. Neural Energy Utilization: Handbook of Neurochemistry and Molecular Biology. 3rd ed. New York: Springer. pp. 401–427. [Google Scholar]
- 19. Igarashi M, Ma K, Gao F, Kim HW, Rapoport SI, et al. (2011) Disturbed choline plasmalogen and phospholipid fatty acid concentrations in Alzheimer's disease prefrontal cortex. Journal of Alzheimer's disease: JAD 24: 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao JS, Kim EM, Lee HJ, Rapoport SI (2007) Up-regulated arachidonic acid cascade enzymes and their transcription factors in post-mortem frontal cortex from bipolar disorder patients. Soc Neurosci Abstr 797.5/Z4.
- 21. Esposito G, Giovacchini G, Liow JS, Bhattacharjee AK, Greenstein D, et al. (2008) Imaging neuroinflammation in Alzheimer's disease with radiolabeled arachidonic acid and PET. Journal of nuclear medicine: official publication, Society of Nuclear Medicine 49: 1414–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Soderberg M, Edlund C, Kristensson K, Dallner G (1991) Fatty acid composition of brain phospholipids in aging and in Alzheimer's disease. Lipids 26: 421–425. [DOI] [PubMed] [Google Scholar]
- 23. Bazan NG, Colangelo V, Lukiw WJ (2002) Prostaglandins and other lipid mediators in Alzheimer's disease. Prostaglandins Other Lipid Mediat 68–69: 197–210. [DOI] [PubMed] [Google Scholar]
- 24. Igarashi M, Ma K, Gao F, Kim HW, Greenstein D, et al. (2010) Brain lipid concentrations in bipolar disorder. Journal of psychiatric research 44: 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rapoport SI (2008) Brain arachidonic and docosahexaenoic acid cascades are selectively altered by drugs, diet and disease. Prostaglandins, leukotrienes, and essential fatty acids 79: 153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Basselin M, Rosa AO, Ramadan E, Cheon Y, Chang L, et al. (2010) Imaging decreased brain docosahexaenoic acid metabolism and signaling in iPLA(2)beta (VIA)-deficient mice. Journal of lipid research 51: 3166–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dennis EA, Cao J, Hsu YH, Magrioti V, Kokotos G (2011) Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chemical reviews 111: 6130–6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ong WY, Sandhya TL, Horrocks LA, Farooqui AA (1999) Distribution of cytoplasmic phospholipase A2 in the normal rat brain. J Hirnforsch 39: 391–400. [PubMed] [Google Scholar]
- 29. Ong WY, Yeo JF, Ling SF, Farooqui AA (2005) Distribution of calcium-independent phospholipase A2 (iPLA 2) in monkey brain. J Neurocytol 34: 447–458. [DOI] [PubMed] [Google Scholar]
- 30. Pardue S, Rapoport SI, Bosetti F (2003) Co-localization of cytosolic phospholipase A(2) and cyclooxygenase-2 in Rhesus monkey cerebellum. Brain Res Mol Brain Res 116: 106–114. [DOI] [PubMed] [Google Scholar]
- 31. Kaufmann WE, Worley PF, Pegg J, Bremer M, Isakson P (1996) COX-2, a synaptically induced enzyme, is expressed by excitatory neurons at postsynaptic sites in rat cerebral cortex. Proc Natl Acad Sci U S A 93: 2317–2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tay A, Simon JS, Squire J, Hamel K, Jacob HJ, et al. (1995) Cytosolic phospholipase A2 gene in human and rat: chromosomal localization and polymorphic markers. Genomics 26: 138–141. [DOI] [PubMed] [Google Scholar]
- 33.Lands WEM, Crawford CG (1976) Enzymes of membrane phospholipid metabolism. In: Martonosi A, editor. The Enzymes of Biological Membranes. New York: Plenum. pp. 3–85. [Google Scholar]
- 34. Robinson PJ, Noronha J, DeGeorge JJ, Freed LM, Nariai T, et al. (1992) A quantitative method for measuring regional in vivo fatty-acid incorporation into and turnover within brain phospholipids: Review and critical analysis. Brain Res Rev 17: 187–214. [DOI] [PubMed] [Google Scholar]
- 35. Sun GY, MacQuarrie RA (1989) Deacylation-reacylation of arachidonoyl groups in cerebral phospholipids. Ann N Y Acad Sci 559: 37–55. [DOI] [PubMed] [Google Scholar]
- 36. Six DA, Dennis EA (2000) The expanding superfamily of phospholipase A(2) enzymes: classification and characterization. Biochimica et biophysica acta 1488: 1–19. [DOI] [PubMed] [Google Scholar]
- 37. Soupene E, Kuypers FA (2008) Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 233: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamashita A, Sugiura T, Waku K (1997) Acyltransferases and transacylases involved in fatty acid remodeling of phospholipids and metabolism of bioactive lipids in mammalian cells. J Biochem (Tokyo) 122: 1–16. [DOI] [PubMed] [Google Scholar]
- 39. Shimizu T, Wolfe LS (1990) Arachidonic acid cascade and signal transduction. J Neurochem 55: 1–15. [DOI] [PubMed] [Google Scholar]
- 40. Serhan CN (2005) Novel eicosanoid and docosanoid mediators: resolvins, docosatrienes, and neuroprotectins. Curr Opin Clin Nutr Metab Care 8: 115–121. [DOI] [PubMed] [Google Scholar]
- 41. Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, et al. (2011) Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 478: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Balsinde J, Balboa MA, Dennis EA (1998) Functional coupling between secretory phospholipase A2 and cyclooxygenase-2 and its regulation by cytosolic group IV phospholipase A2. Proc Natl Acad Sci USA 95: 7951–7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Murakami M, Kambe T, Shimbara S, Kudo I (1999) Functional coupling between various phospholipase A2s and cyclooxygenases in immediate and delayed prostanoid biosynthetic pathways. J Biol Chem 274: 3103–3105. [DOI] [PubMed] [Google Scholar]
- 44. Scott KF, Bryant KJ, Bidgood MJ (1999) Functional coupling and differential regulation of the phospholipase A2-cyclooxygenase pathways in inflammation. Journal of leukocyte biology 66: 535–541. [DOI] [PubMed] [Google Scholar]
- 45. Ueno N, Murakami M, Tanioka T, Fujimori K, Tanabe T, et al. (2001) Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2. J Biol Chem 276: 34918–34927. [DOI] [PubMed] [Google Scholar]
- 46. Naraba H, Murakami M, Matsumoto H, Shimbara S, Ueno A, et al. (1998) Segregated coupling of phospholipases A2, cyclooxygenases, and terminal prostanoid synthases in different phases of prostanoid biosynthesis in rat peritoneal macrophages. J Immunol 160: 2974–2982. [PubMed] [Google Scholar]
- 47. Neufeld EJ, Majerus PW, Krueger CM, Saffitz JE (1985) Uptake and subcellular distribution of [3H]arachidonic acid in murine fibrosarcoma cells measured by electron microscope autoradiography. J Cell Biol 101: 573–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laposata M, Majerus PW (1987) Measurement of icosanoid precursor uptake and release by intact cells. Methods Enzymol 141: 350–355. [DOI] [PubMed] [Google Scholar]
- 49. Casey BJ, Giedd JN, Thomas KM (2000) Structural and functional brain development and its relation to cognitive development. Biological psychology 54: 241–257. [DOI] [PubMed] [Google Scholar]
- 50. Dayan J, Bernard A, Olliac B, Mailhes AS, Kermarrec S (2010) Adolescent brain development, risk-taking and vulnerability to addiction. Journal of physiology, Paris 104: 279–286. [DOI] [PubMed] [Google Scholar]
- 51. Colantuoni C, Henry G, Zeger S, Pevsner J (2002) SNOMAD (Standardization and NOrmalization of MicroArray Data): web-accessible gene expression data analysis. Bioinformatics 18: 1540–1541. [DOI] [PubMed] [Google Scholar]
- 52. de Hoon MJ, Imoto S, Nolan J, Miyano S (2004) Open source clustering software. Bioinformatics 20: 1453–1454. [DOI] [PubMed] [Google Scholar]
- 53.Eisen M (1998) Cluster 3.0 Manual. In: Hoon Md, editor. University of Tokyo: Human Genome Center.
- 54. Saldanha AJ (2004) Java Treeview—extensible visualization of microarray data. Bioinformatics 20: 3246–3248. [DOI] [PubMed] [Google Scholar]
- 55. Quackenbush J (2002) Microarray data normalization and transformation. Nature genetics 32 Suppl: 496–501. [DOI] [PubMed] [Google Scholar]
- 56. Xu LZ, Sanchez R, Sali A, Heintz N (1996) Ligand specificity of brain lipid-binding protein. The Journal of biological chemistry 271: 24711–24719. [DOI] [PubMed] [Google Scholar]
- 57. Leslie JB, Watkins WD (1985) Eicosanoids in the central nervous system. Journal of neurosurgery 63: 659–668. [DOI] [PubMed] [Google Scholar]
- 58. O'Banion MK (1999) COX-2 and Alzheimer's disease: potential roles in inflammation and neurodegeneration. Expert Opin Investig Drugs 8: 1521–1536. [DOI] [PubMed] [Google Scholar]
- 59. Kam PC, See AU (2000) Cyclo-oxygenase isoenzymes: physiological and pharmacological role. Anaesthesia 55: 442–449. [DOI] [PubMed] [Google Scholar]
- 60. Duplus E, Forest C (2002) Is there a single mechanism for fatty acid regulation of gene transcription? Biochemical pharmacology 64: 893–901. [DOI] [PubMed] [Google Scholar]
- 61. Di Nunzio M, Danesi F, Bordoni A (2009) n-3 PUFA as regulators of cardiac gene transcription: a new link between PPAR activation and fatty acid composition. Lipids 44: 1073–1079. [DOI] [PubMed] [Google Scholar]
- 62. Cunnane SC, Chouinard-Watkins R, Castellano CA, Barberger-Gateau P (2013) Docosahexaenoic acid homeostasis, brain aging and Alzheimer's disease: Can we reconcile the evidence? Prostaglandins, leukotrienes, and essential fatty acids 88: 61–70. [DOI] [PubMed] [Google Scholar]
- 63. Giovacchini G, Lerner A, Toczek MT, Fraser C, Ma K, et al. (2004) Brain incorporation of [11C]arachidonic acid, blood volume, and blood flow in healthy aging: a study with partial-volume correction. J Nucl Med 45: 1471–1479. [PubMed] [Google Scholar]
- 64. Ibanez V, Pietrini P, Furey ML, Alexander GE, Millet P, et al. (2004) Resting state brain glucose metabolism is not reduced in normotensive healthy men during aging, after correction for brain atrophy. Brain research bulletin 63: 147–154. [DOI] [PubMed] [Google Scholar]
- 65. Yu YQ, Tao R, Wei J, Xu Q, Liu SZ, et al. (2004) No association between the PTGS2/PLA2G4A locus and schizophrenia in a Chinese population. Prostaglandins, leukotrienes, and essential fatty acids 71: 405–408. [DOI] [PubMed] [Google Scholar]
- 66. Furth EE, Laposata M (1988) Mass quantitation of agonist-induced arachidonate release and icosanoid production in a fibrosarcoma cell line. Effect of time of agonist stimulation, amount of cellular arachidonate, and type of agonist. J Biol Chem 263: 15951–15956. [PubMed] [Google Scholar]
- 67. Rapoport SI (2008) Arachidonic acid and the brain. The Journal of nutrition 138: 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Rao JS, Rapoport SI, Kim HW (2011) Altered neuroinflammatory, arachidonic acid cascade and synaptic markers in postmortem Alzheimer's disease brain. Translational psychiatry 1: e31. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 69. Moses GS, Jensen MD, Lue LF, Walker DG, Sun AY, et al. (2006) Secretory PLA2-IIA: a new inflammatory factor for Alzheimer's disease. J Neuroinflammation 3: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ho L, Purohit D, Haroutunian V, Luterman JD, Willis F, et al. (2001) Neuronal cyclooxygenase 2 expression in the hippocampal formation as a function of the clinical progression of Alzheimer disease. Arch Neurol 58: 487–492. [DOI] [PubMed] [Google Scholar]
- 71. Stephenson DT, Lemere CA, Selkoe DJ, Clemens JA (1996) Cytosolic phospholipase A2 (cPLA2) immunoreactivity is elevated in Alzheimer's disease brain. Neurobiol Dis 3: 51–63. [DOI] [PubMed] [Google Scholar]
- 72. Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, et al. (2002) Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. Journal of neuroscience research 70: 462–473. [DOI] [PubMed] [Google Scholar]
- 73. Bazan NG (2009) Neuroprotectin D1-mediated anti-inflammatory and survival signaling in stroke, retinal degenerations, and Alzheimer's disease. Journal of lipid research 50 Suppl: S400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bazan NG (2007) Omega-3 fatty acids, pro-inflammatory signaling and neuroprotection. Current opinion in clinical nutrition and metabolic care 10: 136–141. [DOI] [PubMed] [Google Scholar]
- 75. Saiz L, Klein ML (2001) Structural properties of a highly polyunsaturated lipid bilayer from molecular dynamics simulations. Biophysical journal 81: 204–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mukherjee PK, Marcheselli VL, Serhan CN, Bazan NG (2004) Neuroprotectin D1: a docosahexaenoic acid-derived docosatriene protects human retinal pigment epithelial cells from oxidative stress. Proceedings of the National Academy of Sciences of the United States of America 101: 8491–8496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445: 168–176. [DOI] [PubMed] [Google Scholar]
- 78. Loerch PM, Lu T, Dakin KA, Vann JM, Isaacs A, et al. (2008) Evolution of the aging brain transcriptome and synaptic regulation. PloS one 3: e3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hawrylycz MJ, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, et al. (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Somel M, Liu X, Khaitovich P (2013) Human brain evolution: transcripts, metabolites and their regulators. Nature reviews Neuroscience 14: 112–127. [DOI] [PubMed] [Google Scholar]
- 81. Numata S, Ye T, Hyde TM, Guitart-Navarro X, Tao R, et al. (2012) DNA methylation signatures in development and aging of the human prefrontal cortex. American journal of human genetics 90: 260–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wang Y, Chen T, Yan H, Qi H, Deng C, et al. (2013) Role of histone deacetylase inhibitors in the aging of human umbilical cord mesenchymal stem cells. Journal of cellular biochemistry 114: 2231–2239. [DOI] [PubMed] [Google Scholar]
- 83. Rhie BH, Song YH, Ryu HY, Ahn SH (2013) Cellular aging is associated with increased ubiquitylation of histone H2B in yeast telomeric heterochromatin. Biochemical and biophysical research communications 439: 570–575. [DOI] [PubMed] [Google Scholar]
- 84. Hodzic M, Naaldijk Y, Stolzing A (2013) Regulating aging in adult stem cells with microRNA. Zeitschrift fur Gerontologie und Geriatrie 46: 629–634. [DOI] [PubMed] [Google Scholar]
- 85. Persengiev S, Kondova I, Bontrop R (2013) Insights on the functional interactions between miRNAs and copy number variations in the aging brain. Frontiers in molecular neuroscience 6: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kim HW, Rao JS, Rapoport SI, Igarashi M (2011) Regulation of rat brain polyunsaturated fatty acid (PUFA) metabolism during graded dietary n-3 PUFA deprivation. Prostaglandins, leukotrienes, and essential fatty acids 85: 361–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Rapoport SI, Basselin M, Kim HW, Rao JS (2009) Bipolar disorder and mechanisms of action of mood stabilizers. Brain research reviews 61: 185–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kim HW, Rapoport SI, Rao JS (2011) Altered arachidonic acid cascade enzymes in postmortem brain from bipolar disorder patients. Molecular psychiatry 16: 419–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The brain arachidonic and docosahexaenoic acid cascades. After AA within phospholipid is released by cPLA2 or sPLA2, a portion is converted to prostaglandin H2 (PGH2) by COX-1 or COX-2, to hydroxyeicosatetraenoic acid (20-HETE), to hydroperoxyeicosatetraenoic acids (HPETES) by lipoxygenase (LOX) subtypes 5, 12 or 15. PGH2 is converted to prostaglandin E2 (PGE2) by membrane prostaglandin synthase-1 and 2 (mPGES-1, 2) or cytosolic prostaglandin synthase (cPGES). PGH2 also can be converted to thromboxane A2 (TXA2) by thromboxane synthase (TXS). In brain, the COX-1 is constitutively expressed, whereas COX-2 is inducible. cPGES uses PGH2 produced by COX-1, whereas mPGES-1 uses COX-2-derived endoperoxide. Unconverted AA has a Co-A group added by ACSL4 and is re-incorporated into the membrane by LPCAT3. After DHA within the phospholipids is released by iPLA or sPLA, some is metabolized to neuroprotectins (NPD1) and resolvins (RvD1). DHA can also be activated to DHA-CoA by ACSL6 and be re-esterified into membrane phospholipids by LPCAT4. Modified from Kim et al [88].
(TIF)