Abstract
Objectives
This study was designed to investigate the activation of the unfolded protein response (UPR) in tumor associated endothelial cells (TECs) and its association with chemoresistance during acidic pH stress.
Materials and Methods
Endothelial cells from human oral squamous cell carcinomas (OSCC) were excised by laser capture microdissection (LCM) followed by analysis of UPR markers (Grp78, ATF4 and CHOP) using quantitative PCR. Grp78 expression was also determined by immunostaining. Acidic stress was induced in primary human dermal microvascular endothelial cells (HDMECs) by treatment with conditioned medium (CM) from tumor cells grown under hypoxic conditions or by adjusting medium pH to 6.4 or 7.0 using lactic acid or hydrochloric acid (HCl). HDMEC resistance to the anti-angiogenic drug Sunitinib was assessed with SRB assay.
Results
UPR markers, Grp78, ATF4 and CHOP were significantly upregulated in TECs from OSCC compared to HDMECs. HDMECs cultured in acidic CM (pH 6.0–6.4) showed increased expression of the UPR markers. However, severe acidosis led to marked cell death in HDMECs. Alternatively, HDMECs were able to adapt when exposed to chronic acidosis at pH 7.0 for 7 days, with concomittant increase in Grp78 expression. Chronic acidosis also confers drug resistance to HDMECs against Sunitinib. Knockdown of Grp78 using shRNA resensitizes HDMECs to drug treatment.
Conclusions
UPR induction in ECs under acidic pH conditions is related to chemoresistance and may contribute to therapeutic failures in response to chemotherapy. Targeting Grp78, the key component of the UPR pathway, may provide a promising approach to overcome ECs resistance in cancer therapy.
Introduction
Antiangiogenic therapy has emerged as a promising adjuvant approach to cancer treatment. However, evidence has shown that tumor associated endothelial cells (TECs) are more resistant to antiangiogenic therapies compared to endothelial cells from normal tissues (NECs) [1]. This resistant phenotype might be acquired by adaptation to stressful conditions in the tumor microenvironment (TME) [2].
TME is characterized by acidosis, low glucose, amino acid deficiency and hypoxia [3], [4]. A common response of cells to stress in the TME is the activation of the unfolded protein response (UPR), a protective mechanism that cells use to overcome endoplasmic reticulum (ER) stress. Different pathological insults can result in either acute or chronic ER stress [5]. Acute stress, if not resolved promptly, can lead to cell apoptosis. In contrast, chronic stress requires extended activation of the UPR, which enables cells to adapt and survive [6], [7].
The UPR is mediated through the activation of three ER transmembrane sensors.
PKR-like endoplasmic reticulum (ER) kinase (PERK), activating transcription factor 6 (ATF6) and inositol-requiring enzyme 1 (IRE1). These proteins are maintained in an inactive state through interaction with the chaperone, glucose-regulated protein 78 kDa (Grp78). Accumulation of unfolded proteins within the ER prompts dissociation of Grp78 from these sensors, and consequent activation of the UPR [8]. PERK activation leads to eukaryotic initiation factor 2α (eIF2α) phosphorylation, which inhibits general protein translation and reduces the load of nascent proteins being directed into the ER. PERK also induces ATF4, a transcription factor which upregulates ER chaperones (such as Grp78 and Grp94). Additionally, C/EBP homologous protein (CHOP) may be induced, which is known to promote apoptotic cell death [9], [10]. After being dissociated from Grp78, ATF6 translocates to the Golgi, where it is cleaved to the p50 active form. Active ATF6 translocates to the nucleus and regulates expression of genes, such as Grp78, protein disulphide isomerase (PDI) and X box-binding protein 1 (XBP1). The third ER stress sensor, IRE1, is responsible for the unconventional splicing of XBP1 mRNA. This alternatively spliced XBP1 encodes a transcription factor that targets diverse genes including chaperones [11]. Therefore, each one of the ER sensors activates a transcription factor, which induces proteins that aim to counteract ER stress and promote survival.
A common finding in a variety of malignant tumors is an increased expression of Grp78 [12], [13], [14]. Upregulation of Grp78 has been associated with chemoresistance to different drugs, such as adriamycin in squamous carcinoma cells [15]; temozolomide in glioma cells [16]; cisplatin in melanoma cells [17] and etoposide and temozolomide in TECs [18].
Although the relationship between UPR activation and chemoresistance in cancer cells is well-established, the effects of UPR on drug resistance of TECs need to be further investigated. The aim of this study was to assess the levels of UPR in ECs from the microvasculature of oral squamous cell carcinomas (OSCC) and to investigate how an acidic environment contributes to the chemoresistance of ECs. We report here that the UPR pathway is activated in ECs in response to acidic environment. Also, the key component of the pathway, Grp78 is, at least partially responsible for the drug resistance of ECs. Targeting Grp78 may provide a promising regimen towards sensitizing TECs to antiangiogenic drugs.
Materials and Methods
cell culture
Primary human dermal microvascular endothelial cells HDMECs (purchased from Lonza, Walkersville, MD) were cultured in endothelial growth medium EGM2-MV (Lonza, Walkersville, MD) at 37°C with 5% CO2. Medium pH was adjusted with 1 M lactic acid (Sigma-Aldrich, St. Louis, MO) or 1 M HCL (Sigma-Aldrich, St. Louis, MO). Tumor cell lines used in this study were received as gifts, human oral squamous cell carcinoma cell line UM-SCC-81B from Dr. Thomas E. Carey (Departments of Otorhinolaryngology and Pharmacology, University of Michigan Medical School, Ann Arbor, MI) and glioblastoma cell line U-87 from Dr. Yi Sun (Department of Radiation Oncology, University of Michigan, Ann Arbor, MI). Cell lines were mantained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS), 1% L-glutamine and 0.05% penicillin/streptomycin (Invitrogen) at 37°C with 5% CO2 [19], [20], [21], [22], [23], [24]. Conditioned medium (CM) was collected from tumor cells cultured in endothelial cell basal medium (EBM) for 48-hours under low oxygen condition. Collected CM was supplemented with glucose at 1.0g/L (5.55 mM) to rule out glucose deprivation-induced UPR. The antiangiogenic drug Sunitinib was purchased from LC Laboratories (Woburn, MA).
Lentivirus infection
Lentiviruses expressing short hairpin RNA (shRNA) constructs for Grp78 (Vector Core, University of Michigan) were generated in human embryonic kidney cells (293T) transfected by the calcium phosphate method. Scrambled oligonucleotide sequences (scshRNA) were used as controls. Virus containing supernatants were collected 48 hours post-transfection and used to infect HDMECs.
Sulforhodamine B assay
Sulforhodamine B (SRB) cytotoxicity assays were done as previously described [25]. Briefly, cells were seeded at 2×103 cells per well of 96-well plates, allowed to attach overnight, and treated with Sunitinib for 72 hours. Cells were fixed with 10% trichloroacetic acid, stained with 0.4% SRB (Sigma-Aldrich, St. Louis, MO) in 1% acetic acid, and plates were read in a microplate reader at 560 nm. Test results were normalized against drug-free controls. Data were obtained from triplicate wells per condition and is representative of at least three independent experiments.
Western Blot
Cell lysates were resolved by SDS-PAGE and transferred to a PVDF membrane. Membranes were probed overnight at 4°C with the following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA): rabbit anti-Grp78 (clone Sc-13968), rabbit anti-ATF4 (clone Sc-22800), rabbit anti-CHOP (clone Sc-575), mouse anti-β-actin (clone Sc-47778), αTubulin (Sc-8035); and from Cell Signaling (Danvers, MA): rabbit anti-p-eIF2α (clone 3597), rabbit anti-elF2α (clone 9722), mouse anti-caspase 7 (clone c7). Proteins were detected using peroxidase conjugated secondary antibodies and bands were visualized using enhanced chemiluminescence (Amersham, Sunnyvale, CA).
Cell cycle analysis
HDMECs were trypsinized, washed with PBS twice and then fixed with 70% ethanol on ice for 1 h. The fixed cells were spun down and resuspended in PBS. After incubation with ribonuclease RNase A (25 µg/mL) at 37°C for 30 minutes, the cell suspension was stained with propidium iodide and followed by analysis on a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA) in the Flow Cytometry Core, University of Michigan Comprehensive Cancer Center.
Laser capture microdissection
Tissue sections of OSCCs and normal oral mucosa were mounted on glass foiled pen slides for laser capture microdissection (LCM) and stained with Hematoxylin. A two-step process was used for cell collection using a microdissection microscope (Leica AS LMD, Leica) with a pulsed 337-nm UV laser by a trained pathologist (FV). ECs were dissected after removal of blood cells from capillaries. Approximately 5000 ECs were retrieved from each sample. The RNA from independent tumors was analyzed by RT-PCR and real-time PCR for comparison to normal mucosa and HDMEC samples.
Real-Time PCR
Total RNA of ECs from tissue sections obtained by LCM method was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA) and purified with RNeasy Minikits (Qiagen, Valencia, CA) according to the manufacturers’ protocols. A cDNA library was prepared with WTA2, TransPlex Complete Whole Transcriptome Amplification Kit (Sigma-Aldrich, St. Louis, MO), which was then amplified using a universal end primer.
To harvest RNA from HDMECs, the RNeasy Minikit (Qiagen, Valencia, CA) was used and cDNA was made according to the manufacturer’s instructions with Verso cDNA kit (ThermoScientific, Waltham, MA).
Quantitative real-time (qRT-PCR) was performed with the following primers: Grp78 (Hs99999174_m1), ATF4 (Hs00909569_g1), CHOP (Hs01090850_m1) and β-tubulin (Hs 03929064_g1) using Gene Expression Assays reagents (Applied Biosystems, Carlsbad, CA). All reactions were done in triplicates and normalized to β-tubulin.
RT- PCR
Reverse transcription PCR (RT-PCR) analysis of spliced and unspliced XBP1 was performed with a single human-specific primer pair: ACACGCTTGGGAATGGACAC (sense) and CCATGGGAAGATGTTCTGGG (antisense). Also, the following primer sequences were used: GAPDH, CATGGCCTCCAAGGAGTAAG (sense) and AGGGGTCTACAGGCAACTG (antisense); VEGFR2, AGCGATGGCCTCTTCTGTAA (sense) and ACACGACTCCATGTTGGTCA (antisense); E-cadherin, TGCCCAGAAAATGAAAAAGG (sense) and GGATGACACAGCGTGAGAGA (antisense). For XBP1 splicing analysis, amplicons were visualized with a Qiaxcel (Qiagen, Valencia, CA) automated nucleic acid fragment analyzer using a high resolution cartridge on the M500 setting using a 15 bp–1 kb alignment marker and a 50 bp–800 bp size marker.
Immunofluorescence
Sections from the same tumors and samples from normal oral mucosa biopsies used for LCM were deparaffinized and rehydrated, and incubated in low pH antigen-retrieval solution (Dakocytomation; Dako, Carpinteria, CA) for 20 minutes at 95°C. Double staining was performed using antibodies against CD31 (Clone JC70A, Dako, Carpinteria, CA) and Grp78 (clone H-129, SantaCruz Biotechnology, Santa Cruz, CA). Alexa Fluor 488 or Alexa Fluor 647 (Molecular Probes, Eugene, OR) conjugated sencondary antibodies were used. Slides were counterstained with 3.5 µM DAPI (Sigma-Aldrich, St. Louis, MO), and mounted with Fluorescent Mounting Media (Dako, Carpinteria, CA). Images were taken with Olympus IX70 microscope (Tokyo, Japan) and analyzed by ImageJ software (National Institute of Health, Bethesda, MD, USA). Only CD31 positive cells lining tubular structures were considered blood vessels. Tissue samples were obtained from the University of Michigan School of Dentistry tissue core [19].
Statistical analyses
For statistical comparison of experimental groups, t-test, one-way ANOVA and two-way ANOVA were used, followed by Bonferroni post-hoc test, using Software Statistical Package for the Social Sciences version 19.0 (SPSS Inc., Chicago, IL, EUA) and GraphPad Prism 5.0 (La Jolla, CA). Statistical significance was determined when p<0.05.
Results
UPR is activated in the vasculature of human tumors
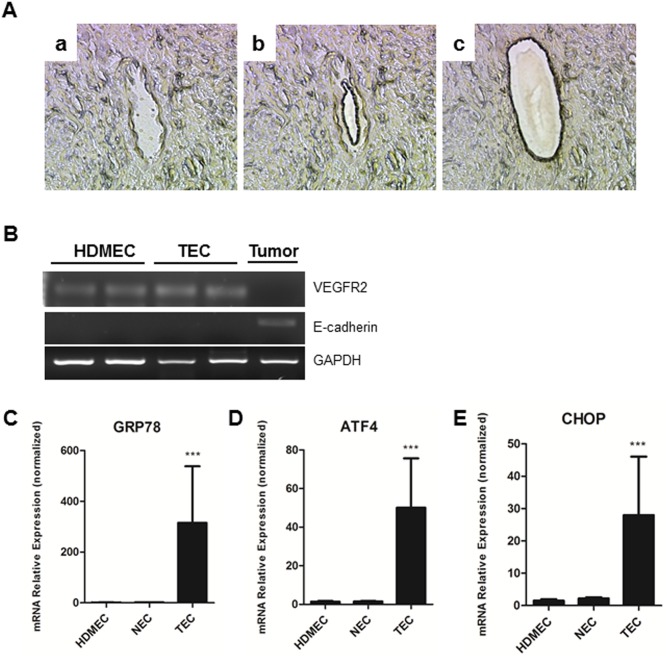
To assess the UPR in tumor associated endothelial cells (TECs) in vivo, LCM was used to retrieve endothelial cells (EC) lining the blood vessels from human oral squamous cell carcinoma (OSCC) samples (Figure 1A). To confirm the purity of the TECs collected, RT-PCR was performed using VEGFR2 as a marker for ECs and E-cadherin as a marker for tumor cells. As it is shown in Figure 1B, VEGFR2 was detected in TECs, while there was no E-cadherin band suggesting that contamination from tumor cells is neglectable. Notably, Grp78 mRNA expression levels were ∼300-fold higher in TECs compared to endothelial cells from normal oral mucosa (NEC) and primary human dermal microvascular endothelial cells (HDMECs) (Figure 1C). To confirm if UPR is active in TECs, two other UPR makers (ATF4 and CHOP) were evaluated with qPCR and the results showed there was about 45-fold increase for ATF4 and 25-fold increase for CHOP (Figures 1D and E, respectively).
Figure 1.
In A, LCM retrieval of endothelial cells from paraffin embedded tissue sections (a - before dissection, b - removal of blood cells, c- capture of endothelial cells). B, RNA purity control, using VEGFR2, a marker for endothelial cells, and E-cadherin, a marker for cells of ectodermal origin. C, D, E, real-time PCR to quantify Grp78, ATF4 and CHOP expression in TEC compared to NEC and primary endothelial cells. Specimens were obtained from human oral squamous cell carcinoma biopsies. Data presented from real-time PCR experiments reflect the expression levels of Grp78, ATF4 and CHOP normalized to β-tubulin. Values are means ± SD. Columns, means of individual experiments; Bars, SD. ***p<0.001.
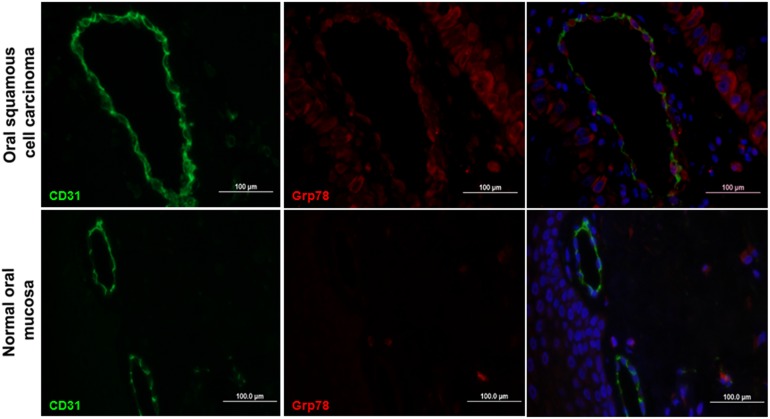
To address whether this observation is valid in situ, i.e. the vasculature of tumor tissues, sections of OSCC were immunostained for both Grp78 (red) and the endothelial cell marker CD31 (green), with DAPI (blue) staining the nuclei. Merged images confirmed that Grp78 was highly expressed in both the tumor vasculature and carcinoma cells. By contrast, normal oral mucosa tissues exhibited CD31-positive (green) blood vessels but minimal labeling for Grp78 (Figure 2). Merged images showed no Grp78 expression in vasculature of normal oral mucosa. These data confirm that Grp78 is preferably expressed in the tumor cells and TECs.
Figure 2. Grp78 expression in tumor vasculature of human oral squamous cell carcinomas.
Histological sections of oral squamous cell carcinoma tissues (top) or normal oral mucosa tissues (bottom) were stained with anti-Grp78 antibody (red), anti-CD31 antibody (green), and 4,6-diamidino-2-phenylindole (DAPI; blue) nuclear staining; the images are merged in the last panel. Bar = 100 µm.
Acidic tumor conditioned medium induces UPR in ECs
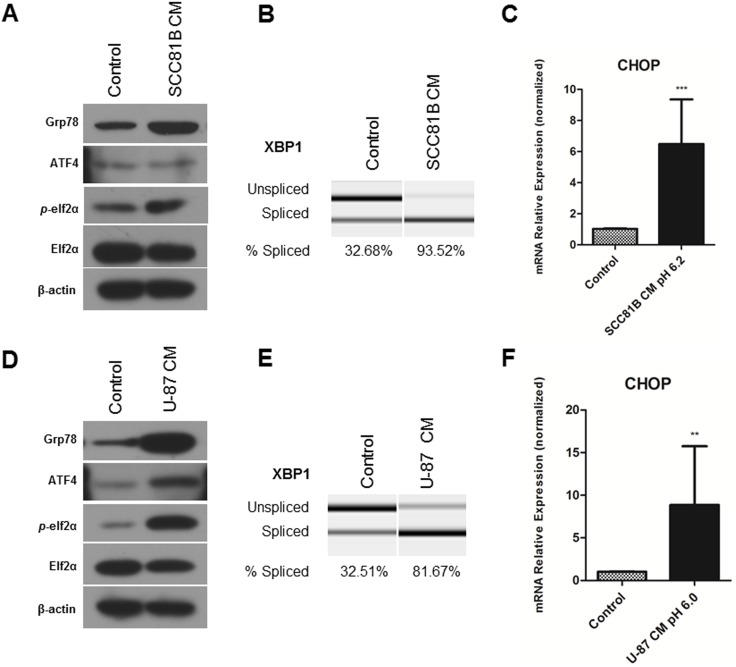
To investigate if increased UPR levels in TECs are caused by the tumor microenvironment, we analyzed the effects of tumor conditioned medium (CM) on primary human dermal microvascular endothelial cells (HDMECs). The CM was prepared from the UM-SCC-81B oral squamous cell carcinoma cell line cultured under hypoxia to mimic the tumor microenvironment conditions. The CM was supplemented with glucose to 5.55 mM, the same concentration of regular endothelial medium to avoid glucose deprivation in ECs. After treatment with CM for 48 hours, HDMECs showed strong upregulation of UPR markers: Grp78, phosphorylation of elF2α and increased cleavage of XBP1 mRNA (Figures 3A and B) compared to ECs kept at regular medium for the same time. CHOP was also increased in ECs treated with CM from tumor cells as shown from qPCR in Figure 3C. These experiments were repeated with a second tumor model, the U-87 glioblastoma cell line, to exclude a cell-specific response (Figures 3D, E and F).
Figure 3. Tumor conditioned medium induces UPR in endothelial cells.
Primary human endothelial cells, HDMECs, are maintained in conditioned medium obtained from oral squamous cell carcinoma UM-SCC-81B cell line (A, B, C) or U-87 glioblastoma cell line (D, E, F), HDMECs kept at regular medium served as control. Cell lysates were collected for western blot analysis, β-actin expression was used as a loading control. A, D, Treatment with CM increased protein expression of UPR markers: Grp78, ATF4, p-elf2α at 48 hours (western blot analysis). B, E, Exposure to CM increased XBP1 mRNA splicing levels at 48 hours. C, F, CM treatment upregulated CHOP mRNA levels. RNA was collected using Qiagen RNAeasy kit, reverse transcribed and analyzed using RT-PCR. Values are means ± SEM. Columns, means of individual experiments; Bars, SEM.**p<0.01, ***p<0.001.
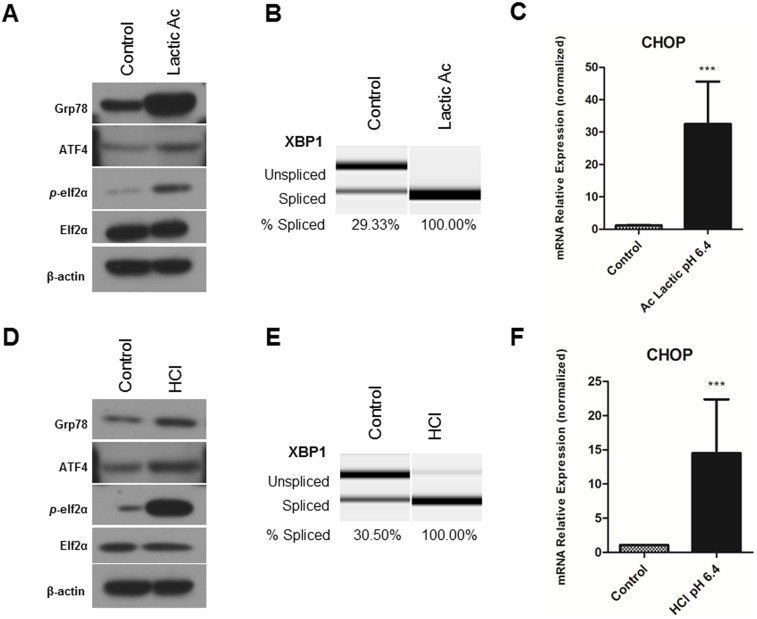
CM produced under hypoxia was found to be strongly acidic. The final pH of CM was 6.2 from UM-SCC-81B cells and 6.0 from U-87 cells. Considering that the extracellular milieu of solid malignant tumors is commonly acidic, we next analyzed specificity of endothelial cell response to acidic pH. HDMECs were treated in medium with a pH of 6.4 adjusted with lactic acid. After 48 hours in pH 6.4 medium, HDMECs showed strong upregulation of UPR markers proteins: Grp78, ATF4, phosphorylation of elF2α and increased cleavage of XBP1 mRNA (Figures 4A and B). Again, mRNA levels of CHOP were increased in acidic medium (Figure 4C). In addition, acidification with hydrochloric acid (HCl) was done to exclude cell-specific response and to confirm the results obtained from lactic acid treatments. As shown in Figure 4D to 4F, the response of ECs to acidic condition induced with HCl is similar to that induced with lactic acid.
Figure 4. Acidic stress induces UPR in endothelial cells.
Primary human endothelial cells, HDMECs, were maintained in acidic cultures for 48 hours; pH was adjusted by lactic acid (A, B, C) or HCl (D, E, F), HDMECs kept at regular pH 7.5 medium served as control. Cell lysates were collected for western blot analysis, β-actin expression was used as a loading control. A, D, Treatment with low pH increased protein expression of UPR markers: Grp78, ATF4, and p-elf2α at 48 hours (western blot analysis). B, E, Exposure to low pH increased XBP1 mRNA splicing levels at 48 hours. C, F. Acidic stress upregulated CHOP mRNA levels. RNA was collected using Qiagen RNAeasy kit, reverse transcribed and analyzed using RT-PCR. Values are means ± SEM. Columns, means of individual experiments; Bars, SEM.***p<0,001.
Mild acidic stress induces Grp78 expression and allows ECs to adapt and survive
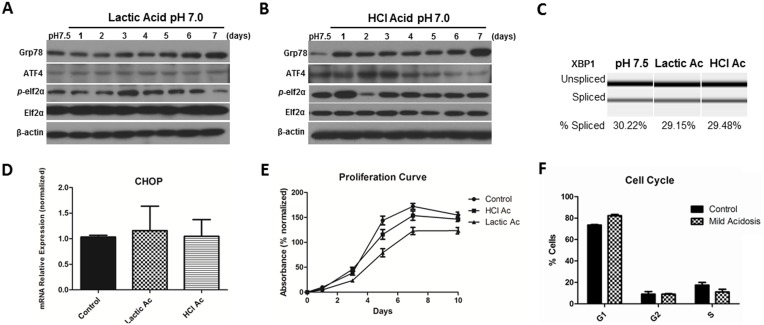
We have demonstrated that acidic pH is a UPR inducer, however HDMECs were not able to survive under such acute stress and massive cell death occurred after 48 hours at pH 6.0–6.4 (data not shown). Therefore, the effects of mild acidic stress (which reflects conditions in the tumor microenvironment) were used during the following experiments. Mild acidic stress was induced by adjusting medium pH to 7.0 with lactic acid or HCl. During the 7-day treatment, Grp78 protein levels increased over time (Figures 5A and B). No significant changes were observed in XBP1 mRNA splicing levels (Figure 5C) or CHOP mRNA levels (Figure 5D). Under mild acidic stress HDMECs were able to proliferate slightly slower than cells in regular pH medium (Figure 5E), but no significant changes were observed in cell cycle distribution (Figure 5F). These results suggest that HDMECs may adapt to mild acidic stress by upregulation of Grp78 levels.
Figure 5. Mild acidic stress induces Grp78 expression mainly and allows endothelial cell adaptation.
HDMECs were maintained in acidic cultures (pH 7.0) for seven days; pH was adjusted by lactic acid (A) or HCl (B), HDMECs kept at regular pH 7.5 served as control. Cell lysates were collected for western blot analysis, β-actin expression was used as a loading control; Grp78 expression increased over time. C, D, RNA was collected using Qiagen RNAeasy kits. XBP1 mRNA splicing and CHOP mRNA levels remained unaltered after exposure to mild acidic stress for seven days. Values are means ± SEM. Columns, means of individual experiments; Bars, SEM. E, Proliferation rates were determined by SRB assay. Values are means ± SD. Columns, means of individual experiments; Bars, SD. F, HDMECs exposed to pH 7.0 for seven days and HDMECs kept at pH 7.5 for same period were stained with propidium iodide and cell cycle was analyzed by flow cytometry. No difference in cell cycle distribution was observed. Values are means ± SD. Columns, means of individual experiments; Bars, SD.
Exposure to mild acidic stress results in a chemoresistance phenotype
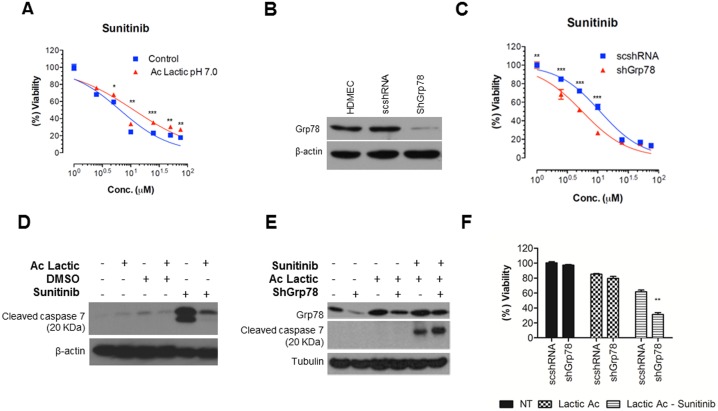
It has been shown that Grp78 plays a role in chemoresistance of tumor cells [15]–[17], so we hypothesized that Grp78 upregulation under mild acidic stress may contribute to chemoresistance of ECs. To test this hypothesis, the sensitivity of HDMECs to an antiangiogenic drug was studied under acidic stress. After exposing HDMECs to pH 7.0 for up to 7 days, cells were treated with the antangiogenic drug Sunitinib (1–75 µmol/L) and DMSO (vehicle; 0.1%) for 72 hours. Cell survival was determined using SRB assay. The results show that pretreatment of HDMECs with chronic acidic stress is able to increase EC50 from 6.48 µM to 11.31 µM for Sunitinib treatment, suggesting that chronic stress confers protective effects (Figure 6A).
Figure 6. Chronic exposure to mild acidic stress results in a chemoresistance phenotype through Grp78 expression.
A, HDMECs cultured under acidic stress for 7 days, with medium pH adjusted to 7.0 with lactic acid and HDMECs maintained at regular pH 7.5 for seven days (controls) were exposed to Sunitinib at different doses or the vehicle control (DMSO) for further 72 h and then examined for cell viability using the SRB assay. Vehicle treatment represents 100% viability. B, HDMECs were infected with control ShRNA (scshRNA) or ShRNA specifically targeted against human Grp78 (ShGrp78) or left uninfected; Four days after infection, cells were harvested for western blot analysis and probed for Grp78 and β-actin. C, Seven days after mild acidic stress (pH 7.0), scshRNA and ShGrp78 cells were treated with either vehicle or Sunitinib for further 72 h and then examined for cell viability using the SRB assay. Vehicle treatment represents 100% viability. D, HDMECs were maintained in acidic cultures (pH 7.0) for 7 days, HDMECs at regular pH 7.5 served as control. At day 7, cells were treated with either DMSO (Vehicle, 1%), or Sunitinib (0.25 µM) for an additional 48 hours after which cell lysates were collected for western blot analysis and probed for caspase 7 and β-actin. Previous exposure to acidic stress resulted in lower levels of cleaved caspase 7. E, ScshRNA and ShGrp78 cells were treated with Sunitinib (0.25 µM) for 48 hours after which cell lysates were collected for western blot analysis and probed for caspase 7 and α-tubulin. We observed increased caspase 7 activation in ShGrp78 cells treated with Sunitinib. F, Survival analysis showed increased cell death in ShGrp78 cells treated with Sunitinib. Values are means ± SEM. Columns, means of individual experiments; Bars, SEM. Significance is calculated by comparing drug-treated cells and vehicle control-treated cells. *p<0.05, **p<0.01, ***p<0.001.
To determine whether Grp78 overexpression contributes to chemoresistance, HDMECs were infected with lentivirus expressing shRNA against human Grp78 (shGrp78) with scrambled shRNA as control (scshRNA). Four days after infection, cell samples were analyzed for Grp78 expression by western blot. As shown in Figure 6B, the expression of Grp78 was significantly reduced, and as expected scshRNA showed no effect (Figure 6B). To determine the role of Grp78 in regulating chemoresistance of HDMECs, both shGrp78 and scshRNA cells were kept in acidic medium for 7 days and then treated with Sunitinib (1–75 µmol/L) for addtional 72 hours. We observed that Grp78 knockdown makes HDMECs more sensitive to drug treatment, EC50 of Sunitinib was decreased from 11 µM to 5.5 uM (Figure 6C). In conclusion, knocking down Grp78 resensitizes HDMECs to Sunitinib treatment. These results corroborate the notion that chronic stress confers drug resistance to HDMECs by increasing Grp78 expression.
Acidic stress suppresses drug-mediated cleavage of caspase 7
It has been shown that caspase 7 is associated with genotoxic drug-induced cell death and Grp78 can directly inhibit its activation [26], [27]. Upon induction of apoptosis, procaspase 7 is processed into an active 20-kDa subunit. To study the mechanisms involved in Grp78-induced chemoresistance, we analyzed the impact of preconditioning with mild acidic stress on caspase 7 cleavage. We found that cleavage of caspase 7 was significantly inhibited in HDMECs preexposed to chronic mild acidic stress prior to drug treatment, suggesting that chronic stress can protect HDMECs from apoptosis by inhibiting the production of active caspase 7 (Figure 6D).
Futhermore, shGrp78 cells showed increased caspase 7 activation along with decreased survival after Sunitinib treatment as compared to scshRNA cells (figure 6E and F). These results strengthen the central role of GRP78 in promoting chemoresistance through reducing caspase 7 cleavage.
Discussion
The tumor microenvironment (TME) is characterized by extracellular acidosis, glucose deprivation (GD) and low oxygen tension [3], [4]. We have previously demonstrated that GD promotes angiogenesis by inducing expression of angiogenic mediators, such as VEGF, FGF and IL6 through the activation of UPR [19]. Here, expression of UPR markers in ECs from human OSCC was evaluated using LCM to analyze endothelium gene expression in situ. The data obtained shows that Grp78 mRNA expression levels were ∼300-fold higher in ECs retrieved from human tumors, compared to normal human ECs. Immunofluorescence staining with CD31 and Grp78 confirmed the overexpression of Grp78 in TECs and carcinoma cells compared to the vasculature and epithelium from normal oral mucosa.
Recently it has been reported that TECs are more chemoresistant to drugs when compared to NECs [1]. Virrey and his colleagues reported increased expression of Grp78 in human ECs derived from blood vessels of malignant glioblastoma tumors, and Grp78 expression showed positive correlation with chemoresistance to CPT-11, etoposide, and temozolomide [18]. Since the emergence of angiogenesis as a potential target for tumor therapy, several antiangiogenic therapies have been developed. However, antiangiogenic therapies have not produced the sustained antitumor benefit, once envisioned. This is in part, due to the fact that the biology of TECs is more complex than previously thought [2], and that many early antiangiogenesis studies had not taken into account the influence of the TME on the angiogenic phenotype. Acidic pH, a feature of the TME, can potentially modify the endothelial phenotype. We found that the increased expression of Grp78 in ECs after exposure to chronic acidic stress is related to a drug-resistant phenotype. The acidic pH of the TME is well known to increase chemoresistance in tumor cells [28], [29], [30]. To our best knowledge this is the first report of acidic stress contributing to EC chemoresistance through Grp78 induction. Exposing ECs to a pH of 6.4 induces acute UPR and ultimately results in cell death at 48 hours of treatment, while mild acidic stress triggers an adaptive UPR with progressive increase in Grp78 expression.
In our study, different stimuli produced distinct UPR responses; UM-SCC-81B CM was not able to increase ATF4 expression, unlike U-87 CM treatment. During chronic stress induction, lactic acid adjusted medium did not increase ATF4 levels, in comparison to HCl treatment. In addition, increased expression of Grp78 after HCl treatment occurred after 24 hours, whereas medium adjusted with lactic acid took longer to induce significant Grp78 expression. While the reasons for these disparaties are unknown, a striking common response to both stimuli is increased Grp78 levels.
Depending on the intensity and duration of stress, the UPR response is structured to enable cells to either adapt to stress or to undergo apoptosis. Considering that the UPR is linked to several pathological conditions, it is important to understand how the molecular decisions, which allow cells to adapt to stress rather than undergo apoptosis, are made. This knowledge is essential in developing therapies that on the one hand allow cells to avoid apoptosis in diseases such as neurodegenerative disorders and diabetes, while on the other hand, program cells to undergo apoptosis for diseases such as cancer. Rutkowski et al. [7] demonstrated that a possible mechanism of adaptation to mild stress is the consistent expression of proteins that facilitate survival, in particular ER chaperones, without persistence of proapoptotic proteins such as CHOP, possibly due to selective intrinsic instabilities of mRNAs and proteins. Our results are in agreement with Rutkowski et al. [7], because HDMECs seem to adapt to chronic stress induced by acidic pH by upregulating Grp78, an ER chaperone with pro-survival functions.
Chemoresistance was abrogated by Grp78 knockdown, demonstrating that Grp78 plays a key role in the resistant phenotype of ECs under acidic stress. The exact mechanism by which Grp78 protects cells from death is not known. Whereas the majority of Grp78 resides in the ER lumen, it has been shown that a fraction of Grp78 exists as an ER transmembrane protein, which could explain how Grp78 can directly inhibit the activation of caspase 7, an executor caspase activated by both ER stress and genotoxic drugs [26], [27]. Ours results support the notion that pretreatment with acidic stress decreases the cleavage of caspase 7 in HDMECs after drug treatment and protects HDMECs from apoptosis.
The effects of acidic stress on ER homeostasis are poorly understood. There are several possible ways by which acidic pH may induce ER stress, including direct inactivation of enzymes involved in ER protein processing, denaturation of proteins, and inhibition of the ER Ca2þ/ATP-ase [31], [32]. Previous reports have shown the effects of acidic pH in coronary ECs, in the context of myocardial ischemia. The main pathway of acidity-induced apoptosis consists of Ca2+ leak from the ER [33]. Therefore, acidic stress may cause ER stress by decreasing ER Ca2+ levels, which is required for optimum protein folding since many of chaperones are Ca2+-dependent [13]. Additionally, Kumar et al. reported that acidic pre-conditioning (APC) suppresses apoptosis in coronary ECs [34]. The concept of stress promoting resistance to cell death is in accordance with the preconditioning paradigm: almost any stress factor that is potentially harmful for cells can elicit a preconditioned state, i.e. increased resistance to the stress, when applied in small quantities [35]. The mild acidic stress conferring chemoresistance to HDMECs further supports this notion.
This work illustrates the complexities of effects of the TME on tumor angiogenesis, highlighting acidic stress as an important feature of the TME that modifies the EC phenotype. Also, this work suggests that UPR-mediated endothelial resistance may results in therapeutic failures. The results indicate that UPR response promoted by the TME is in favor of tumor growth and resistance to chemotherapy. Targeting UPR components, such as Grp78 may provide a promising alternative in cancer treatment.
Acknowledgments
The authors thank members from Jacques E. Nör’s laboratory (University of Michigan School of Dentistry, Ann Arbor, MI), Andrew Fribley and Justin R. Miller (Department of Pediatrics, Division of Hematology/Oncology, Wayne State University, Detroit, MI) for helpful discussions and assistance. The authors also thank University of Michigan core facilities for their technical assistance.
Funding Statement
This work was supported by CAPES (Brazil) and by Sharon and Larry Daniels Cancer Biology Fellowship. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Xiong Y, Sun H, Zhang W, Zhu X, Zhuang P, et al. (2009) Human hepatocellular carcinoma tumor–derived endothelial cells manifest increased angiogenesis capability and drug resistance compared with normal endothelial cells. Clin Cancer Res 15: 4838–4846. [DOI] [PubMed] [Google Scholar]
- 2. Dudley AC, Klagsbrun M (2009) Tumor endothelial cells join the resistance. Clin Cancer Res 15: 4787–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gillies RJ, Raghunand N, Karczmar GS, Bhujwalla ZM (2002) MRI of the tumor microenvironment. J Magn Reson Imaging 16: 430–450. [DOI] [PubMed] [Google Scholar]
- 4. Becelli R, Renzi G, Morello R, Altieri F (2007) Intracellular and extracellular tumor ph measurement in a series of patients with oral cancer. J Craniofacial Surg 18: 1051–1054. [DOI] [PubMed] [Google Scholar]
- 5. Kaufman RJ (2002) Orchestrating the unfolded protein response in health and disease. J Clin Invest 110: 1389–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rutkowski DT, Kaufman RJ (2007) That which does not kill me makes me stronger: adapting to chronic ER stress. TRENDS Bio Sci 32: 469–476. [DOI] [PubMed] [Google Scholar]
- 7. Rutkowski DT, Arnold SM, Miller CN, Wu J, Li J, et al. (2006) Adaptation to ER stress is mediated by differential stabilities of pro-survival and pro-apoptotic mRNAs and proteins. PLOS Biol 4: 2024–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schroder M, Kaufman RJ (2005) The mammalian unfolded protein response. Annu Rev Biochem 74: 739–789. [DOI] [PubMed] [Google Scholar]
- 9. Scheuner D, Song B, McEwen E, Liu C, Laybutt R, et al. (2001) Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell 7: 1165–1176. [DOI] [PubMed] [Google Scholar]
- 10. Marciniak SJ, Yun CY, Oyadomari S, Novoa I, Zhang Y, et al. (2004) CHOP induces death by promoting protein synthesis and oxidation in the stressed endoplasmic reticulum. Genes Dev 18: 3066–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee AH, Iwakoshi NN, Glimcher LH (2003) XBP-1 regulates a subset of endoplasmic reticulum resident chaperone genes in the unfolded protein response. Mol Cell Biol 23: 7448–7459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee AS (2007) GRP78 induction in cancer: therapeutic and prognostic implications. Cancer Res 67: 3496–3499. [DOI] [PubMed] [Google Scholar]
- 13. Healy SJM, Gorman AM, Mousavi-Shafaei P, Gupta S, Samali A (2009) Targeting the endoplasmic reticulum-stress response as an anticancer strategy. Eur J Pharmacol 625: 234–246. [DOI] [PubMed] [Google Scholar]
- 14. Suzuki T, Lu J, Zahed M, Kita K, Suzuki N (2007) Reduction of GRP78 expression with siRNA activates unfolded protein response leading to apoptosis in HeLa cells. Arch Biochem Biophys 468: 1–14. [DOI] [PubMed] [Google Scholar]
- 15. Ranganathan AC, Zhang L, Adam AP, Aguirre-Ghiso JA (2006) Functional coupling of p38-induced upregulation of BiP and activation of RNA-dependent protein kinase-like endoplasmic reticulum kinase to drug resistance of dormant carcinoma cells. Cancer Res 66: 1702–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pyrko P, Schönthal AH, Hofman FM, Chen TC, Lee AS (2007) The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res 67: 9809–9816. [DOI] [PubMed] [Google Scholar]
- 17. Jiang CC, Mao ZG, Avery-Kiejda KA, Wade M, Hersey P, et al. (2009) Glucose-regulated protein 78 antagonizes cisplatin and adriamycin in human melanoma cells. Carcinogenesis 30: 197–204. [DOI] [PubMed] [Google Scholar]
- 18. Virrey JJ, Dong D, Stiles C, Patterson JB, Pen L, et al. (2008) Stress chaperone GRP78/BiP confers chemoresistance to tumor-associated endothelial cells. Mol Cancer Res 6: 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Alam GN, Ning Y, Visioli F, Dong Z, et al. (2012) The unfolded protein response induces the angiogenic switch in human tumor cells through the PERK/ATF4 pathway. Cancer Res 72: 5396–5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brenner JC, Graham MP, Kumar B, Saunders LM, Kupfer R, et al. (2010) Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head Neck 32: 417–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Misawa K, Kanazawa T, Misawa Y, Uehara T, Imai A, et al. (2013) Galanin has tumor suppressor activity and is frequently inactivated by aberrant promoter methylation in head and neck cancer. Transl Oncol 6: 338–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Misawa K, Ueda Y, Kanazawa T, Misawa Y, Jang I, et al. (2008) Epigenetic inactivation of galanin receptor 1 in head and neck cancer. Clin Cancer Res 14: 7604–7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jia L, Soengas MS, Sun Y (2009) ROC1/RBX1 E3 ubiquitin ligase silencing suppresses tumor cell growth via sequential induction of G2-M arrest, apoptosis, and senescence. Cancer Res 69: 4974–4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pamarthy D, Tan M, Wu M, Chen J, Yang D, et al. (2007) p27 degradation by an ellipticinium series of compound via ubiquitin-proteasome pathway. Cancer Biol Ther 6: 360–366. [DOI] [PubMed] [Google Scholar]
- 25. Zeitlin BD, Joo E, Dong Z, Warner K, Wang G, et al. (2006) Antiangiogenic effect of TW37, a small-molecule inhibitor of bcl-2. Cancer Res 66: 8698–8706. [DOI] [PubMed] [Google Scholar]
- 26. Rao RV, Peel A, Logvinova A, Rio G, Hermel E, et al. (2002) Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Let 514: 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, et al. (2003) Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem 278: 20915–20924. [DOI] [PubMed] [Google Scholar]
- 28. Singh AK, Manns MP, Seidler U (2011) Cytoprotective effects of acidosis via heat shock protein HSP27 against the anticancer drug doxorubicin. Cell Mol Life Sci 68: 1041–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sauvant C, Nowak M, Wirth C, Schneider B, Riemann A, et al. (2008) Acidosis induces multi-drug resistance in rat prostate cancer cells (AT1) in vitro and in vivo by increasing the activity of the p-glycoprotein via activation of p38. Int J Cancer 123: 2532–2542. [DOI] [PubMed] [Google Scholar]
- 30. Thews O, Gassner B, Kelleher DK, Schwerdt G, Gekle M (2006) Impact of extracellular acidity on the activity of p-glycoprotein and the cytotoxicity of chemotherapeutic drugs. Neoplasia 8: 143–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wolosker H, Rocha JB, Engelender S, Panizzutti R, De Miranda J, et al. (1997) Sarco/endoplasmic reticulum Ca2+-ATPase isoforms: diverse responses to acidosis. Biochem J 321: 545–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aoyama K, Burns DM, Suh SW, Garnier P, Matsumori Y, et al. (2005) Acidosis causes endoplasmic reticulum stress and caspase-12-mediated astrocyte death. J Cereb Blood Flow Metab 25: 358–370. [DOI] [PubMed] [Google Scholar]
- 33. Kumar S, Kasseckert S, Kostin S, Abdallah Y, Schafer C, et al. (2007) Ischemic acidosis causes apoptosis in coronary endothelial cells through activation of caspase-12. Cardiovasc Res 73: 172–180. [DOI] [PubMed] [Google Scholar]
- 34. Kumar S, Reusch HP, Ladilov Y (2008) Acidic pre-conditioning suppresses apoptosis and increases expression of Bcl-xL in coronary endothelial cells under simulated ischaemia. J Cell Mol Med 12: 1584–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bolli R (2007) Preconditioning: a paradigm shift in the biology of myocardial ischemia. Am J Physiol Heart Circ Physiol 292: H19–H27. [DOI] [PMC free article] [PubMed] [Google Scholar]