Abstract
Background and Aims
Salvia is the largest genus in Lamiaceae and it has recently been found to be non-monophyletic. Molecular data on Old World Salvia are largely lacking. In this study, we present data concerning Salvia in Africa. The focus is on the colonization of the continent, character evolution and the switch of pollination systems in the genus.
Methods
Maximum likelihood and Bayesian inference were used for phylogenetic reconstruction. Analyses were based on two nuclear markers [internal transcribed spacer (ITS) and external transcribed spacer (ETS)] and one plastid marker (rpl32-trnL). Sequence data were generated for 41 of the 62 African taxa (66 %). Mesquite was used to reconstruct ancestral character states for distribution, life form, calyx shape, stamen type and pollination syndrome.
Key Results
Salvia in Africa is non-monophyletic. Each of the five major regions in Africa, except Madagascar, was colonized at least twice, and floristic links between North African, south-west Asian and European species are strongly supported. The large radiation in Sub-Saharan Africa (23 species) can be traced back to dispersal from North Africa via East Africa to the Cape Region. Adaptation to bird pollination in southern Africa and Madagascar reflects parallel evolution.
Conclusions
The phenotypic diversity in African Salvia is associated with repeated introductions to the continent. Many important evolutionary processes, such as colonization, adaptation, parallelism and character transformation, are reflected in this comparatively small group. The data presented in this study can help to understand the evolution of Salvia sensu lato and other large genera.
Keywords: Salvia, Lamiaceae, Canary Islands, character evolution, ITS, ETS, Madagascar, ornithophily, pollination, rpl32-trnL, Sub-Saharan Africa
INTRODUCTION
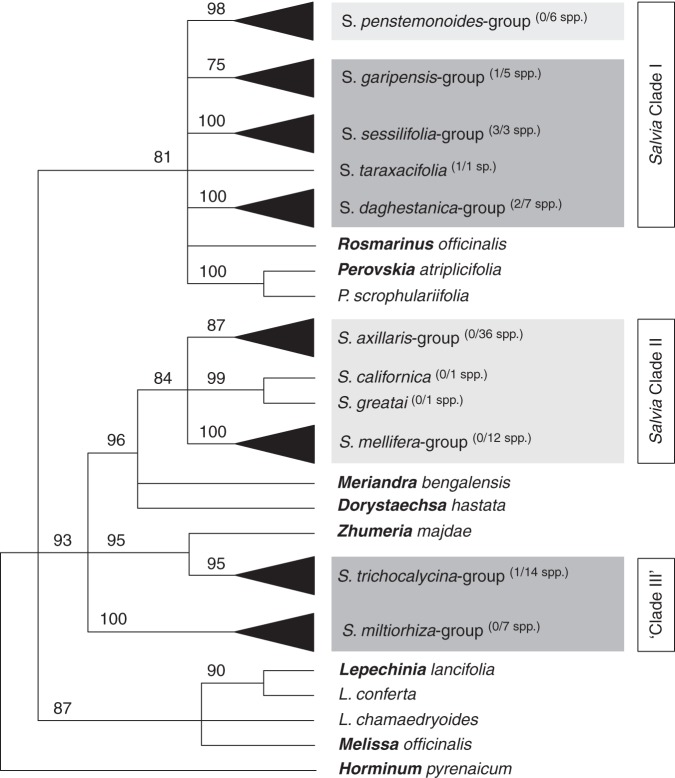
Throughout the world, many Salvia spp. (Lamiaceae) are known as ornamental (e.g. S. coccinea, S. patens, S. viridis), medicinal (S. officinalis, S. miltiorhiza) and even hallucinogenic plants (S. divinorum) (Clebsch, 2008; Froissart, 2008). Salvia is the largest genus in the mint family, with 900–1000 species distributed worldwide (Alziar, 1988–1993; Harley et al., 2004). Molecular studies have shown many large genera to be non-monophyletic, and this is also true for Salvia (Walker et al., 2004), with respect to Dorystaechas, Meriandra, Perovskia, Rosmarinus and Zhumeria (Walker and Sytsma, 2007). Major clades containing Salvia spp. were named Salvia Clade I, II and ‘III’ Salvia (s.l.). So far, previous molecular studies have focused on New World (NW) Salvia. In contrast, Old World (OW) species have been largely disregarded (Walker et al., 2004; Sudarmono, 2007, 2008; Walker and Sytsma, 2007; Jenks et al., 2011, 2012; Li et al., 2013). Only seven African species were included in the genus-wide study of Walker and Sytsma (2007), which revealed that they are members of two of the three major clades proposed by the authors (Fig. 1; Clade I, ‘Clade III’). In this study, Salvia ‘Clade III’ is paraphyletic with respect to Zhumeria majdae. Thus, we accept the south-west (SW) Asian species and Zhumeria as one clade (S. trichocalycina group; Clade III) and the East Asian species as the fourth independent evolutionary lineage (S. miltiorhiza group; Clade IV). According to Walker and Sytsma (2007), Salvia Clade I is monophyletic and covers the type species of the genus (S. officinalis; Jarvis, 2007). We therefore refer to it as Salvia sensu stricto (s.s.).
Fig. 1.
Phylogenetic tree of Salvia s.l. defining the major clades (Walker and Sytsma, 2007, simplified, i.e. nodes with bootstrap values <75 % were collapsed). Strict consensus tree based on the combined analysis of trnL-F and ITS data; MP. Non-Salvia genera are highlighted (bold); NW Salvia is highlighted in light grey and OW taxa in dark grey. Numbers in parentheses, separated by slashes, indicate the number of African species and the total number of taxa in the corresponding clade in their analyses.
Sixty-two Salvia spp. occur in Africa and adjacent areas (Fig. 2; Hedge, 1974; Santos and Fernández, 1986; Van Jaarsveld, 1999). Most are endemics distributed in North Africa (25 species including the Canary Island endemics), southern Africa (23 species), Madagascar (six species), East Africa and on the Arabian Peninsula (eight species). Similar distribution ranges are known for other plant genera such as Androcymbium, Senecio and Zygophyllum (Caujapé-Castells, 2001; Colemann et al., 2003; Bellstedt et al., 2008; Del Hoyo et al., 2009) and for animals, e.g. Diptera (Kirk-Spriggs and McGregor, 2009). Salvia thus appears to be another genus adapted to the similar climatic conditions occurring in North (Mediterranean area), East (East African mountains) and southern (Cape Region) Africa.
Fig. 2.
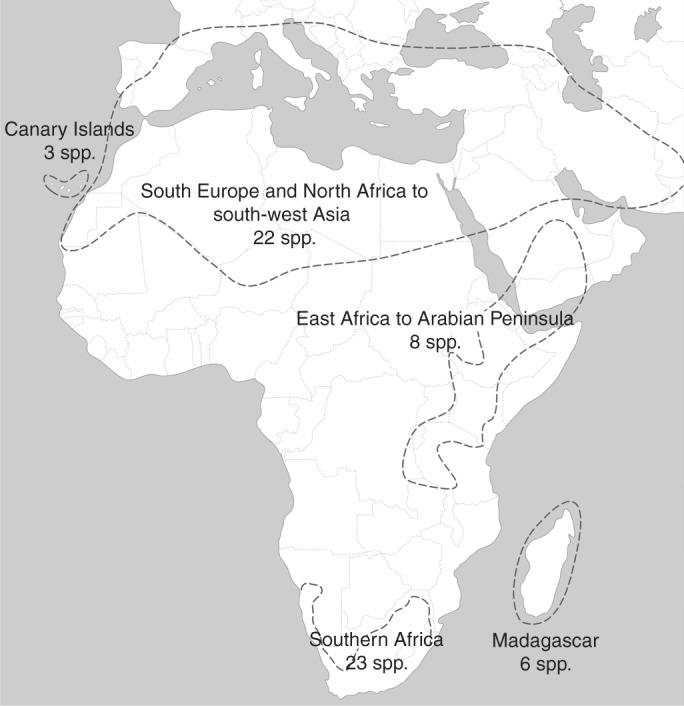
Distribution of Salvia s.l. on the African continent. Five regions on the continent and adjacent areas are recognized with 62 Salvia spp.: (1) the Canary Islands; (2) southern Europe/North Africa to south-west Asia; (3) East Africa/Arabian Peninsula; (4) southern Africa; and (5) Madagascar. Distributions are based on: Hedge (1974), Codd (1985), Santos and Fernández (1986), Thulin (1993, 2009) and Van Jaarsveld (1999).
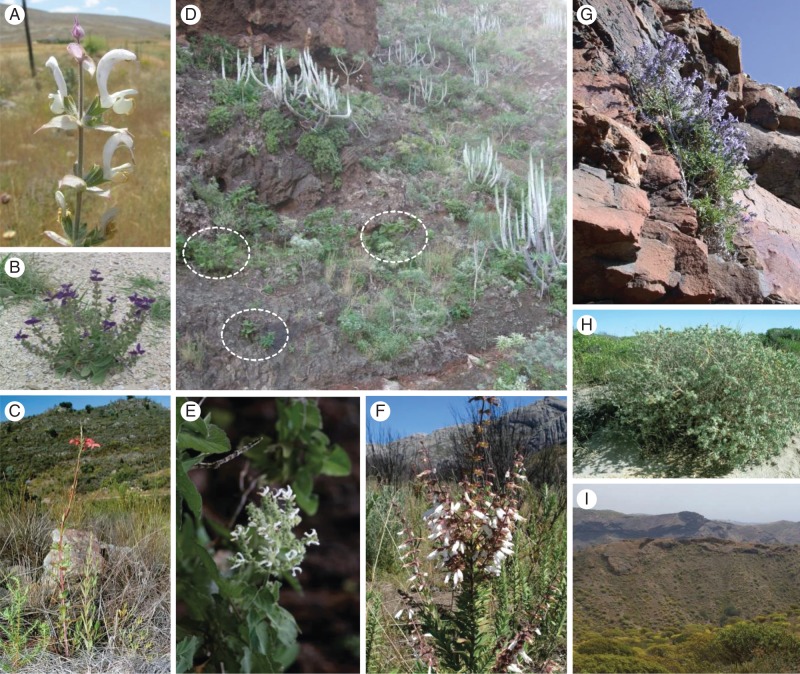
African Salvia is of special interest for addressing evolutionary questions. Species are highly diverse in habitat preferences (Fig. 3), floral morphology (shape, size, colour and stamen construction) and pollination (Fig. 4). The only bird-pollinated species known from the Old World evolved in Sub-Saharan Africa (SSA) (Scott-Elliot, 1890; Van Jaarsveld, 1999; Wester and Claßen-Bockhoff, 2006, 2007). The most recent classification of African Salvia was presented by Hedge (1974). Based on distribution and morphology, he arranged 59 species in 23 species groups. However, it is not known to which degree these species groups represent natural lineages.
Fig. 3.
Habitat diversity of Salvia s.l. in Africa. (A) Salvia sclarea, Anatolian steppe; (B) S. viridis, Anatolia; (C) S. thermarum, fynbos Western Cape, South Africa; (D, E) S. broussonetii, basalt maritime cliffs on Tenerife (D, dotted circles), flowering plant (E); (F) S. leucodermis, Madagascar; (G) S. herbanica, rocky habitat south-east Fuerteventura, Canary Islands; (H) S. africana-lutea, coastal sand dunes in the Western Cape; (I) S. aegyptiaca, semi-arid habitat on Gran Canaria, Canary Islands. Photographs: (A, B) F. Celep, (C, H) P. Wester, (D, E, G) R. Claßen-Bockhoff, (F) B. Bytebier, and (I) M. Thulin.
Fig. 4.
Phenotypic diversity of Salvia s.l. in Africa. (A) Salvia verticillata; (B) S. nilotica; (C) S. disermas; (D) S. argentea; (E) S. aegyptiaca; (F) S. canariensis; (G) S. broussonetii; (H) S. herbanica; (I) S. lanceolata with Cinnyris chalybeus (southern double-collared sunbird; formerly Nectarinia chalybeus); (J) S. geminata; (K) S. taraxacifolia; (L) S. interrupta; (M) S. dolomitica; (N) S. chamelaeagnea with Xylocopa caffra (carpenter bee); (O) S. scabra; (P) S. leucodermis; (Q) S. sessilifolia; (R) S. thermarum; and (S) S. africana-lutea. Note the typical falcate upper corolla lip in bee-pollinated species (e.g. S. argentea, D) in contrast to the straight upper lip in bird-pollinated taxa (I, P–R) and the diverse floral morphologies in the Canary Islands endemics (F–H). Photographs: (A, B, C, D, F, L) M. Will, (E, H) R. Claßen-Bockhoff, (G, K, O, R, S) P. Wester, (I) R. Groneberg, (J) M. Thulin, (N) H. Technau, (P) P. B. Phillipson and (Q) D. Hannon.
In the present study, we examine the monophyly of Hedge's (1974) species groups based on a phylogenetic analysis that includes two-thirds of the African species. We intend to determine the number of independent origins of African Salvia. Furthermore, the colonization of the African continent and the evolution of African Salvia, e.g. the origin of bird pollination, are reconstructed.
MATERIALS AND METHODS
Plant material
Our analyses include 84 Salvia spp., 46 of them sequenced for the first time. We cover 41 (66 %) of the 62 African species: two endemics from Madagascar, all endemics from the Canary Islands, eight species from eastern Africa and the Arabian Peninsula, 18 southern African species and 12 species restricted to North Africa, the circum-Mediterranean area and SW Asia. Accessions derived from GenBank were used to complement the data set. Voucher information and GenBank accession numbers are provided in the Appendix. Due to the lack of suitable plant material and/or successful PCR, some species are only represented by a sub-set of the three molecular markers.
Our sampling covers all major lineages previously identified in Salvia (Walker and Sytsma, 2007; Fig. 1). Well-supported clades that are not the focus of this study (Clades II and IV; see Walker and Sytsma, 2007; Will, 2013) are represented by a sub-set of species only. Independent accessions are included, especially for taxonomically critical and polymorphic species. Hyptis laniflora was used as the outgroup in all analyses. Nomenclature is in accordance with Alziar (1988–1993) and with the International Plant Names Index (http://www.ipni.org/ipni/, accessed 30 April 2013). The term Sub-Saharan Africa is used in the sense of Linder (2001) but additionally includes the Namib–Kalahari region.
DNA extraction, amplification and sequencing
For Salvia, new sequence data are presented for the internal transcribed spacer (ITS; 39 species), the external transcribed spacer (ETS; 38 species) and the plastid marker rpl32-trnLUGA (57 species). The latter was selected based on the results of previous primer screening (trnL-F and rpl32-ndhF). Total genomic DNA was obtained from silica-dried or herbarium leaf material. DNA was extracted according to the manufacturer's protocol for the NucleoSpin® plant DNA extraction kit (Macherey-Nagel, Düren, Germany). The standard 25 μL PCR mix consisted of 2 mm MgCl2, 200 μm dNTPs, 1 pm primer, 0·025 U μL–1 Taq polymerase and 0·5–1·0 μL of DNA extract in the reaction buffer provided by the manufacturer of the polymerase.
The PCRs were carried out in a Biometra T3 or a PTC 100 MJ Research thermocycler using the following program: 60 s at 94 °C; followed by 35 cycles of 20 s at 94 °C, 30 s at 55 °C and 60 s at 72 °C; and a post-treatment of 80 s at 55 °C and 8 min at 72 °C for each marker. The whole ITS region was sequenced as a single piece using the ITS-A (Noyes and Rieseberg, 1999) and ITS-4 primers (White et al., 1990). The ETS region was sequenced using 18S-E (Baldwin and Markos, 1998) and ETS-B (Beardsley and Olmstead, 2002). For plastid sequences, we used the rpl32 and trnLUGA primers (Shaw et al., 2007). PCR products were purified according to the manufacturers' protocols using ExoSAP-IT PCR Product Clean-up (Affymetrix UK Ltd, Wooburn Green, UK) or NucleoSpin®Extract II-kit (Macherey-Nagel).
Cycle sequencing was performed using ABI Prism Big DyeReady Reaction Mix (Perkin Elmer/Applied Biosystems, Foster City, CA, USA) using the primers listed above and following the manufacturer's protocol. Products were purified with Sephadex™ G50 (VWR International GmbH, Darmstadt, Germany) and sequenced on a 16-capillary ABI 3130 xl automated sequencer (Life Technologies GmbH, Darmstadt, Germany).
DNA sequence alignment and phylogenetic analyses
Sequencing was straightforward for each marker. Forward and reverse sequences were edited manually, merged into consensus sequences using Sequencer™ 4.1.2. (GeneCodeCorp., Ann Arbor, MI, USA) and aligned manually in McClade4.1 (Maddison and Maddison, 2000). Ambiguously alignable regions (identified manually) were excluded from analyses. The three data sets were analysed separately. In order to increase resolution, we combined nuclear and plastid data (combined data set). Partitions were defined for the combined data set before the best-fit models of nucleotide substitution were selected with jModeltest 2.1.1 (Darriba et al., 2012). Under the Akaike information criterion (AIC), the GTR+I+G model was selected for the ITS data set and TVM+G for ETS and the plastid marker. Two tree searches, one under maximum likelihood (ML) with bootstrapping (BS; RAxML-HPC BlackBox v.7.4.4; Stamatakis, 2006; Stamatakis et al., 2008) and one under Bayesian inference (BI; MrBayes v.3.1.2 on XSEDE; Ronquist and Huelsenbeck, 2003), were performed on the CIPRES Science Gateway v.3.3 server (Miller et al., 2010). Since MrBayes does not allow nst = 5, required for TVM+G, we chose the more complex model (nst = 6). For BI, we ran four Markov chains simultaneously for 10 million generations analysing the plastid and ETS data sets. Two independent runs of 40 million generations were performed for the ITS and combined data sets. Every thousandth generation was sampled. The burn-in was determined with Tracer v.1.5 implemented in BEAST. We generated 50 % majority rule consensus trees with posterior probabilities (PPs) using MrBayes v.3.1.2.
The ITS, ETS and plastid data were analysed separately to identify incongruences. To combine data without conflict, strongly supported (PP = 1·00 or ≥92 % ML BS) incongruences were dealt with by the duplication of the corresponding individuals, with one duplicate having only ITS and ETS sequences, and one only having plastid sequences (Pirie et al., 2008). The absent sequences were coded as missing data (‘?’). Sequences from the same or different species that were completely identical were reduced to one haplotype. In the text or figures, sequence identity is indicated by a slash separating the corresponding accessions. The existing concept of clades sensu Walker and Sytsma (2007) was adopted, except for ‘Clade III’. The latter was split into two independent clades, i.e. the S. trichocalycina group (Clade III) and the S. miltiorrhiza group (Clade IV) (Fig. 1).
Ancestral character state reconstruction
Ancestral character states were reconstructed using the Mesquite software package v.2.75 with Fitch parsimony optimization (Maddison and Maddison, 2011). Five characters, i.e. distribution area, life form, calyx morphology, stamen type and pollination system, were coded (Supplementary Data Table S1). Character states are based on literature research and observations. Stamen classification is based on stamen types and intermediate forms introduced by Hedge (1974, 1982): A, lower lever arm with fertile thecae; B, lower lever arm sterile; and C, lower lever arm reduced. Information on pollinators is based on the literature or personal communications, or was postulated according to character syndromes (e.g. Wester and Claßen-Bockhoff, 2011). In all cases, reconstructions are performed on 100 randomly sampled trees with branch lengths from the BI analyses, as well as the consensus tree (50 % majority rule) of the combined data BI analysis (Pirie et al., 2009). Statistical support was calculated under maximum parsimony (MP).
RESULTS
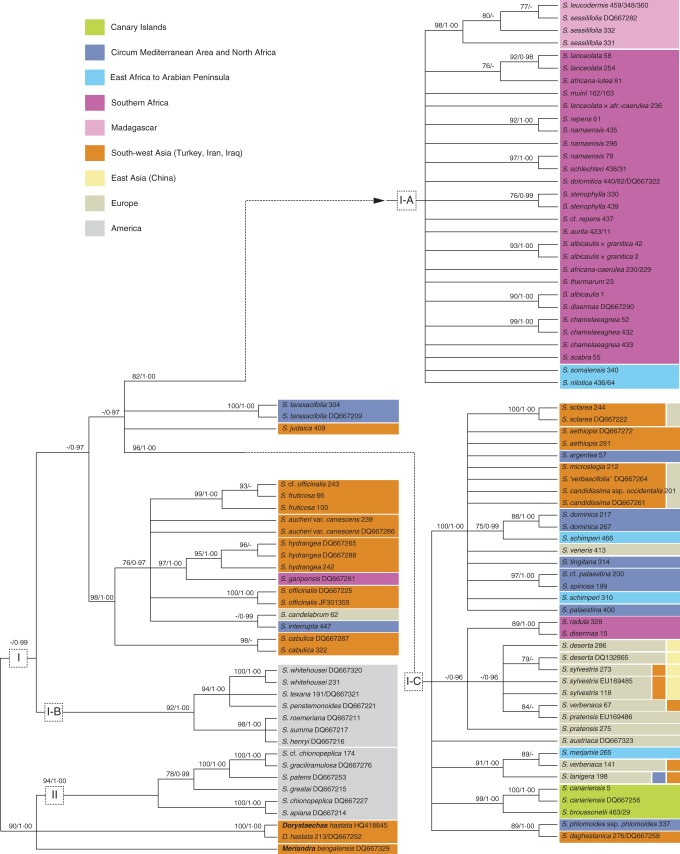
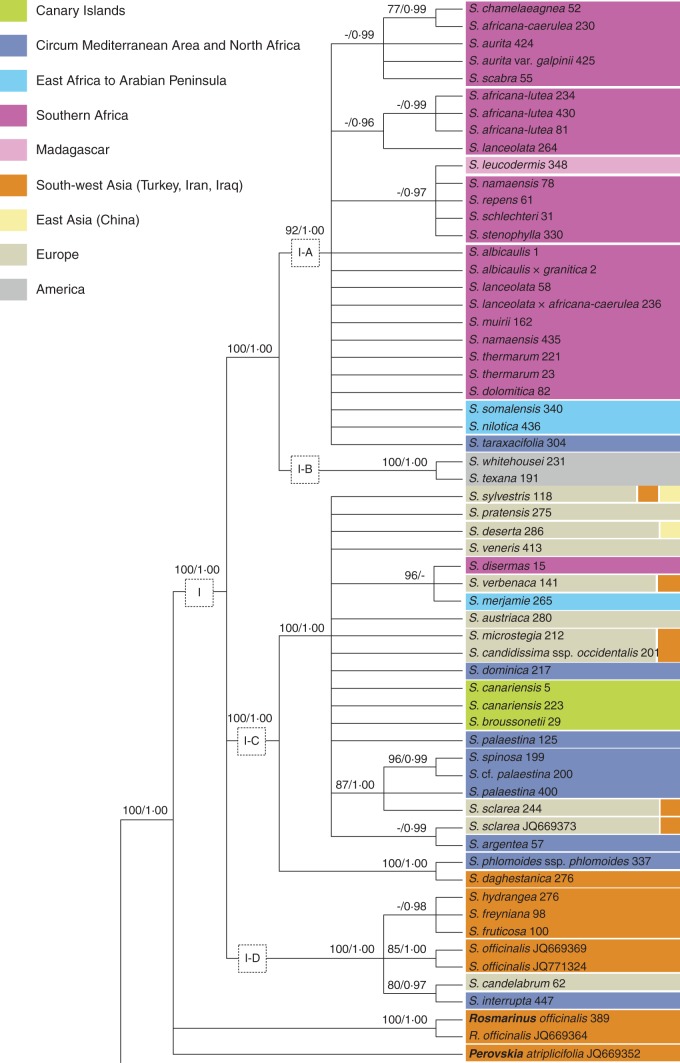
Phylogenetic analyses: nrITS (Fig. 5)
Fig. 5.
Analyses of the nrITS data set. Non-Salvia genera are highlighted (bold); names of accessions with identical sequences are separated by slashes; only support values ≥75 % (BS) and ≥0·95 (PP) are illustrated. Species distribution is indicated by different colours.
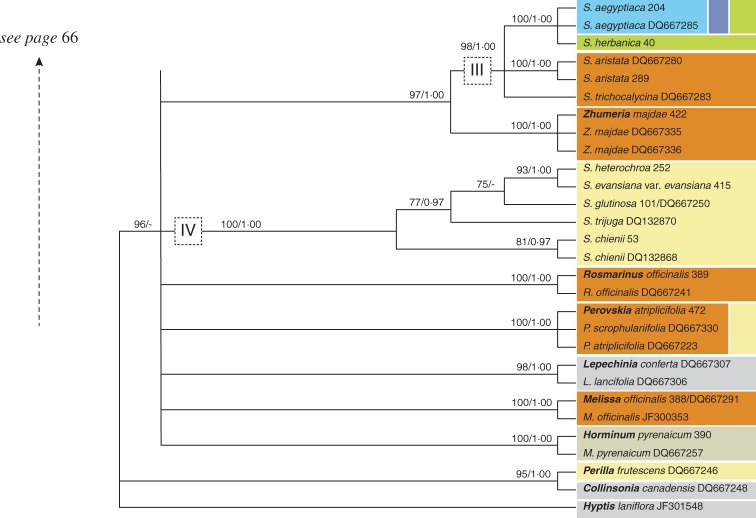
The aligned length of the nuclear data set is 637 bp, 255 (40 %) of which are potentially parsimony informative. Hyptis laniflora is found in a polytomy with (1) Collinsonia and Perilla and (2) an unresolved, weakly supported clade. Within the last of these, nine lineages are strongly supported by BI: (1) Horminum; (2) Melissa; (3) Lepechinia; (4) Perovskia; (5) Rosmarinus; (6) Clade IV; (7) Clade III plus the genus Zhumeria; (8) a trichotomy including Meriandra, Dorystaechas and NW Salvia species (Clade II); and (9) Clade I (Salvia s.s.).
Clade IV is a well-supported clade including four East Asian and one European species (S. glutinosa). Clade III consists of a trichotomy composed of (1) S. trichocalycina, (2) S. aristata and (3) S. aegyptiaca plus S. herbanica. The American Clade II is strongly supported as part of a trichotomy with the two OW genera Meriandra and Dorystaechas. It is divided into two sub-clades, with S. chionopeplica and S. cf. chionopeplica in separate sub-clades. Clade I is only supported by BI (PP = 0·99). Sub-clade I-C forms a polytomy with Clade I-A, S. judaica and S. taraxacifolia. Sub-clade I-D is sister to this clade, and the American sub-clade I-B is in turn sister to the remainder of Clade I. Within sub-clade I-D (seven species), S. cabulica is sister to a polytomy consisting of six lineages (Fig. 5: I-D). The two accessions of S. fruticosa do not cluster together. Six major lineages are recognized within sub-clade I-C. One lineage contains the two Canary Island endemics S. broussonetii and S. canariensis (Fig. 5: I-C; green). A second lineage includes the South African endemics S. radula and S. disermas (Fig. 5: I-C; magenta) along with four European species (Fig. 5: I-C; beige; monophyly for each of these species is not confirmed). Sub-clade I-A is only poorly resolved. It includes exclusively African taxa restricted to SSA. Within sub-clade I-A, one strongly supported clade includes all six Madagascan accessions (2 spp.) (Fig. 5: I-A; pink). The ML analysis weakly indicates that S. sessilifolia is paraphyletic with respect to S. leucodermis. Sequences of S. leucodermis and S. sessilifolia differ only in one position (A or C) which is ambiguous (IUPAC code: M) in S. sessilifolia accession DQ667282.
ETS (Supplementary Data Fig. S1)
The alignment contains 69 accessions (53 species), 60 (44 species) representing the genus Salvia. The aligned length of the data set is 466 bp, 230 (40·4 %) of which are potentially parsimony informative. There are few major conflicts with the ITS topology. Differences concern a clade which is moderately supported by BI (PP = 0·98) including: (1) Clade II plus Meriandra and Dorystaechas; (2) Clade III without S. aristata (here called Clade III-A); (3) Zhumeria plus S. aristata; and (4) S. przewalskii (Clade IV represented by only one species in this data set). This clade is not supported (but also not contradicted) in the ITS analyses. The incongruence between the ITS and ETS data sets is in the position of Zhumeria. However, this difference might be based on the slightly different sampling in Clade III. Furthermore, support for a monophyletic Clade II is lacking. Instead, the three lineages of NW Salvia spp. form a polytomy with Meriandra and Dorystaechas (Supplementary Data Fig. S1].
Compared with the ITS data set, support for Clade I (Salvia s.s.) is low (PP = 0·96). The four sub-clades (sub-clades I-A through I-D) form a polytomy. Differences in the topology of sub-clade I-C are mainly based on additional accessions in the ETS data set, e.g. S. canariensis 464. The latter renders S. canariensis paraphyletic with respect to S. broussonetii, but this relationship is not strongly supported. Similarly, adding S. disermas 454 causes S. disermas to be paraphyletic with respect to S. radula. Sub-clade I-A is better resolved in the ETS than in the ITS data set. The former supports sister relationships for (1) S. cf. repens 437 and S. stenophylla and (2) S. aurita and S. dolomitica. Furthermore, S. africana-caerulea, S. albicaulis, S. chamelaeagnea and S. lanceolata × africana-caerulea appear in a moderately supported clade not supported in ITS data (PP = 0·98).
rpl32-trnL (Fig. 6)
Fig. 6.
Analyses of the rpl32-trnL data set. Non-Salvia genera are highlighted (bold); only support values ≥75 % (BS) and ≥0·95 (PP) are illustrated. Species distribution is indicated by different colours.
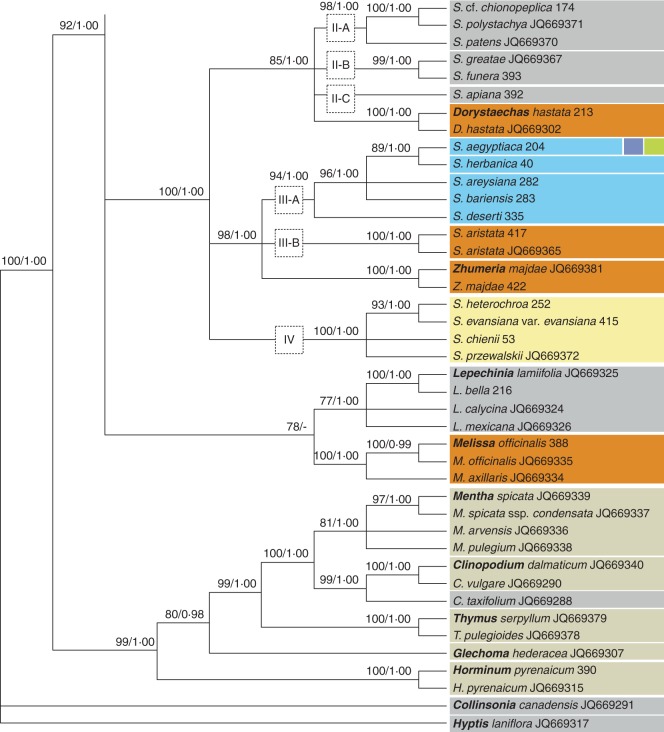
The aligned length of the plastid data set is 929 bp, with 246 (26·5 %) potentially informative nucleotide positions. Hyptis is again found in a trichotomy with Collinsonia and a strongly supported clade that includes all other accessions. The latter splits into two major lineages one only including non-Salvia samples (Horminum, Glechoma, Thymus, Clinopodium and Mentha). The second clade contains all Salvia samples and six additional genera. Melissa and Lepechinia are moderately supported (ML) as sister genera. They are found in a polytomy with two strongly supported clades.
The first includes Clade IV, sub-clades III-A, III-B, Zhumeria, Clade II and Dorystaechas, as found in the ETS data set. The position of Zhumeria differs from the ETS data; the genus is placed in a trichotomy with S. aristata (III-B) and the S. aegyptiaca group (III-A) based on plastid data. As in the ETS data set, the monophyly of Clade II is not supported. Instead, three sub-clades (II-A, II-B and II-C) are found in a polytomy with Dorystaechas. The most striking incongruence between nuclear and plastid data is the position of S. deserti (Fig. 6; III-A). It is sister to S. aegyptiaca based on ETS data (Supplementary Data Fig. S1) but strongly supported in a sister relationship to all species nesting in sub-clade III-A based on the plastid data set (Fig. 6). In the ITS data set, S. deserti is not represented.
The second major clade consists of a trichotomy composed of Rosmarinus, Perovskia and a strongly supported Clade I (Salvia s.s.). Compared with nuclear data, the latter is better resolved, splitting into three major lineages (sub-clades): (1) I-D (topology corresponding to nuclear data); (2) I-C; and (3) I-B plus I-A. Within sub-clade I-C, neither S. sclarea (244 and JQ669373) nor S. palaestina (400 and 200) is supported as monophyletic. Sister grouping of sub-clades I-A (Africa) and I-B (America) is in conflict with ITS data (Fig. 5). Sub-clade I-A was strongly supported by the nuclear data, but the relationships among its species were largely unresolved. However, its topology slightly differs. Instead of being part of a basal polytomy, S. chamelaeagnea 52 and S. africana-caerulea 230 are sister species in the plastid data set, closely related to S. aurita and S. scabra. The ITS data do not suggest any relationships for S. aurita, whereas the ETS data weakly support a sister relationship to S. dolomitica. Furthermore, in contrast to the ITS topology, different accessions of S. namaensis (78 and 435) and S. lanceolata (264 and 58) are not supported to be monophyletic based on plastid data.
Combined analyses of the nuclear and plastid data sets (Fig. 7)
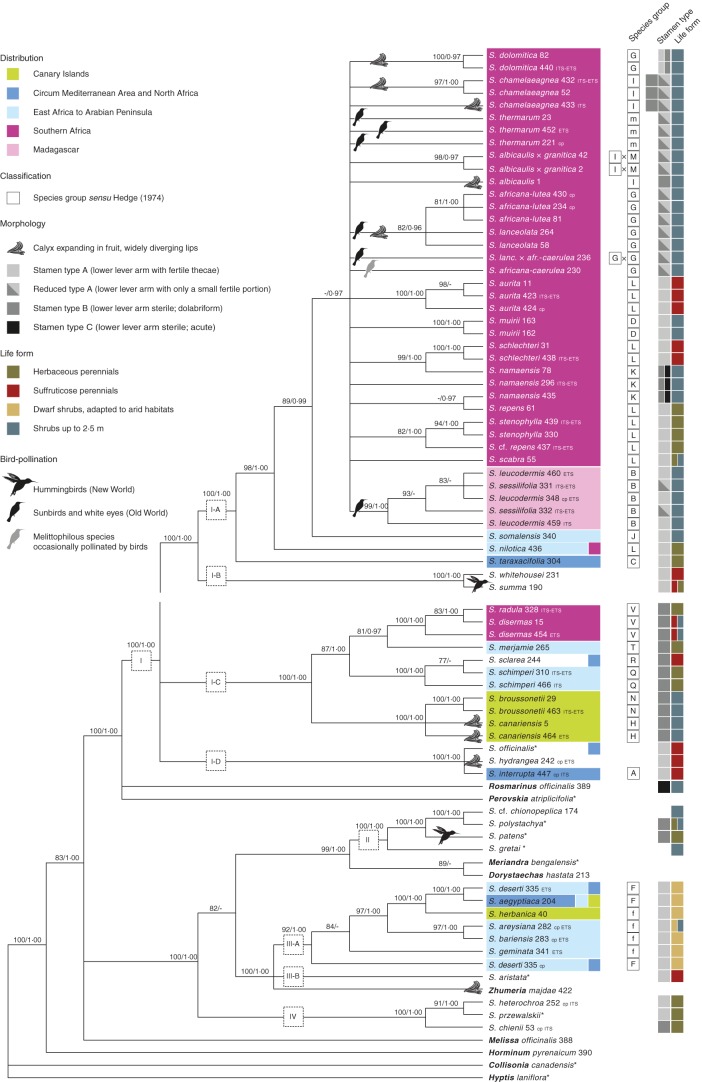
Fig. 7.
Analyses of the combined data set. Non-Salvia genera are highlighted (bold); GenBank accessions are marked with an asterisk (*); for taxa with only one or two markers, the corresponding marker is given after the taxon name and extraction number. Uncapitalized letters (m, f) indicate taxa described after the revision of African Salvia (Hedge, 1974); classification in the corresponding species group is based on morphology and the corresponding species description (Santos and Fernández, 1986; Thulin, 1993, 2009; Van Jaarsveld, 1999). For detailed information about ancestral character reconstruction see also Supplementary Data Table S1 and Figs S2–S6.
The aligned length of the combined data set is 1982bp, of which 582 (29·4 %) are potentially parsimony informative. The sequence duplication approach was not suitable to resolve the conflicting placement of sub-clade I-B within Salvia s.s., which was placed either at the base of Clade I (ITS; Fig. 5) or as sister to sub-clade I-A (plastid; Fig. 6). Thus, we used the tree with the best topology for illustration, being aware of the unresolved conflict for the two clades.
The combined tree largely reflects the topology of the plastid data set (e.g. sister relationship of sub-clade I-A and I-B), but shows better resolution and higher support within sub-clade I-A. Melissa is in a trichotomy with two clades containing Salvia spp. The first covers (1) Clade II, Dorystaechas and Meriandra, and the latter two moderately supported as sister; (2) sub-clades III-A, III-B and Zhumeria; and (3) Clade IV. The second includes Perovskia, Rosmarinus, and Salvia s.s. (Clade I). The latter is strongly supported and falls into the same three major lineages as in the plastid data set. As to sub-clade I-A, S. taraxacifolia is sister to all remaining taxa. Salvia nilotica splits next, followed by S. somalensis, which is sister to a large clade including only taxa from southern Africa and Madagascar. Monophyly of S. namaensis, S. repens, S. sessilifolia and S. leucodermis, each of which was represented by more than one accession, is not confirmed.
Ancestral character state reconstruction (Supplementary Data Figs S2–S6; Fig. 7)
All African areas, except Madagascar, were colonized more than once (Supplementary Data Fig. S2). The ancestral area is reconstructed as East Africa and the Arabian Peninsula for sub-clades of Salvia sensu lato (s.l.) (III-A; MP 100 %) and Salvia s.s. (sub-clade of I-C covering S. sclarea, S. schimperi, S. merjamie, S. disermas and S. radula; MP 89 %). Within the latter, S. disermas and S. radula point to the colonization (migration and/or dispersal) of southern Africa. A second colonization is supported for the largest lineage of sub-clade I-A, which only contains accessions from southern Africa (MP 94 %). Madagascar was most probably colonized from southern Africa (MP 64 %).
Reconstruction of the ancestral life form (Supplementary Data Fig. S3) revealed that three of the four defined growth forms evolved several times in parallel. Only some crown groups can clearly be characterized by this character, e.g. Clade IV (MP 100 %: perennial herbs), Clade III-A (MP 100 %; dwarf shrubs) and the clade consisting of S. canariensis and S. broussonetii (sub-clade I-C; MP 100 %; shrubs). For the clade containing Clades II, III and IV, herbaceous growth most probably reflects the ancestral state (MP 72 %). Clade IV includes exclusively herbaceous perennials, while its sister group is characterized by shrubby growth (MP 94 %). Within this clade, dwarf shrubby growth is a synapomorphy for sub-clade III-A (Fig. 7). A character transformation from herbaceous to shrubby growth is also found in Salvia s.s. (sub-clade I-A), and a reversal back to herbaceous growth is found for S. repens and S. stenophylla nesting within the same clade. Clade II (including Dorystaechas hastata and Meriandra bengalensis) appears to be originally shrubby. However, our study underrepresents the diversity of growth forms in this large NW clade, which includes shrubs, perennials, annuals and trees. The SW Asian species (sub-clade III-A, S. aristata and Zhumeria) show two different trends, one towards a suffruticose life form (S. aristata) and one towards dwarf shrubby growth (III-A).
A thick-textured, non-expanding calyx represents the ancestral state in each of the major Salvia clades (MP 100 %) (Supplementary Data Fig. S4). Expanding calyxes with widely diverging lips, which are papery and often coloured, evolved several times in parallel, not only in Salvia (sub-clades I-A, I-C and I-D) but also in Zhumeria (Fig. 7).
For stamen construction, type A with a fertile theca at the lower lever arm is strongly supported as the ancestral state in sub-clade III-A and Salvia s.s. (Supplementary Data Fig. S5). Within the latter, stamen type B evolved in sub-clade I-C and in I-A. Furthermore stamen type C is found in S. namaensis in sub-clade I-A (Fig. 7). The same reduction of the lower lever arm is found in Rosmarinus officinalis, which is not closely related to S. namaensis.
Melittophily, bee pollination, is reconstructed as the ancestral pollination system for each clade, except for sub-clade I-B (Supplementary Data Fig. S6). The latter is represented by only two species, one of them assumed to be ornithophilous and the other psychophilous (Wester and Claßen-Bockhoff, 2011). Thus, its ancestral character state remains ambiguous. Bird-pollinated flowers evolved repeatedly in the NW (Clade II and sub-clade I-B) and in the OW sub-clade I-A (Fig. 7). Within the latter, at least two pollinator shifts are suggested (Supplementary Data Fig. S6; Fig. 7), one by the Madagascan sub-clade and another by two South African species forming a clade (S. lanceolata and S. africana-lutea). A third switch to bird pollination might be represented by S. thermarum from South Africa. Two of the sampled accessions are found in a derived position of a melittophilous lineage (MP 60 %; Fig. 7).
DISCUSSION
Our study confirms the non-monophyly of both Salvia s.l. and the African Salvia spp. (Walker et al., 2004). We also confirm that all African species are restricted to two of the four major lineages of Salvia (Clades I and III; Fig. 7).
Interspecific relationships in African Salvia: species groups sensu Hedge
Subgeneric classification of the genus is based on morphology and distribution (Bentham, 1832–1836, 1848, 1876; Briquet, 1897). Hedge (1974) established 23 ‘species groups’ to address relationships among African Salvia spp. and their affinities beyond the continent. For four of these groups, molecular data can be used to discuss their monophyly.
The two taxa placed in species group V (S. disermas and S. radula) form a clade within sub-clade I-C (Fig. 7). Both species occur in southern Africa but do not overlap in their distribution (Fig. 9E: 26, 34). They have similar flower morphology but differ in flowering time and indumentum (Hedge, 1974). We confirm monophyly of this group and, based on ETS and combined data sets, find some support for Hedge's (1974) idea that S. radula could be a subspecies of S. disermas (Supplementary Data Table S1).
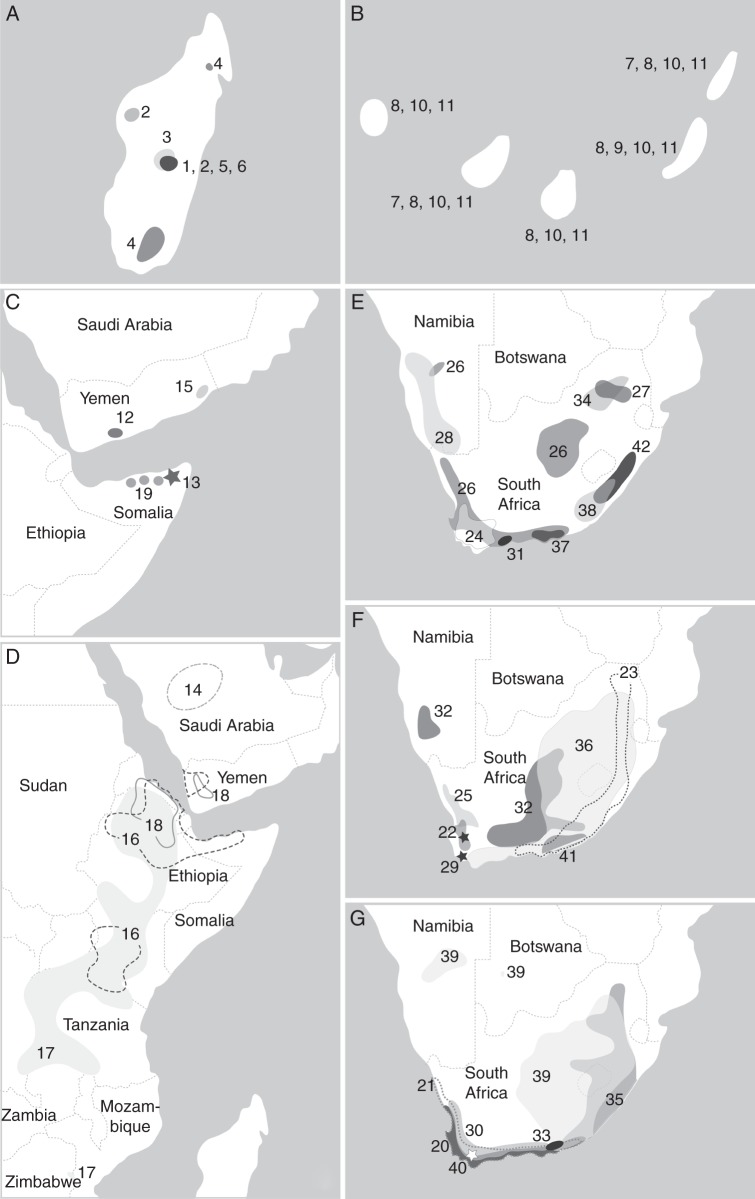
Fig. 9.
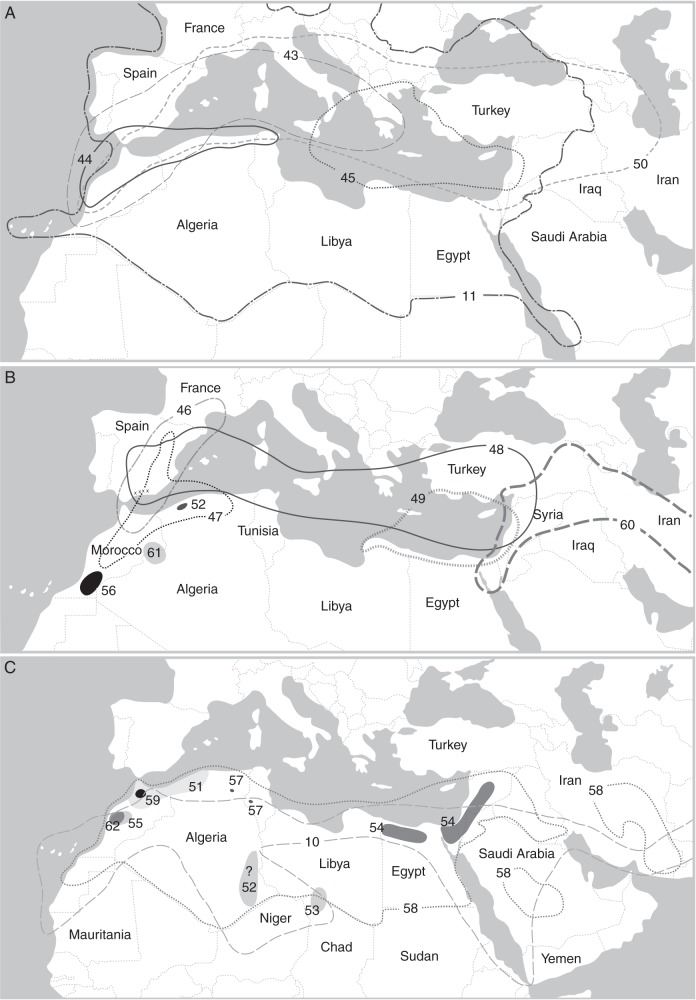
Distribution of African Salvia spp. (except North Africa and circum-Mediterranean area). (A) Madagascar: (1) S. cryptoclada; (2) S. leucodermis; (3) S. parvifolia; (4) S. perrieri; (5) S. porphyrocalyx; (6) S. sessilifolia. (B) Canary Islands: (7) S. broussonetii; (8) S. canariensis; (9) S. herbanica; (10) S. aegyptiaca; (11) S. verbenaca. (C, D) East Africa and Arabian Peninsula: (12) S. areysiana; (13, black asterisk) S. bariensis; (14) S. deserti; (15) S. geminata; (16) S. merjamie; (17) S. nilotica; (18) S. schimperi; (19) S. somalensis. (E–G) southern Africa: (20) S. africana-caerulea; (21) S. africana-lutea; (22) S. albicaulis; (23) S. aurita; (24) S. chamelaeagnea; (25) S. dentata; (26) S. disermas; (27) S. dolomitica; (28) S. garipensis; (29, black asterisk) S. granitica; (30) S. lanceolata; (31) S. muirii; (32) S. namaensis; (33) S. obtusata; (34) S. radula; (35) S. repens; (36) S. runcinata; (37) S. scabra; (38) S. schlechteri; (39) S. stenophylla; (40, white asterisk) S. thermarum; (41) S. triangularis; (42) S. tysonii. Based on Codd (1985), Hedge (1974), Santos and Fernández (1986), Thulin (1993, 2009) and Van Jaarsveld (1999). Note the overlapping distributions of species in southern Africa (Fig. 7E–G) and the disjunct area of S. stenophylla (39) and S. disermas (26).
Species group F originally included three African species (Hedge, 1974). The two species included in this study (S. aegyptiaca and S. deserti) form a clade with four more recently described species (Fig. 7; III-A), all of which are adapted to arid or semi-arid habitats. Except for the widespread S. aegyptiaca (Fig. 10C: 10), they are all local endemics, e.g. in East Africa and the Arabian Peninsula (S. areysiana, S. bariensis and S. geminata) or Fuerteventura (S. herbanica) (Fig. 9B: 9; C: 12, 13, 15, D: 14). As the clade is well supported by synapomorphies, e.g. growth as dwarf shrubs with simple, revolute leaves, straight upper corolla lips with exposed stamens and minute flowers (Hedge, 1974; Santos and Fernández, 1986; Scholz, 1993; Thulin, 1993, 2009), we not only confirm monophyly of species group F but extend it to include at least the six sampled species. Based on the unique character syndrome, more species are likely to be included in this species group (Bokhari and Hedge, 1977; M. Will and R. Claßen-Bockhoff, unpubl. data). Furthermore, our data confirm the close relationship proposed for African and SW Asian Salvia (Davis and Hedge, 1971; Hedge, 1974).
Fig. 10.
Distribution of Salvia spp. from North Africa and the circum-Mediterranean area. (A–C) (10) S. aegyptiaca; (11) S. verbenaca; (43) S. argentea; (44) S. barrelieri; (45) S. fruticosa; (46) S. lavandulifolia; (47) S. phlomoides; (48) S. sclarea; (49) S. spinosa; (50) S. viridis; (51) S. algeriensis; (52) S. balansae; (53) S. chudaei; (54) S. dominica; (55) S. gattefossei; (56) S. interrupta; (57) S. jaminiana; (58) S. lanigera; (59) S. mouretii; (60) S. palaestina; (61) S. pseudojaminiana; (62) S. taraxacifolia. Based on Hedge (1974); the distribution of S. sclarea on the Iberian Peninsula is based on Rosúa and Blanca (1986). Note the overlapping distribution of S. phlomoides (47) and S. candelabrum (endemic to the Iberian Peninsula, indicated by ‘x’; based on Rosúa and Blanca (1986).
In contrast, relationships for the monospecific species groups H and N were not predicted before. While Hedge (1974) supposed that the allies of S. canariensis (H) occur in southern Africa, he described S. broussonetii (N) as a relict species without any close ally. However, our data point to a close relationship between these two endemics from the Canary Islands (Fig. 7).
Morphological characters used for classification: stamen types
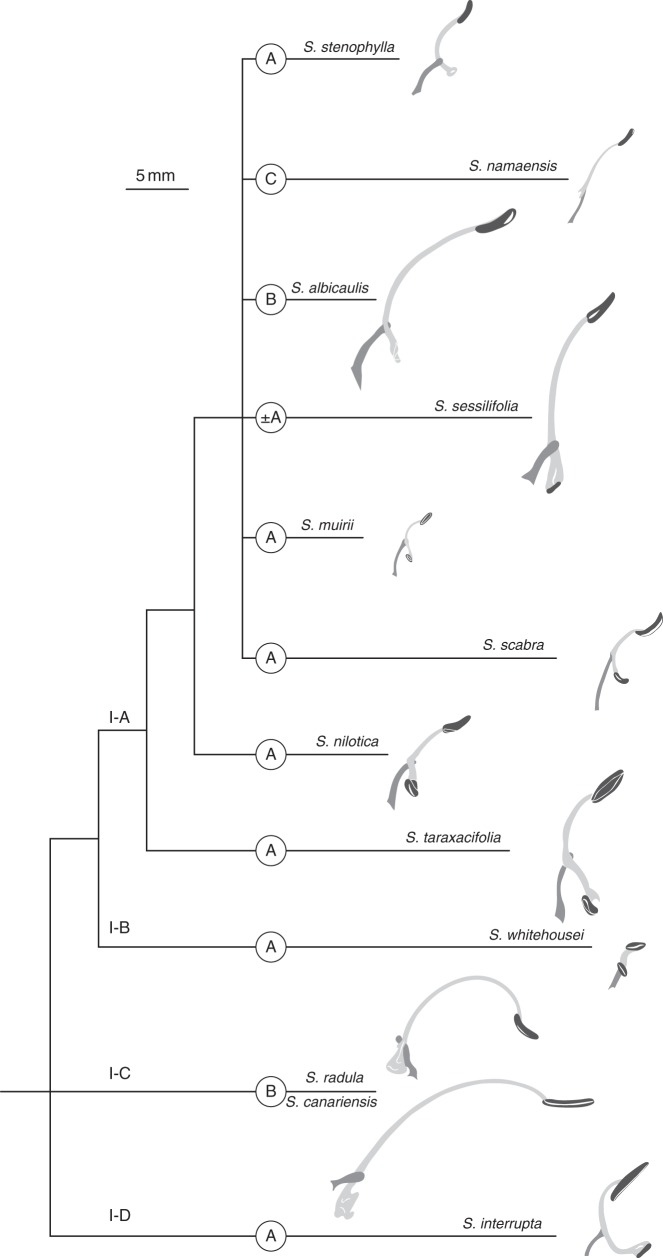
Stamen morphology was used by Walker and Sytsma (2007) to distinguish two major lineages within Clade I. However, our analysis clearly shows that stamen morphology is much more variable. Character state reconstruction revealed that stamen type A (Supplementary Data Fig. S5) is the ancestral state for Clade III-A and Salvia s.s. (Clade I). Consequently, the reduction of the lower lever arm evolved several times in parallel. In Salvia s.s. (Fig. 8), sub-clade I-A covers the whole range of stamen modifications described in African Salvia (Fig. 8; Hedge, 1974), including the rare stamen type C (S. namaensis). In the OW, this stamen modification is only known from the Eurasian S. verticillata group (four species), which is also part of Salvia s.s. (Clade I; Will, 2013), but not closely related to S. namaensis from SSA. A third species with stamen type C and the same ontogeny as S. verticillata is Rosmarinus officinalis (unpubl. res.). Parallel evolution is thus evident (Supplementary Data Fig. S5), restricting the use of stamen types to lower taxonomic levels.
Fig. 8.
Trends in the evolution of stamen types in African representatives of Salvia s.s. (Clade I). Proceeding from the ancestral stamen type A, the hypothetical stamen evolution is illustrated. Stamen of S. whitehousei modified after Whitehouse (1949), S. interrupta modified after Rosúa and Blanca (1986), schemata of all other stamens modified after Hedge (1974); filament (medium grey); connective (light grey); theca (dark grey); stamen types (in circles; ± A = reduced type A); and clades (I-A to I-D) represented by the species are given above the branches; scale bar = 5 mm.
Floristic links of Salvia spp. in North Africa
Salvia spp. distributed in North Africa are clearly members of two different clades (Fig. 7; I-A and III-A). Based on the molecular data, they show many floristic links to southern Europe, SW Asia, East and southern Africa (Fig. 5). One example is S. taraxacifolia, a relict species endemic to the High Atlas, Morocco (Hedge, 1974). It is most closely related to East (S. nilotica and S. somalensis) and southern African species (Fig. 7; I-A). Our data suggest dispersal from North to East Africa followed by a second dispersal to southern Africa. Salvia taraxacifolia and the two East African species are adapted to mesic habitats, whereas their southern African relatives prefer arid habitats (Fig. 9E–G). This indicates that the common ancestor of sub-clade I-A might have been adapted to mesic conditions and that within the large SSA radiation (sub-clade I-A), adaptation to arid localities evolved in southern Africa.
The floristic links between North Africa and southern Europe already proposed by Hedge (1974) (Fig. 7; sub-clade I-C) were confirmed by the close relationship of S. interrupta (SW Morocco; Fig. 10B: 56) and S. candelabrum (southern Spain; Fig. 10B). Both are thermophilic and partly overlap in their distribution (Fig. 10B; Rosúa and Blanca, 1986, 1990). They have a similar habit (divided leaves, most of them at the base of the stem), conspicuous, elongated inflorescences and the same chromosome number (2n = 14) (Hedge, 1974; Rosúa and Blanca, 1985, 1990). Salvia interrupta is considered as a Tertiary relict which was more widely distributed when the climate was more mesic (Rosúa and Blanca, 1990). Both species are obviously derived from a common, probably mesic-adapted, ancestor. Their relationship might reflect allopatric speciation probably triggered by different edaphic factors in the corresponding habitats.
The strongly supported sister relationship of S. daghestanica (Caucasus) and S. phlomoides subsp. phlomoides (North Africa and southern Europe; Fig. 10B: 47) reflects floristic links between the Mediterranean and SW Asia. Since contact between the African and Eurasia floras should have increased during the Messinian Salinity Crisis in the late Miocene [5·96–5·33 million years ago (Mya)], plant colonization across the Mediterranean is expected to have occurred often during this time frame (Caujapé-Castells and Jansen, 2003). We assume that this ‘route’ was also used repeatedly by Salvia. We thus support the hypothesis of Davis and Hedge (1971) that the SW Asian origin of some NW African species was triggered by a westward shift of Irano-Turanian elements.
Repeated colonization of the Canary Islands and long-distance dispersal in Salvia
The Macaronesian flora is composed of endemics derived from an ancient Tertiary relict flora and more recently introduced species (e.g. Helfgott et al., 2000; Manen et al., 2002; Carine et al., 2004). This general pattern also appears to hold for the Macaronesian S. canariensis, S. broussonetii and S. herbanica (Fig. 9B: 7–9). Since the three species clearly differ in their morphology (Fig. 4F–H) and habitat preference (Fig. 3D, G), they were never expected to be closely related (Hedge, 1974; Carine et al., 2004). However, S. canariensis and S. broussonetii are sister species forming one clade within sub-clade I-C (Figs 5 and 7). The proposed allies of S. canariensis (Hedge, 1974) are not closely related to these two species, thus contradicting Hedge's (1974) hypothesis of a link between the Canary Island and southern African Salvia.
In contrast, S. herbanica is found in a different clade (Fig. 7; III-A) and has clear links to species from East Africa and the Arabian Peninsula. Our findings indicate the non-monophyly of the Canary Island endemics and support the hypothesis of repeated dispersals to the archipelago from different mainland sources (Emerson et al., 2000; Arnedo et al., 2001; Fuertes-Aguilar et al., 2002; Carine et al., 2004, and references therein; Vargas, 2007).
The colonization of the Canary Islands raises the question of how dispersal might have taken place. The question of how (if at all) Salvia might be adapted to long-distance dispersal (LDD) is not yet answered. For the Canary Islands, the proximity to the African continent might have eased dispersal. The oldest islands, Lanzarote and Fuerteventura, are presently 100 km from the coast of North Africa (Francisco-Ortega et al., 2000; Acosta et al., 2005), but at some periods during the last 20 million years they were probably much closer (García-Talavera, 1997). García-Talavera (1997) suggested that the volcanic sea mounts served as ‘stepping stones’ when the sea level dropped during glacial periods. In addition, recent studies have assumed convective updraft to be the key mechanism for LDD of even heavy diaspores (Nathan et al., 2002; Tackenberg et al., 2003). Long-distance dispersal to the archipelago mediated by wind is conceivable for S. herbanica, S. aegyptiaca and their potential common ancestor. Salvia aegyptiaca is distributed within the area of the Saharan Air Layer, a westward-directed wind of comparably high velocity (Carlson and Prospero, 1972; Tackenberg et al., 2003)
East Africa and the Arabian Peninsula as ‘melting pots’ for Salvia
East African Salvia is found in three independent clades (Fig. 7; III-A, I-C and I-A) and consequently has various floristic links beyond the continent.
Salvia merjamie (sub-clade I-C; Fig. 9D: 16) is a frequent and extremely variable species in the montane forest belts from Ethiopia to Zimbabwe (Hedge, 1974). It is part of a strongly supported (ITS) clade with two widely distributed species from SW Asia. Salvia merjamie is moderately supported (ML) as the sister to S. verbenaca 141 from Turkey. Both are polymorphic, occasionally have cleistogamous flowers and share the chromosome number of 2n = 42, which is uncommon in Salvia (Reese, 1957; Gadella et al., 1966; Hedge, 1974; Hedberg and Hedberg, 1977; Haque and Ghoshal, 1980; Codd, 1985; Vogt and Aparicio, 1999; Foley et al., 2008).
Salvia nilotica (sub-clade I-A; Fig. 9D: 17) has a broader distribution range than S. merjamie. Hedge (1974) assumed that S. nilotica was a distinct, taxonomically isolated species but also discussed its similarities with species restricted to the eastern Cape. With S. taraxacifolia and S. somalensis, S. nilotica is found in a basal position of sub-clade I-A, suggesting a dispersal from North to southern Africa via East Africa. The relationship of S. nilotica and the African members of section Heterosphace Benth. proposed by Hedge (1974) is confirmed, since all of these species are placed in the same sub-clade (I-A).
Salvia deserti (sub-clade III-A; Fig. 9D: 14) is an endemic of the Egyptian and Arabian deserts (Boulos, 2008; Hedge, 1974; Migahid, 1978). It is morphologically and genetically distinct from Salvia s.s. Incongruences detected in nuclear and plastid data support a hybrid origin of this species.
Pollinator diversity and evolution of bird pollination in Sub-Saharan Africa
Based on flower morphology, bees are seen as the most important pollinators in Salvia overall (Wester and Claßen-Bockhoff, 2006, 2011). This was also assumed for the African and in particular for the southern African species (Hedge, 1974). However, only a limited number of field observations confirm this view, e.g. the first report of small bees on S. africana-caerulea (Marloth, 1908), data on Anthophora diversipes (Goldblatt et al., 2000a, b), and observations of Xylocopa caffra, Amegilla spp., further solitary bees and honey-bees by P. Wester, R. Claßen-Bockhoff and H. Technau (pers. comm.).
Flowers with long tubes and freely accessible pollen were expected to be pollinated by long-tongued insects, e.g. flies. This pollinator guild is characteristic for South Africa, especially in the western Cape Region (Goldblatt and Manning, 2000). Potgieter and Edwards (2001, 2005) assumed that S. scabra and S. repens are pollinated by long-tongued flies (Stenobasipteron wiedemanni and Prosoeca spp.), but field observations are still lacking.
Although bird pollination is quite frequent in NW Salvia (Wester and Claßen-Bockhoff, 2007), the only known ornithophilous Salvia spp. in the OW appear in southern Africa (Wester and Claßen-Bockhoff, 2006). Three, S. lanceolata, S. thermarum and S. africana-lutea, are restricted to southern Africa (Fig. 4I, R, S). Probably two more (S. leucodermis and S. sessilifolia; Fig. 4P, Q) occur in Madagascar. Our data confirm that ornithophily evolved repeatedly in the NW (Fig. 7), but also indicate at least two pollinator shifts from bee to bird pollination in Africa (Supplementary Data Fig. S6). Most probably, this pollination system evolved three times in parallel within sub-clade I-A: (1) in the Madagascan sub-clade; (2) in the S. africana-lutea and S. lanceolata clade; and (3) in S. thermarum. Including species adapted to bee, bird and, most probably, long-tongued fly pollination, sub-clade I-A represents a further example of a monophyletic lineage having undergone pollinator-driven diversification in southern African (e.g. Van der Niet and Johnson, 2012; Sun et al., 2014; Van der Niet et al., 2014).
In bird-pollinated species, bees are largely excluded from nectar access but not from collecting pollen. They might therefore trigger hybridization between bird- and bee-pollinated species (Van Jaarsveld, 2002; P. Wester, University of Düsseldorf, Germany, pers. comm.), e.g. in S. africana-caerulea. The species is morphologically well adapted to bee pollination but is also occasionally pollinated by birds (Wester, 2013). It might be a species in which the exploitation of a food plant by pollinators (birds) can be observed even though both species are not yet perfectly adapted to each other (Thomson and Wilson, 2008). Thus, S. africana-caerulea might represent an example of a pollinator shift in progress (Rodríguez-Gironés and Santamaría, 2004).
Conclusions
African Salvia is non-monophyletic. Continental Africa, the Canary Islands, East Africa and the Arabian Peninsula were each colonized repeatedly. The morphological diversity of African sage results from independent dispersals from different mainland sources and diversification in the diverse African environment. Parallel evolution played an important role for the evolution of stamen types, calyx enlargement and pollination syndrome. These data can even be transferred to Salvia s.l. for which similar trends have been observed.
SUPPLEMENTARY DATA
ACKNOWLEDGEMENTS
We thank Dirk Albach (Oldenburg), Safi Bagherpour (Ankara), Benny Bytebier (KwaZulu-Natal), Ferhat Celep (Nevşehir), Ahmet El-Banhawy (Ismailia/Redding), Ahmed Kahraman (Ankara), Alexander P. Sukhorukov (Moscow), Mats Thulin (Uppsala) and Petra Wester (Düsseldorf) for sampling, and the following herbaria for offering plant material: ACECR (Iran), B, E, EA, GOET, HUH, M, MJG, MO, MPU and MW. Photographs were kindly provided by Ferhat Celep, Rafael N. B. Groneberg (Mainz), Dylan Hannon (San Marino, CA), Peter B. Phillipson (Paris), Hen Technau, Mats Thulin and Petra Wester. We thank Berit Gehrke and Michael D. Pirie (both Mainz) for assistance with data analyses, and Natalie Schmalz, Abigail J. Moore (Providence, RI) and two anonymous reviewers for helpful comments to improve the manuscript. This work was supported by the DFG (Deutsche Forschungsgemeinschaft; Cl 81/10–1) and the Fachbereich Biologie (Universität Mainz).
APPENDIX: PLANT MATERIAL INCLUDED IN THIS STUDY
For all sequences generated by the first author, a DNA accession number is given
| Taxon | Locality | Voucher collector with collection no. (herbarium) | DNA acc. no. | GenBank accession no. |
||
|---|---|---|---|---|---|---|
| rpl32-trnL | nrITS | ETS | ||||
| Clinopodium dalmaticum (Benth.) Bräuchler & Heubl | M. Kintgen s.n. | – | JQ669340 | – | – | |
| Clinopodium taxifolium (Kunth) Govaerts | B. Drew 228 | – | JQ669288 | – | – | |
| Clinopodium vulgare L. | Riina 1579 | – | JQ669290 | – | – | |
| Collinsonia canadensis L. | JBW 958 | – | – | DQ667248 | – | |
| Raiche s.n. UCBG 1984.0696 | – | JQ669291 | – | JQ669157 | ||
| Dorystaechas hastata Boiss. & Heldr. ex Benth. | Anatolia | Albach D6–4 (OLD) | 213 | KJ747319 | KJ584248 | KJ584275 |
| Cult. RBGE1972–0177D | – | JQ669302 | DQ667252 | – | ||
| – | – | HQ418845 | – | |||
| Glechoma hederacea L. | B. Drew 69 | – | JQ669307 | – | – | |
| Horminum pyrenaicum L. | Cult. RBGE 1997–2109a | – | JQ669315 | DQ667257 | – | |
| M. Will 63 (MJG 003069) | 390 | KJ747327 | KJ584247 | KJ584279 | ||
| Hyptis laniflora Benth. | B. Drew 41 | – | JQ669317 | – | – | |
| Isolate K36721 | – | – | JF301548 | JF404259 | ||
| Lepechinia bella Epling | Bolivia | P. Wester 145 (MJG 009474) | 216 | KJ747326 | – | – |
| Lepechinia calycina (Benth.) Epling ex Munz | Drew 197 | – | JQ669324 | – | – | |
| Lepechinia conferta Epling | Alonso 8376 (F) | – | – | DQ667307 | – | |
| Lepechinia lamiifolia (Benth.) Epling | B. Drew 178 | – | JQ669325 | – | – | |
| Lepechinia lancifolia (Rusby) Epling | Smith 444 (F) | – | – | DQ667306 | – | |
| Lepechinia mexicana (S.Schauer) Epling | B. Drew 164 | – | JQ669326 | – | – | |
| Melissa axillaris (Benth.) Bakh.f. | D. E. Boufford et al. 24526 | – | JQ669334 | – | – | |
| Melissa officinalis L. | M. Will 64 (MJG 003068) | 388 | KJ747325 | KJ584249 | KJ584285 | |
| JBW 2575 (cult. USA/WIS) | – | – | DQ667291 | – | ||
| B. Drew 70 | – | JQ669335 | JF301353 | – | ||
| Mentha arvensis L. | B. Drew 82 | – | JQ669336 | – | – | |
| Mentha pulegium L. | Riina 1574 | – | JQ669338 | – | – | |
| Mentha spicata L. | J. Walker 2566 | – | JQ669339 | – | – | |
| Mentha spicata subsp. condensata (Briq.) Greuter & Burdet | Riina 1575 | – | JQ669337 | – | – | |
| Meriandra bengalensis (Konig ex Roxb.) Benth. | Lavranus & Newton 15796 (MO 2633828) | – | – | DQ667329 | – | |
| Perilla frutescens (L.) Britton | JBW 1078 (cult. USA/WIS) | – | – | DQ667246 | JF301326 | |
| Perovskia atriplicifolia Benth. | M. Will 65 (MJG 003070) | 472 | – | KJ584242 | – | |
| JBW 2524 (cult. USA/WIS) | – | JQ669352 | DQ667223 | JF301328 | ||
| Perovskia scrophulariifolia Bunge | Kinziraeva 6751 (MO 5201778) | – | – | DQ667330 | – | |
| Rosmarinus officinalis L. | M. Will 66 (MJG 003071) | 389 | KJ747310 | KJ584197 | KJ584296 | |
| JBW 2558 (cult. USA/WIS) | – | JQ669364 | DQ667241 | – | ||
| Salvia aegyptiaca L. | M. Kuschewitz s.n. (cult. BG HH) | 204 | KJ747314 | KJ584245 | KJ584254 | |
| McLeish 3728 (E) | – | – | DQ667285 | – | ||
| S. aethiopis L. | Armenia | J. Hellwig s.n. 26/6/02 (MJG 009919) | 281 | – | KJ584163 | – |
| Armenia | J. Hellwig s.n. 26/6/02 (MJG 009919) | – | – | DQ667272 | – | |
| S. africana-caerulea L. | S Africa | P. Wester & R. Claßen-Bockhoff 319 (MJG 041401) | 230 | KJ747271 | KJ584204 | KJ584255 |
| S Africa | P. Wester & R. Claßen-Bockhoff 317 (MJG 041402) | 229 | – | KJ584203 | – | |
| S. africana-lutea L. | S Africa | P. Wester 342 (MJG 041393) | 81 | KJ747273 | KJ584205 | KJ584256 |
| S Africa | P. Wester 708 (MJG) | 234 | KJ747259 | – | – | |
| S Africa | P. Wester 708 (MJG) | 430 | KJ747272 | – | – | |
| S. albicaulis Benth. | S Africa | P. Wester, R. Claßen-Bockhoff & E. v. Jaarsveld 340 (MJG 041403) | 1 | KJ747274 | KJ584206 | KJ584257 |
| S. albicaulis × granitica | S Africa | P. Wester, R. Claßen-Bockhoff & E. v. Jaarsveld 341 (MJG 041404) | 2 | KJ747275 | KJ584207 | KJ584258 |
| S Africa | P. Wester, R. Claßen-Bockhoff & E. v. Jaarsveld 341 (MJG 041404) | 42 | – | KJ584215 | KJ584277 | |
| S. apiana Jeps. | California | P. Wester 411 (MJG 041452) | 392 | KJ747321 | – | – |
| JBW 2509 USA (WIS) | – | – | DQ667214 | – | ||
| S. areysiana Deflers | Yemen | Thulin, Eriksson, Gifri & Långström 8472 (UPS) | 282 | KJ747315 | – | KJ584259 |
| S. argentea L. | Italy | R. Claßen-Bockhoff s.n. Mai 2002 (MJG) | 57 | KJ747299 | KJ584164 | – |
| S. aristata Aucher | Iran | K.H. Rechinger s.n. 1974 (M) | 289 | – | KJ584244 | – |
| Iran | Y. Ajani 1569 (ACECR) | 417 | KJ747264 | – | – | |
| Wedelbo & Assadi s.n. (E) | – | JQ669365 | DQ667280 | JF301336 | ||
| S. aucheri var. canescens Boiss. & Heldr. | Anatolia | F. Celep 1245 (PSL METU) | 239 | – | KJ584193 | – |
| Archibald 7670 (E) | – | – | DQ667286 | – | ||
| S. aurita L.f. | P. Wester & R. Claßen-Bockhoff 324 (MJG 041405) | 11/424 | KJ747276 | KJ584218 | KJ584261 | |
| M. Will 26 (MJG 041563) | 423 | – | KJ584219 | KJ584260 | ||
| S. aurita var. galpinii (Skan) Hedge | P. Wester 472 (MJG) | 425 | KJ747269 | – | – | |
| S. austriaca Jacq. | R. Claßen-Bockhoff s.n. 2004 (cult. BG Mz) | – | – | DQ667323 | – | |
| Austria | R. Claßen-Bockhoff s.n. 12.06.2003 (MJG) | 280 | KJ747261 | – | – | |
| S. bariensis Thulin | Somalia | M. Thulin, A. Dahir & A. Osman 9429 (UPS) | 283 | KJ747316 | – | KJ584262 |
| S. broussonetii Benth. | M. Will 33 (MJG 041537) | 29 | KJ747293 | KJ584226 | KJ584263 | |
| Tenerife | R. Claßen-Bockhoff 2/10 (MJG 009887) | 463 | – | KJ584225 | KJ584264 | |
| S. cabulica Benth. | Afghanistan | H. Freitag 4683 (MSB 137713) | 322 | – | KJ584189 | – |
| Ghafoor & Goodman 5148 (E) | – | – | DQ667287 | – | ||
| S. canariensis L. | M. Will 46 (MJG 041565) | 5 | KJ747295 | KJ584227 | KJ584266 | |
| Tenerife | R. Claßen-Bockhoff 3/10 (MJG) | 464 | – | – | KJ584265 | |
| Tenerife | R. Claßen-Bockhoff 1/03 (MJG) | 223 | KJ747294 | – | – | |
| Cult. RBGE 1986–0478 | – | – | DQ667256 | – | ||
| S. candelabrum Boiss. | M. Will 42 (MJG 041557) | 62 | KJ747255 | KJ584190 | – | |
| S. candidissima subsp. occidentalis Hedge | Anatolia | F. Celep 1487 (PSL METU) | 201 | KJ747300 | KJ584165 | – |
| S. candidissima Vahl. | Cult. RBGE 1999–2202A | – | – | DQ667261 | – | |
| S. chamelaeagnea Berg. | P. Wester & R. Claßen-Bockhoff 314 (MJG 041407) | 52 | KJ747289 | KJ584210 | KJ584268 | |
| S Africa | P. Wester, R. Claßen-Bockhoff & E. Van Jaarsveld 313 (MJG 041406) | 432 | – | KJ584211 | KJ584267 | |
| M. Will 47 (MJG 041541) | 433 | – | KJ584212 | – | ||
| S. chienii E. Peter | M. Will 61 (MJG 003066) | 53 | KJ747322 | KJ584250 | – | |
| AnH0305–21 | – | – | DQ132868 | – | ||
| S. chionopeplica Epling | JBW 2545 (cult. USA/WIS) | – | – | DQ667227 | – | |
| S. cf. chionopeplica Epling | P. Wester 485 (MJG 041435) | 174 | KJ747318 | KJ584188 | KJ584269 | |
| S. daghestanica Sosn. | M. Will 34 (MJG 041551) | 276 | KJ747308 | KJ584187 | – | |
| Cult. RGB E 1988–2283A | – | – | DQ667258 | – | ||
| S. deserta Schang | M. Will 96 (MJG 003100) | 286 | KJ747263 | KJ584176 | – | |
| XingJ0305–1 | – | – | DQ132865 | – | ||
| S. deserti Dcne. | Egypt | E. Gamal Eldin s.n. 3.5.1991 (GOET) | 335 | KJ747312 | – | KJ584270 |
| S. disermas L. | P. Wester & R. Claßen-Bockhoff 326 (MJG 041413) | 15 | KJ747296 | KJ584179 | KJ584271 | |
| M. Will 80 (MJG 003116) | 454 | – | – | KJ584272 | ||
| S. disermas L. (syn. S. rugosa in GenBank) | Goldblatt 7500 (E) | – | – | DQ667290 | – | |
| S. dolomitica Codd | P. Wester & R. Claßen-Bockhoff 321 (MJG 041411) | 82 | KJ747290 | KJ584214 | KJ584274 | |
| F. Brusse 5610 (M) | 440 | – | KJ584213 | KJ584273 | ||
| JBW 3200 (cult. USA/WIS) | – | – | DQ667322 | – | ||
| S. dominica L. | Cyprus | A. Seregin, D. Sokoloff & M. Remizova A-211 (MW) | 267 | – | KJ584167 | – |
| M. Kuschewitz s.n. (MJG 009323) | 217 | KJ747262 | KJ584166 | – | ||
| S. evansiana Hand.-Mazz. var. evansiana | M. Will 55 (MJG 003060) | 415 | KJ747323 | KJ584251 | – | |
| S. freyniana Bornm. | Anatolia | S. Bagherpour 493 (PSL METU) | 98 | KJ747266 | – | – |
| S. fruticosa Miller | Anatolia | F. Celep 1373 (PSL METU) | 100 | KJ747256 | KJ584195 | – |
| G. Hausner GR 31 (MJG 003078) | 66 | – | KJ584194 | – | ||
| S. funerea Jones | P. Wester 490 (MJG 041430) | 393 | KJ747320 | – | – | |
| S. garipensis E.Meyer ex Benth. | Strohbach 149 (E) | – | – | DQ667281 | – | |
| S. geminata Thulin | Yemen | M. Thulin, Beier & M. Hussein 9629 (UPS) | 341 | – | – | KJ584276 |
| S. glutinosa L. | Anatolia | F. Celep 1196 (PSL METU) | 101 | – | KJ584253 | – |
| JBW 2568 (cult. USA/WIS) | – | – | DQ667250 | – | ||
| S. graciliramulosa Epling & Játiva | Bolivia | P. Wester 14 (MJG 041090) | – | – | DQ667276 | – |
| S. gretai Brandegee | USA | JBW 2511 (WIS) | – | JQ669367 | DQ667215 | JF301331 |
| S. henryi Gray | USA | JBW 2516 (WIS) | – | – | DQ667216 | – |
| S. herbanica A.Santos & M.Fernández | Fuerteventura | R. Claßen-Bockhoff 1/05 (MJG 009888) | 40 | KJ747313 | KJ584246 | KJ584278 |
| S. heterochroa Fern. | Yunnan, China | D. E. Boufford, J. H. Chen, S. L. Kelley, R. H. Ree, H. Sun, B. Xü, J. P. Yue, L. L. Yue, D. C. Zhang & W. D. Zhu 35205 (HUH 286716) | 252 | KJ747324 | KJ584252 | – |
| S. hydrangea Benth. | Anatolia | A. Kahraman 1468 (PSL METU) | 242 | KJ747257 | KJ584192 | – |
| Rechinger 47123 (E) | – | – | DQ667288 | – | ||
| S. hydrangea Benth. (syn. S. dracocepha-loides Boiss. in GenBank) | Armenia | Hellwig s.n. (MJG 009920) | – | – | DQ667265 | – |
| S. interrupta Schousb. | M. Will 30 (MJG 041550) | 447 | KJ747265 | KJ584191 | – | |
| S. judaica Boiss. | M. Will 57 (MJG 003061) | 409 | – | KJ584241 | – | |
| S. lanceolata Lam. | P. Wester 316 (MJG 041396) | 58 | KJ747278 | KJ584202 | KJ584281 | |
| S Africa | P. Wester 1129 (NBG) | 264 | KJ747277 | KJ584201 | KJ584280 | |
| S. lanceolata × africana-caerulea | P. Wester & R. Claßen-Bockhoff 315 (MJG 041400) | 236 | KJ747279 | KJ584216 | KJ584282 | |
| S. lanigera Poir. | Sinai | A. El-Banhawy 11 (University of Ismailia, Egypt) | 198 | – | KJ584185 | – |
| S. leucodermis Baker | Madagascar | R. A. Clement, P. B. Phillipson & G. Rafamantanantsoa 2137 (E 00161484) | 348 | KJ747280 | KJ584220 | KJ584284 |
| Madagascar | P. Wester 1131 (TAN) | 459 | – | KJ584221 | – | |
| Madagascar | B. Bytebier 3193 (TAN) | 460 | – | KJ584222 | KJ584283 | |
| S. merjamie Forsk. | M. Will 83 (MJG 003113) | 265 | KJ747297 | KJ584184 | KJ584286 | |
| S. microstegia Boiss. & Bal. | Armenia | J. Hellwig s.n. (MJG 009884) | 212 | KJ747307 | KJ584171 | – |
| S. microstegia Boiss. & Bal. (S. verbascifolia M.Bieb.in GenBank) | Armenia | J. Hellwig s.n. (MJG 009884) | – | – | DQ667264 | – |
| S. muirii L.Bolus | P. Wester & R. Claßen-Bockhoff 328 (MJG 041409) | 162 | KJ747283 | KJ584208 | KJ584287 | |
| P. Wester & R. Claßen-Bockhoff 318 (MJG 041410) | 163 | – | KJ584209 | KJ584288 | ||
| S. namaensis Schinz | M. Will 28 (MJG 041552) | 435 | KJ747284 | KJ584217 | KJ584290 | |
| P. Wester & R. Claßen-Bockhoff 330 (MJG 041415) | 78 | KJ747281 | KJ584234 | KJ584291 | ||
| SW Africa | W. Giess & M. Müller 14319 (M) | 296 | – | KJ584200 | KJ584289 | |
| S. nilotica Juss. ex Jacq. | U. Hecker g3186 (MJG 003079) | 64 | – | KJ584230 | – | |
| M. Will 49 (MJG 041538) | 436 | KJ747258 | KJ584229 | KJ584292 | ||
| S. officinalis L. | JBW 2580 (cult. USA/WIS) | – | – | DQ667225 | – | |
| Voucher 2160 | – | JQ771324 | – | – | ||
| M. Palma s.n. UCBG 7·0083 | – | JQ669369 | JF301355 | JF301332 | ||
| N Africa | A. El-Banhawy 16 (University of Ismailia, Egypt) | 243 | – | KJ584196 | – | |
| S. palaestina Benth. | Anatolia | F. Celep 1083 (PSL METU) | 400 | KJ747304 | KJ584175 | – |
| Anatolia | A. Kahraman 1443 (PSL METU) | 125 | KJ747301 | – | – | |
| S. cf. palaestina Benth. | N Africa | A. El-Banhawy 6 (University of Ismailia, Egypt) | 200 | KJ747302 | KJ584172 | – |
| S. patens Cav. | Cult RBGE 1973–9197 | – | JQ669370 | DQ667253 | JF301333 | |
| S. penstemonoides Kunth & Bouché | JBW 2578 (cult. USA/WIS) | – | – | DQ667221 | – | |
| S. phlomoides ssp. phlomoides Asso | Morocco | R. Vogt 10336 & Ch. Oberprieler 4784 (B 100145114) | 337 | KJ747309 | KJ584186 | – |
| S. polystachya Ort. | Breedlove & Mahoney 72286 (UC) cult. UCBG 92·052 | – | JQ669371 | – | JF301334 | |
| S. pratensis L. | C Russia | A. Suchorukow s.n. VII 2003 (MW) | 275 | KJ747291 | KJ584178 | – |
| Isolate S0628 | – | – | EU169486 | – | ||
| S. przewalskii Maxim. | Cult. RBGE 1993–2067A | – | JQ669372 | DQ667254 | JF301339 | |
| S. radula Benth. | S Africa | Germishuizen 3950 (MO 4385830) | 328 | – | KJ584180 | KJ584293 |
| S. repens Burch. ex Benth. | M. Will 50 (MJG 041931) | 61 | KJ747282 | KJ584232 | KJ584295 | |
| S. cf. repens Burch. ex Benth. | P. Wester & R. Claßen-Bockhoff 325 (MJG 041412) | 437 | – | KJ584231 | KJ584294 | |
| S. roemeriana Scheele | USA | JBW 2515 (WIS) | – | – | DQ667211 | – |
| S. scabra L. | M. Will 37 (MJG 041549) | 55 | KJ747285 | KJ584233 | KJ584297 | |
| S. schimperi Benth. | Yemen | D. Podlech 36057 (M 55003) | 310 | – | KJ584174 | KJ584298 |
| M. Will 72 (MJG 003099) | 466 | – | KJ584168 | – | ||
| S. schlechteri Briq. | P. Wester & R. Claßen-Bockhoff 323a (MJG 041416) | 31 | KJ747286 | KJ584235 | KJ584299 | |
| M. Will 51 (MJG 041539) | 438 | – | KJ584236 | KJ584300 | ||
| S. sclarea L. | Anatolia | F. Celep 1492 (PSL METU) | 244 | KJ747305 | KJ584162 | KJ584301 |
| JBW 2527 (cult. USA/WIS) | – | JQ669373 | DQ667222 | – | ||
| S. sessilifolia Baker | Madagascar | R. A. Clement, P. B. Phillipson & G. Rafamantanantsoa 2001 (MO 4328854) | 331 | – | KJ584223 | KJ584302 |
| Madagascar | C. H. Jongkind & S. Rapanarivo 929 (MO 4870099) | 332 | – | KJ584224 | KJ584303 | |
| Madagascar | Jongkind & Rapanarivo 929 (E) | – | – | DQ667282 | – | |
| S. somalensis Vatke | M. Will 77 (MJG 003119) | 340 | KJ747311 | KJ584240 | KJ584304 | |
| S. spinosa L. | N Africa | A. El-Banhawy 14 (University of Ismailia, Egypt) | 199 | KJ747303 | KJ584173 | – |
| S. stenophylla Burch. ex Benth. | S Africa | Burgoyne & Snow 4805 (MO 5649981) | 330 | KJ747260 | KJ584237 | KJ584305 |
| Giess & Hübsch 11607 (M) | 439 | – | KJ584238 | KJ584306 | ||
| S. summa A. Nelson | Texas | JBW 1972 USA (WIS) | – | – | DQ667217 | – |
| Texas | P. Wester 373 (MJG 041338) | 190 | – | – | KJ584307 | |
| S. sylvestris L. | Anatolia | A. Kahraman 1568 (PSL METU) | 118 | KJ747292 | KJ584181 | – |
| (syn. S. tesquicola Kiok. & Pobed.) | C Russia | A. Suchorukow s.n. VIII 1994 (MW) | 273 | – | KJ584177 | – |
| Isolate S0626 | – | – | EU169485 | – | ||
| S. taraxacifolia Hook.f. | Morocco | W. Lippert 25355 (M) | 304 | KJ747270 | KJ584228 | KJ584308 |
| JBW 2521 (cult. USA/WIS) | – | – | DQ667209 | – | ||
| S. texana (Scheele) Torr. | Texas | P. Wester 362 (MJG 041477) | 191 | KJ747267 | KJ584199 | – |
| Texas | P. Wester 362 (MJG 041477) | – | – | DQ667321 | – | |
| S. thermarum Van Jaarsv. | P. Wester & R. Claßen-Bockhoff 336 (MJG 041398) | 23 | KJ747288 | KJ584239 | KJ584309 | |
| M. Will 52 (MJG 041933) | 452 | – | – | KJ584310 | ||
| S Africa | P. Wester 312 (MJG 041397) | 221 | KJ747287 | – | – | |
| S. tingitana Etling | Morocco | D. Podlech 43384 (M 54979) | 314 | – | KJ584169 | – |
| S. trichocalycina Benth. | Breckle 4963 (E) | – | – | DQ667283 | – | |
| S. trijuga Diels | YunN0309–5 | – | – | DQ132870 | – | |
| S. veneris Hedge | Cyprus | M. J. Y. Foley 1701 (E 00147797) | 413 | KJ747306 | KJ584170 | – |
| S. verbenaca L. | Syria | W. Licht SYR 307 (MJG 003082) | 67 | – | KJ584182 | – |
| Anatolia | F. Celep 1408 (PSL METU) | 141 | KJ747298 | KJ584183 | – | |
| S. whitehousei Alziar | Texas | P. Wester 352 (MJG 041389) | 231 | KJ747268 | KJ584198 | KJ584311 |
| Texas | P. Wester 352 (MJG 041389) | – | – | DQ667320 | – | |
| Thymus pulegioides L. | Riina 1577 | – | JQ669378 | – | – | |
| Thymus serpyllum L. | J. Walker 2564 (cult. USA/WIS) | – | JQ669379 | – | – | |
| Zhumeria majdae Rech.f. & Wendelbo | Iran | F. Sharififar 1651 (ACECR) | 422 | KJ747317 | KJ584243 | KJ584312 |
| Wendelbo 15793 (V 21730) | – | – | DQ667336 | – | ||
| Ghazi s.n. (V 01176) | – | – | DQ667335 | – | ||
acc. no., accession number; locality: origin from natural habitats confirmed; ACECR, Iranian Academic Center for Education Culture and Research; BG HH, Botanical Garden Hamburg (Germany); cult., cultivated; PSL METU, Plant Systematics Lab. Department of Biological Sciences, Middle East Technical University, Ankara (Turkey); RBGE, Royal Botanical Garden Edinburgh (UK); herbarium acronyms according to Index Herbariorum.
LITERATURE CITED
- Acosta J, Uchupi E, Muñoz A, Herranz P, Palomo C, Ballesteros M. Geologic evolution of the Canarian Islands of Lanzarote, Fuerteventura, Gran Canaria and La Gomera, and comparison of landslides at these islands with those at Tenerife, La Palma and El Hierro. In: Clift P, Acosta J, editors. Geophysics of the Canary Islands. Dordrecht, The Netherlands: Springer; 2005. pp. 1–40. [Google Scholar]
- Alziar G. Catalogue synonymique des Salvia L. du monde (Lamiaceae). I à VI. Biocosme Mésogéen, Nice. 1988–1993;10:33–117. 5: 87–136; 6: 79–115; 6: 163–204; 7: 59–109; 9: 413–497. [Google Scholar]
- Arnedo MA, Oromí P, Ribera C. Radiation of the spider genus Dysdera (Araneae, Dysderidae) in the Canary Islands: cladistic assessment based on multiple data sets. Cladistics. 2001;17:313–353. [Google Scholar]
- Baldwin BG, Markos S. Phylogenetic utility of the external transcribed spacer (ETS) of 18S–26S rDNA: congruence of ETS and ITS trees of Calycadenia (Compositae) Molecular Phylogenetics and Evolution. 1998;10:449–463. doi: 10.1006/mpev.1998.0545. [DOI] [PubMed] [Google Scholar]
- Beardsley PM, Olmstead RG. Redefining Phrymaceae: the placement of Mimulus, tribe Mimuleae, and Phryma. American Journal of Botany. 2002;89:1093–1102. doi: 10.3732/ajb.89.7.1093. [DOI] [PubMed] [Google Scholar]
- Bellstedt DU, van Zyl L, Marais EM, et al. Phylogenetic relationships, character evolution and biogeography of southern African members of Zygophyllum (Zygophyllaceae) based on three plastid regions. Molecular Phylogenetics and Evolution. 2008;47:932–949. doi: 10.1016/j.ympev.2008.02.019. [DOI] [PubMed] [Google Scholar]
- Bentham G. Salvia. In: Bentham G, editor. Labiatarum genera et species: or, a description of the genera and species of plants of the order Labiatae with their general history, characters, affinities, and geographical distribution. London: J. Ridgway and Sons; 1832–1836. pp. 190–312. [Google Scholar]
- Bentham G. Labiatae. In: De Candolle A, editor. Prodromus systematis naturalis regni vegetabilis 12. Paris: Treuttel and Würtz; 1848. pp. 262–358. [Google Scholar]
- Bentham G. Labiatae. In: Bentham G, Hooker JD, editors. Genera plantarum 2. London: Reeve and Co.; 1876. pp. 1160–1196. [Google Scholar]
- Bokhari MH, Hedge IC. Anatomical observations on a desert group of Salvia species. Notes from the Royal Botanical Garden, Edinburgh. 1977;35:377–389. [Google Scholar]
- Boulos L. Flora and vegetation of the deserts of Egypt. Flora Mediterranea. 2008;18:341–359. [Google Scholar]
- Briquet J. Labiatae. In: Engler A, Prantl K, editors. Die natürlichen Pflanzenfamilien nebst ihrer Gattungen und wichtigen Arten. Leipzig: Engelmann; 1897. pp. 183–380. [Google Scholar]
- Carine MA, Russell SJ, Santos-Guerra A, Francisco-Ortega J. Relationships of the Macaronesian and Mediterranean floras: molecular evidence for multiple colonizations into Macaronesia and back-colonization of the continent in Convolvulus (Convolvulaceae) American Journal of Botany. 2004;91:1070–1085. doi: 10.3732/ajb.91.7.1070. [DOI] [PubMed] [Google Scholar]
- Carlson TN, Prospero JM. The large-scale movement of Saharan Air outbreaks over the Northern Equatorial Atlantic. Journal of Applied Meteorology. 1972;11:283–297. [Google Scholar]
- Caujapé-Castells J, Jansen RK. The influence of the Miocene Mediterranean desiccation on the geographical expansion and genetic variation of Androcymbium gramineum (Cav.) McBride (Colchicaceae) Molecular Ecology. 2003;12:1515–1525. doi: 10.1046/j.1365-294x.2003.01839.x. [DOI] [PubMed] [Google Scholar]
- Caujapé-Castells J, Jansen RK, Membrives N, Pedrola-Monfort J, Montserrat JM, Ardanuy A. Historical biogeography of Androcymbium Willd. (Colchicaceae) in Africa: evidence from cpDNA RFLPs. Botanical Journal of the Linnean Society. 2001;136:379–392. [Google Scholar]
- Clebsch B. The new book of salvias: sages for every garden. Cambridge: Timber Press; 2008. [Google Scholar]
- Codd LE. Salvia. In: Leistner OA, editor. Flora of Southern Africa. Lamiaceae. Pretoria: Botanical Research Institute, Department of Agriculture and Water Supply; 1985. pp. 79–101. [Google Scholar]
- Coleman M, Liston A, Kadereit JW, Abbott RJ. Repeat intercontinental dispersal and Pleistocene speciation in disjunct Mediterranean and desert Senecio (Asteraceae) American Journal of Botany. 2003;90:1446–1454. doi: 10.3732/ajb.90.10.1446. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods. 2012;9:772–772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis PH, Hedge IC. Floristic links between NW Africa and SW Asia. Annalen des Naturhistorischen Museums in Wien. 1971;75:43–57. [Google Scholar]
- Del Hoyo A, García-Marín JL, Pedrola-Monfort J. Temporal and spatial diversification of the African disjunct genus Androcymbium (Colchicaceae) Molecular Phylogenetics and Evolution. 2009;53:848–861. doi: 10.1016/j.ympev.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Emerson BC, Oromí P, Hewitt GM. Interpreting colonisation of the Calathus (Coleoptera: Carabidae) on the Canary Islands and Madeira through the application of the parametric bootstrap. Evolution. 2000;54:2081–2090. doi: 10.1111/j.0014-3820.2000.tb01251.x. [DOI] [PubMed] [Google Scholar]
- Foley MJY, Hedge IC, Möller M. The enigmatic Salvia tingitana (Lamiaceae): a case study in history, taxonomy and cytology. Willdenowia – Annals of the Botanic Garden and Botanical Museum Berlin-Dahlem. 2008;38:41–59. [Google Scholar]
- Francisco-Ortega J, Santos-Guerra A, Kim S-C, Crawford DJ. Plant genetic diversity in the Canary Islands: a conservation perspective. American Journal of Botany. 2000;87:909–919. [PubMed] [Google Scholar]
- Froissart C. La connaissance des sauges. Aix-en-Provence, France: Edisud; 2008. [Google Scholar]
- Fuertes-Aguilar J, Ray MF, Francisco-Ortega J, Santos-Guerra A, Jansen RK. Molecular evidence from chloroplast and nuclear markers for multiple colonizations of Lavatera (Malvaceae) in the Canary Islands. Systematic Botany. 2002;27:74–83. [Google Scholar]
- Gadella TWJ, Kliphuis E, Mennega EA. Chromosome numbers of some flowering plants of Spain and S-France. Acta Botanica Neerlandica. 1966;15:484–489. [Google Scholar]
- García-Talavera F. Las Canarias orientales y la vecina costa Africana en el holoceno Eres. 1997;7:55–63. [Google Scholar]
- Goldblatt P, Manning JC. The long-proboscid fly pollination system in Southern Africa. Annals of the Missouri Botanical Garden. 2000;87:146–170. [Google Scholar]
- Goldblatt P, Bernhardt P, Manning JC. Adaptive radiation of pollination mechanisms in Ixia (Iridaceae: Crocoideae) Annals of the Missouri Botanical Garden. 2000a;87:564–577. [Google Scholar]
- Goldblatt P, Manning JC, Bernhardt P. Adaptive radiation of pollination mechanisms in Sparaxis (Iridaceae: Ixioideae) Adansonia, série 3. 2000b;22:57–70. [Google Scholar]
- Haque MS, Ghoshal KK. Karyotypes and chromosome morphology in the genus Salvia Linn. Cytologia. 1980;45:627–640. [Google Scholar]
- Harley RM, Atkins S, Budantsev AL, et al. Labiatae. In: Kadereit JW, editor. The families and genera of vascular plants 7, Lamiales. Berlin: Springer; 2004. pp. 167–282. [Google Scholar]
- Hedberg I, Hedberg O. Chromosome numbers of afroalpine and afromontane angiosperms. Botaniska Notiser. 1977;130:1–24. [Google Scholar]
- Hedge IC. A revision of Salvia in Africa including Madagascar and the Canary Islands. Notes from the Royal Botanical Garden, Edinburgh. 1974;33:1–121. [Google Scholar]
- Hedge IC. Salvia. In: Davis PH, editor. Flora of Turkey. Edinburgh: Edinburgh University Press; 1982. pp. 400–461. [Google Scholar]
- Helfgott DM, Francisco-Ortega J, Santos-Guerra A, Jansen RK, Simpson BB. Biogeography and breeding system evolution of the woody Bencomia alliance (Rosaceae) in Macaronesia based on ITS sequence data. Systematic Botany. 2000;25:82–97. [Google Scholar]
- Jarvis C. Order out of chaos. Linnaean plant names and their types. London: The Linnean Society of London in association with the Natural History Museum; 2007. [Google Scholar]
- Jenks AA, Walker JB, Kim S-C. Evolution and origins of the Mazatec hallucinogenic sage, Salvia divinorum (Lamiaceae): a molecular phylogenetic approach. Journal of Plant Research. 2011;124:593–600. doi: 10.1007/s10265-010-0394-6. [DOI] [PubMed] [Google Scholar]
- Jenks A, Walker J, Kim S. Phylogeny of New World Salvia subgenus Calosphace (Lamiaceae) based on cpDNA (psbA-trnH) and nrDNA (ITS) sequence data. Journal of Plant Research. 2012;126:483–496. doi: 10.1007/s10265-012-0543-1. [DOI] [PubMed] [Google Scholar]
- Kirk-Spriggs AH, McGregor G. Disjunctions in the Diptera (Insecta) fauna of the Mediterranean province and southern Africa and a discussion of biogeographical considerations. Transactions of the Royal Society of South Africa. 2009;64:32–52. [Google Scholar]
- Li QQ, Li MH, Yuan QJ, Cui ZH, Huang LQ, Gen XP. Phylogenetic relationships of Salvia (Lamiaceae) in China: evidence from DNA sequence datasets. Journal of Systematics and Evolution. 2013;51:184–195. [Google Scholar]
- Linder HP. Plant diversity and endemism in sub-Saharan tropical Africa. Journal of Biogeography. 2001;28:169–182. [Google Scholar]
- Maddison WP, Maddison DR. MacClade: analysis of phylogeny and character evolution, version 4.0. Sunderland, MA: Sinauer Associates; 2000. [Google Scholar]
- Maddison WP, Maddison DR. Mesquite: a modular system for evolutionary analysis. 2011. Version 2.75. http://mesquiteproject.org [accessed March 15th 2013]
- Manen JF, Boulter MC, Naciri-Graven Y. The complex history of the genus Ilex L. (Aquifoliaceae): evidence from the comparison of plastid and nuclear DNA sequences and from fossil data. Plant Systematics and Evolution. 2002;235:79–98. [Google Scholar]
- Marloth R. Some observations on entomophilous flowers. South African Journal of Science. 1908;4:110–113. [Google Scholar]
- Migahid AM. Salvia. In: Migahid AM, editor. Migahid and Hammouda's Flora of Saudi Arabia. Riyadh: Riyadh University; 1978. pp. 464–465. [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. 2010. Proceedings of the Gateway Computing Environments Workshop (GCE), 14 November 2010. New Orleans, LA, 1–8. [available from http://www.phylo.org/portal2/login!input.action. ]
- Nathan R, Katul GG, Horn HS, et al. Mechanisms of long-distance dispersal of seeds by wind. Nature. 2002;418:409–413. doi: 10.1038/nature00844. [DOI] [PubMed] [Google Scholar]
- Noyes RD, Rieseberg LH. ITS sequence data support a single origin for North American Astereae (Asteraceae) and reflect deep geographic divisions in Aster s.l. American Journal of Botany. 1999;86:398–412. [PubMed] [Google Scholar]
- Pirie MD, Humphreys AM, Galley C, et al. A novel supermatrix approach improves resolution of phylogenetic relationships in a comprehensive sample of danthonioid grasses. Molecular Phylogenetics and Evolution. 2008;48:1106–1119. doi: 10.1016/j.ympev.2008.05.030. [DOI] [PubMed] [Google Scholar]
- Pirie MD, Humphreys AM, Barker NP, Linder HP. Reticulation, data combination, and inferring evolutionary history: an example from Danthonioideae (Poaceae) Systematic Biology. 2009;58:612–628. doi: 10.1093/sysbio/syp068. [DOI] [PubMed] [Google Scholar]
- Potgieter CJ, Edwards TJ. The occurrence of long, narrow corolla tubes in southern African Lamiaceae. Systematics and Geography of Plants. 2001;71:493–502. [Google Scholar]
- Potgieter CJ, Edwards TJ. The Stenobasipteron wiedemanni (Diptera, Nemestrinidae) pollination guild in Eastern Southern Africa. Annals of the Missouri Botanical Garden. 2005;92:254–267. [Google Scholar]
- Reese G. Über die Polyploidiespektren in der nordsaharischen Wüstenflora. Flora. 1957;144:608–634. [Google Scholar]
- Rodríguez-Gironés MA, Santamaría L. Why are so many bird flowers red? PLoS Biology. 2004;2:1515–1519. doi: 10.1371/journal.pbio.0020350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rosúa JL, Blanca G. Notas caríosistemáticas de la sección Salvia del género Salvia L. (Lamiaceae) Anales del Jardín Botánico de Madrid. 1985;42:101–112. [Google Scholar]
- Rosúa JL, Blanca G. Revisión del género Salvia L. (Lamiaceae) en el Mediterráneo occidental: la sección Salvia. Acta Botanica Malacitana. 1986;11:227–271. [Google Scholar]
- Rosúa JL, Blanca G. Acerca de la distribucion de la seccion Salvia en la region mediterranea occidental y sus relaciones de vicarianza con el este del mediterraneo. Lagascalia. 1990;15:137–143. [Google Scholar]
- Santos A, Fernández M. Lazaroa. Vol. 9. 51–54; 1986. Salvia herbanica spec. nova (Labiatae) en la flora de Fuerteventura (I. Canarias) [Google Scholar]
- Scholz S. Nuevos datos acerca de Salvia herbanica Santos et Fernandez (Lamiaceae) Vieraea. 1993;22:29–34. [Google Scholar]
- Scott-Elliot GF. Ornithophilous flowers in South Africa. Annals of Botany. 1890;4:265–280. [Google Scholar]
- Shaw J, Lickey EB, Schilling EE, Small RL. Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. American Journal of Botany. 2007;94:275–288. doi: 10.3732/ajb.94.3.275. [DOI] [PubMed] [Google Scholar]
- Sudarmono HO. Speciation process of Salvia isensis (Lamiaceae), a species endemic to serpentine areas in the Ise-Tokai district, Japan, from the viewpoint of the contradictory phylogenetic trees generated from chloroplast and nuclear DNA. Journal of Plant Research. 2007;120:483–490. doi: 10.1007/s10265-007-0075-2. [DOI] [PubMed] [Google Scholar]
- Sudarmono HO. Genetic differentiations among the populations of Salvia japonica (Lamiaceae) and its related species. Hayati Journal of Biosciences. 2008;15:18–26. [Google Scholar]
- Sun M, Gross K, Schiestl FP. Floral adaptation to local pollinator guilds in a terrestrial orchid. Annals of Botany. 2014;113:289–300. doi: 10.1093/aob/mct219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Stamatakis A, Hoover P, Rougemont J. Rapid bootstrap algorithm for the RAxML Web-Servers. Systematic Biology. 2008;75:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- Tackenberg O, Poschlod P, Kahmen S. Dandelion seed dispersal: the horizontal wind speed does not matter for long-distance dispersal – it is updraft! Plant Biology. 2003;5:451–454. [Google Scholar]
- Thulin M. Salvia (Labiatae) in the mountains of northern Somalia. Opera Botanica. 1993;121:145–148. [Google Scholar]
- Thulin M. Salvia geminata sp. nov. with remarkable stamen arrangement from southern Yemen, with notes on S. areysiana (Lamiaceae) Nordic Journal of Botany. 2009;27:336–338. [Google Scholar]
- Thomson JD, Wilson P. Explaining evolutionary shifts between bee and hummingbird pollination: convergence, divergence, and directionality. International Journal of Plant Sciences. 2008;169:23–38. [Google Scholar]
- Van Jaarsveld EJ. Salvia thermara, a new species from the Western Cape, South Africa. Bothalia. 1999;29:100–102. [Google Scholar]
- Van Jaarsveld EJ. South African sages. Veld and Flora. 2002;88:102–104. [Google Scholar]
- Van der Niet T, Johnson SD. Phylogenetic evidence for pollinator-driven diversification of angiosperms. Trends in Ecology and Evolution. 2012;27:353–361. doi: 10.1016/j.tree.2012.02.002. [DOI] [PubMed] [Google Scholar]
- Van der Niet T, Peakall R, Johnson SD. Pollinator-driven ecological speciation in plants: new evidence and future perspectives. Annals of Botany. 2014;113:199–212. doi: 10.1093/aob/mct290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas P. Are Macaronesian islands refugia of relict plant lineages?: a molecular survey. In: Weiss S, Ferrand N, editors. Phylogeography of southern European refugia. Dordrecht, The Netherlands: Springer; 2007. pp. 297–314. [Google Scholar]
- Vogt R, Aparicio A. Chromosome numbers of plants collected during Iter Mediterraneum IV in Cyprus. Bocconea. 1999;11:117–169. [Google Scholar]
- Walker JB, Sytsma KJ. Staminal evolution in the genus Salvia (Lamiaceae): molecular phylogenetic evidence for multiple origins of the staminal lever. Annals of Botany. 2007;100:375–391. doi: 10.1093/aob/mcl176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JB, Sytsma KJ, Treutlein J, Wink M. Salvia (Lamiaceae) is not monophyletic: implications for the systematics, radiation, and ecological specializations of Salvia and tribe Mentheae. American Journal of Botany. 2004;91:1115–1125. doi: 10.3732/ajb.91.7.1115. [DOI] [PubMed] [Google Scholar]
- Wester P. Sunbirds hover at flowers of Salvia and Lycium. Ostrich. 2013;84:27–32. [Google Scholar]
- Wester P, Claßen-Bockhoff R. Bird pollination in South African Salvia species. Flora – Morphology, Distribution, Functional Ecology of Plants. 2006;201:396–406. [Google Scholar]
- Wester P, Claßen-Bockhoff R. Floral diversity and pollen transfer mechanisms in bird- pollinated Salvia species. Annals of Botany. 2007;100:401–421. doi: 10.1093/aob/mcm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wester P, Claßen-Bockhoff R. Pollination syndromes of New World Salvia species with special reference to bird pollination. Annals of the Missouri Botanical Garden. 2011;98:101–155. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis M, Gelfand D, Sninsky J, White T, editors. PCR protocols: a guide to methods and applications. Orlando, FL: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Whitehouse E. Revision of Salvia section Salviastrum Gray. Field and Laboratory. 1949;17:151–165. [Google Scholar]
- Will M. Old World Salvia – morphological and molecular evidence for its evolution and non-monophyly. Mainz, Germany: Johannes Gutenberg University; 2013. PhD thesis. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.