Abstract
Coordinated gene expression is crucial in facilitating proper lymphoid cell development and function. The precise patterns of gene expression during B and T cell development are regulated through a complex interplay between a multitude of transcriptional regulators, both activators and repressors. We have recently identified the Snail family of transcription factors as playing significant and overlapping roles in lymphoid cell development in that deletion of both Snai2 and Snai3 was required to fully impact the generation of mature T and B cells. Analyses using compound heterozygote animals further demonstrated that Snai2 and Snai3 were partially haplo-sufficient and relatively equivalent in their ability to preserve B cell generation in the bone marrow. In this review, we summarize studies elucidating the role(s) of the Snail family in hematopoiesis with a focus on lymphoid cell development. Using the Snail family as an example, we discuss the concepts of functional redundancy and strategies employed to assay transcription factor families for “intra-member” compensation.
Keywords: Snail, transcription factor, hematopoiesis, B cell, lymphocyte, redundancy, leukemia
An introduction to the Snail family
Embryonic development along with the development of cell lineages with specialized functions requires a properly orchestrated network of gene expression. The Snail family is a primordial metazoan collection of transcriptional regulators consisting of three members: Snai1 (Snail), Snai2 (Slug) and Snai3 (Smuc) [1, 2]. Snail family members classically function as transcriptional repressors. To carry out this function, all three members share two cardinal features. Within the C-terminal portions of each protein reside multiple C2H2-type zinc finger DNA-binding domains (DBD). Snai1 possesses four DBDs while both Snai2 and Snai3 possess five [3]. These DBDs recognize the consensus E-box sequence, CANNTG. In particular, Snail proteins show a strong preference for GC-rich central dinucleotides [4]. At the extreme N-terminus, each protein contains a SNAG domain (Snail/Gfi1) [5]. Using this domain, the recruitment of various chromatin co-repressor modifiers leads to the generation of a transcriptionally silent state. Examples of such modifiers include histone deacetylaseses, such as HDACs 1/2, lysine-specific demethylases (LSD1) and methyltransferases (EZH2) [6, 7].
The Snail family has been demonstrated to participate in a wide variety of physiological and pathological processes [8, 9]. The founding member, Snai1, was first described in Drosophila melanogaster and was shown to be essential in the developing embryo for proper ventral-dorsal patterning leading to eventual mesoderm formation [10, 11]. Analogous to Drosophila, deletion of murine Snai1 results in embryonic lethality due to gastrulation defects [12]. This points to an evolutionarily conserved role for Snai1 in the developing embryo. Deletion of Snai1 at the epiblast stage also resulted in embryonic lethality, in this instance due to global defects in vascularization [13]. Continuing to focus on the murine system, deletion of Snai2 did not result in organismal catastrophe. Snai2 germline knockout mice possess impaired physical and hair follicle developmental kinetics most readily observed within the pre-weaning period [14, 15]. On select genetic backgrounds, these mice develop piebaldism (suggestive of defective melanocyte function) and symptoms analogous to Type II Waardenburg syndrome (characterized by hearing loss and skin/hair pigment anomalies) [16]. Of significance, a functional redundancy between Snai1 and Snai2 in both chondrogenesis and cranial-facial development has been previously shown [17, 18]. Recently, two laboratories including our own have described the generation of Snai3 deficient animals. Unlike both Snai1 and Snai2, no phenotypes were apparent upon the deletion of only Snai3 [19, 20]. Our studies, however, additionally analyzed the loss of Snai3 in the context of a Snai2 deficient animal, a germ line double knockout (DKO), which resulted in clear developmental abnormalities [20]. Some of these included severe “runting” with an overall failure to thrive and a definitive skewing towards generation of the male sex. Additionally, multiple lymphopoietic abnormalities were apparent only upon deletion of both genes (discussed below). This data supported a physiological role for Snai3 along with a continued theme of functional redundancy among various Snail members.
The Snail family and hematopoiesis
At this time, there is no data elucidating the role of Snai1 within the hematopoietic system. Recently, we have generated a hematopoietic-specific deletion of Snai1 via utilization of the Vav-Cre deleter strain and a strain possessing a conditionally targeted Snai1 gene. Unlike embryogenesis, Snai1 is not required for hematopoiesis since these conditional Snai1-deleted mice are viable, outwardly healthy, and with no obvious hematopoietic deficiencies (unpublished data). A more detailed analysis of these mice is underway.
For the rest of this article, we shift our focus towards the hematopoietic functions of both Snai2 and Snai3. In an initial report, Inukai et al. demonstrated overexpression of Snai2 in several human B cell leukemia cell lines. Upregulation of Snai2 was dependent upon the E2A-HLF oncoprotein generated from a t(17;19) chromosomal translocation. Usage of the murine IL-3 dependent Baf3 Pro-B cell line demonstrated that overexpression of Snai2 was sufficient to confer resistance to apoptosis induced by growth factor withdrawal which was accompanied by exit from the cell cycle [21]. In regards to cancer progression, Snai2 and Snai1 are most commonly identified for their ability to induce epithelial-to-mesenchymal transition (EMT) resulting in a more migratory and invasive cancer phenotype. This result suggested an alternative mechanism for the survival of transformed cells. Less appreciated, but maybe just as significant; these data may point to a role for Snai2 in chemotherapeutic resistance. This is most relevant for DNA damaging agents such as Doxorubicin, which are most effective in actively cycling cells. Interestingly, Perez-Losada et al., demonstrated the ability of c-Kit signaling to induce Snai2 expression. In vitro studies utilized both Baf3 and LAMA-84 cells overexpressing c-Kit. Of note, LAMA-84 cells were originally derived from a chronic myeloid leukemia (CML) patient undergoing blast crisis [22]. The mechanism of Snai2 upregulation becomes relevant when one considers that c-Kit is highly expressed on the surface of acute myeloid leukemia (AML) cells [23, 24]. Unfortunately, this study did not conduct any experiments to assess the downstream consequences of Snai2 induction in LAMA-84 cells. Overall, these data provided some interesting insights into the potential role(s) of the Snail family in promoting hematological malignancies.
Moving forward, the point of emphasis shifted towards the role of Snai2 in more physiologic hematopoietic settings. Included within the Perez-Losada report was an initial description of hematopoiesis in the Snai2 knockout mouse. They observed multiple defects, which mainly focused on erythropoiesis [14]. Complete blood counts showed a trend towards lower erythroid “output”. Using in vivo models of phenylhydrazine (PHZ)- and pregnancy-induced anemia, a lower percentage of Ter119+ cells was observed in the spleen. This pointed to a role for Snai2 in hematopoietic stress responses and its requirement in reconstituting the erythroid lineage. Intriguingly, Snai2+/− animals showed similar defects as the Snai2−/− suggesting a gene dosage component. Examination of steady state hematopoiesis did not show any derivation from normal myeloid and B cell generation. However, a decreased ratio of developing CD4 and CD8 double positive thymocytes was observed in Snai2 knockout animals. This was apparently a result of increased apoptosis as assayed by TUNEL staining of thymic cross-sections. Almost immediately following this publication, a separate study from the Look lab showed the expression of Snai2 in multiple bone marrow progenitor lineages including the hematopoietic stem cell (HSC), both long term (HSC-LT) and short term (HSC-ST), common lymphoid (CLP) and common myeloid (CMP) progenitors among others. Colony forming assays were performed to evaluate differentiation potential of progenitors in the bone marrow and spleen. While the Snai2−/− progenitors trended towards an increase in colony forming units (CFUs) for various lineages, no clear-cut enhancement over wildtype (WT) was observed. Expanding on this, they assayed the ability of Snai2−/− animals to reconstitute the hematopoietic compartment following an LD50 dose of total body irradiation (TBI), also a stress response [25]. By 13 days post-irradiation, all Snai2−/− animals had died due to pancytopenia. This was in stark contrast to both WT and Snai2+/− animals in which approximately fifty percent survival was achieved when followed out to 29 days. Closer examination of these animals demonstrated increased apoptosis within the Snai2−/− lineage negative (Lin−) bone marrow leading to the pancytopenia. As such, administration of a thrombopoietin analog rescued irradiation-induced lethality of Snai2−/− animals. Using bone marrow chimeras, a follow up study demonstrated that loss of Snai2 in the hematopoietic compartment was sufficient to propagate the irradiation phenotype described above [26]. Importantly, WT bone marrow administered to a Snai2−/− host completely prevented any lethality providing further evidence for a hematopoietically cell intrinsic role for Snai2 that was independent of the surrounding bone marrow stroma. Using a well-designed combination of knockout and transgenic animals, it was shown that Snai2 functioned downstream of p53 to block the activation of Puma, a pro-apoptotic BH3-only family member. This was accomplished via a direct E-box interaction within intron 1 of the Puma gene. Somewhat surprisingly, this effect was specific to Puma as Snai2−/− Puma−/− compound knockout animals were completely radio-resistant. If other apoptotic mediators were playing a role, an intermediate phenotype would have been expected. Finally, a group led by Wen-Shu Wu examined the self-renewal capacity of hematopoietic stem cells with and without Snai2 [27]. Using limiting dilution bone marrow chimera transplants, it was determined that Snai2−/− marrow possessed an approximately 8-fold higher repopulation efficiency as compared to Snai2+/−. Competitive serial transplantation, a gold standard for assessing HSC fidelity, further demonstrated the enhanced ability of the Snai2−/− HSC to reconstitute the entire hematopoietic system. Incorporation of 5-ethynyl-2’-deoxyuridine (Edu) combined with Annexin V cell surface staining demonstrated this result was a consequence of higher proliferative capacity rather than augmented apoptosis within the Snai2−/− Lin− Sca1+ compartment. This result coalesces with the previously identified ability of Snai2 overexpression to induce cell cycle exit by Baf3 cells [21]. While the molecular targets of Snai2 were not identified, they may be similar to what has been previously observed for Snai1. In a non-tumorigenic context, Snai1 is able to inhibit the cell cycle of MDCK cells by repressing cyclin D2 at the transcriptional level [28]. In a non-canonical role for the Snail family, it was also reported that Snai1 co-associated with EGR-1 and SP-1 to upregulate p15INK4b leading to the arrest of HepG2 cells [29].
The function of the Snai3 transcriptional regulator in the hemaotopoietic system was first analyzed in an indirect investigation, the analysis of binding proteins to negative regulator elements of the murine Itgb2l (Pactolus) gene [30]. Using a combination of EMSA and supershift assays, it was shown that Snai3 was capable of binding to these transcriptional regulatory sites. The most intriguing aspect of this work was the hypothesis that in scenarios where diverse cell types, such as B cells and neutrophils, express a common transcriptional activator (e.g. PU.1), differential expression of a negative regulator may potentially augment lineage specific expression of downstream genes. Thinking more globally, Dahlem et al. asked whether retroviral-induced overexpression of Snai3 in a bone marrow chimera model was capable of impairing hematopoiesis. Using c-Kit and Sca-1 based analysis of lineage depleted marrow, it was determined that overexpression of Snai3 did not alter ratios of various subsets of hematopoietic progenitors [31]. However, inspection of peripheral blood demonstrated a clear loss of B and T cell lineages in cells that were infected with the Snai3-expressing retrovirus (as assayed by GFP expression). This effect was dose dependent as B and T cell populations were present when Snai3 overexpression was at a modest amount (GFP low). Importantly, B and T cells were present when the empty vector was expressed at both low and high levels arguing against any retroviral-induced artifacts. These data suggested that Snai3 was either capable of saturating endogenous E-boxes necessary for the proper bifurcation of lymphoid and myeloid lineages or directly repressed gene expression by the recruitment of transcriptional control complexes. This data also alluded to a requirement for precise regulation of Snai3 within the hematopoietic system.
Phenotypic analysis of animals lacking both Snai2 and Snai3 transcription factors
Two groups recently described the deletion of the Snai3 gene using both conditional and germ line deletion models. Upon deletion of Snai3, no effects were observed in any hematopoietic lineage assayed [19, 20]. Indeed the Snai3−/− strains created by both groups displayed little to no phenotypic changes from WT. Gene expression analysis of Snai1, Snai2 and Snai3 in bone marrow derived lineages demonstrated widespread Snai1 expression in T and B precursor and mature cells, as well as cells of the myeloid lineage[20]. Snai2 expression was more limited with highest levels of transcripts found in immature bone marrow cells (B220−, CDllb− cells) and double negative (DN) (CD4− CD8−) T cells of the thymus. Snai3 expression was highest in the DN, the double positive (DP) (CD4+CD8+) and CD8+ cells of the thymus and spleen. The potential of the Snail family of proteins to functionally complement one another led us to create and analyze a germ line DKO animal lacking both the Snai2 and Snai3 genes [20] (the absence of Snai1 in the germ line creates an embryonic lethality) [13]. In this animal we observed multiple hematopoietic and non-hematopoietic phenotypes, all of them greater than that found in the single Snai2−/− or Snai3−/− animals. The DKO animals were distinctly smaller than the single deficient animals and had significantly smaller spleens and thymi, which possessed morphological distortions. The most striking hematologic phenotype of the DKO animals was a loss of B cells in the bone marrow. This was correlated with an expanded myeloid compartment. In the DKO thymus, double positive thymocytes were reduced in favor of enhanced percentages of CD4 single positive T cells. Unlike in the marrow, skewing of thymocyte populations displayed variable penetrance. Analysis of compound heterozygotes suggested that on a per allele basis, one allele of either Snai2 or Snai3 equivalently compensated for the loss of the other three alleles. The DKO animals did possess a normal Snai1 gene; which is transcriptionally active in immature B and T cell subsets. As such, incomplete complementation of the DKO animal via Snai1 may have continued to mask the full dependence of hematopoietic cell development on members of the Snail family.
One of the unanswered questions from our recent study was the level of stromal contribution to the phenotypes observed. Development of both B and T cells is dependent upon proper organization within their respective sites of generation. For example, pre-Pro-B and Pro-B cells congregate in bone marrow niches high in CXCL12 and IL-7, respectively [32]. In contrast, Pre-B cells were not found to associate with either zone suggesting precise localization is required for the proper progression along each stage of B cell maturation. Histological examination of the germline Snai2 and Snai3 DKO bone marrow assessing B cell localization has not been performed. However, as mentioned above, the analysis of thymic cross-sections from DKO mice showed gross alterations in overall architecture [20]. In DKO mice, intense hematoxylin staining was observed in the region corresponding to the thymic medulla, the site of single positive thymocyte negative selection. Normally, the thymic cortex demonstrates the highest intensity of hematoxylin staining due to high numbers of CD4 and CD8 double positive thymocytes. Since double positive thymocytes were still the majority population in the DKO thymus, this argues against preferential staining of an over expanded single positive compartment. Rather this may suggest a dysfunctional organization of the thymus due to altered chemokine signaling. Multiple studies have demonstrated the importance of stromal-derived CCL25 and P-selectin in thymic homing and organization [33, 34]. Interestingly, competitive bone marrow chimeras using WT and CCR9−/− donor marrow revealed deficiencies in CCR9−/− thymocyte development that mirrored many of the phenotypes observed upon thymic stromal-specific deletion of Il7 [35]. This points to a role for CCL25 not only in thymic homing, but perhaps also in positioning developing thymocytes in the proximity of survival/proliferative signals such as IL-7. Given the known role of the Snail family in cellular migration (i.e. gastrulation and EMT), it is possible that the contributions of this family to lymphocyte development include a role in lymphocyte migration. This function would assist in orchestrating lymphocyte maturation by allowing a cell at any given developmental stage to receive signals required for its survival and continued differentiation. Due to the lack of an animal model possessing a Snai2 conditional allele, the examination of stromal contributions in providing organizational and/or survival signals will require the use of bone marrow chimera models. While inconvenient due to such factors as length of time for functional reconstitution, these studies may provide the most flexibility in assaying the stromal versus hematopoietic function of each Snail gene. In this sense, one has the option of “selectively” deleting Snai2 in the stroma while also deleting Snai3 in just the hematopoietic compartment and vice versa. Experiments such as these would be virtually impossible using current recombinase deleter strains.
Functional redundancy within the Snail gene family
The requirement of Snai2 deletion to unveil a role for Snai3 in hematopoiesis was somewhat of a surprise. Classically, the thought has been: delete Gene X, deduce the function of Gene X based on the resulting phenotype(s). Clearly, nature does not operate in such a simple and linear fashion especially when considering multigene families of transcription factors. So can the need for compound knockouts be predicted? In reality, the answer is most likely not a simple yes or no. When dealing with a multigene family such as Snail, one might transcriptionally profile the cell/tissue type of interest for overlapping expression. While coexpression of multiple Snail members may suggest a potential for redundancy, it does not rule out of the possibility of completely independent functions for each factor within a given cell. Conversely, a situation lacking coexpression rules out nothing. This was demonstrated for Snai1 and Snai2 during chondrogenesis [18]. In the steady state, Snai1 and Snai2 were expressed mutually exclusively within hypertrophic and proliferating chondrocytes, respectively. However, deletion of Snai1 resulted in expression of Snai2 in hypertrophic chondrocytes and vice versa. It was later shown that Snai1 and Snai2 could repress one another at the level of transcription [36]. In a circumstance such as this, analysis of single knockouts provided the evidence for a compound knockout approach.
Within hematopoiesis, functional redundancy among transcription factors is not a new concept. For example, the GATA family of transcriptional regulators possesses six members, three of which (GATA1-3) are highly expressed in hematopoietic lineages. Using mouse deficiency models, the lack of both GATA-1 and GATA-2 has a more profound impact upon primitive hematopoiesis during development than seen with animals lacking only a single such factor [37]. Additionally, all three Runx family members (Runx1, Runx2 and Runx3) are expressed in the hematopoietic stem cells and may play redundant roles in influencing stem cell quiescence and cycling [38].
Functional redundancy for transcription factors implies common gene target recognition. However unique protein sequences present within highly conserved family members can alter target site specificity by recruiting additional targeting factors, as demonstrated by the highly conserved members of the ETS family [39].
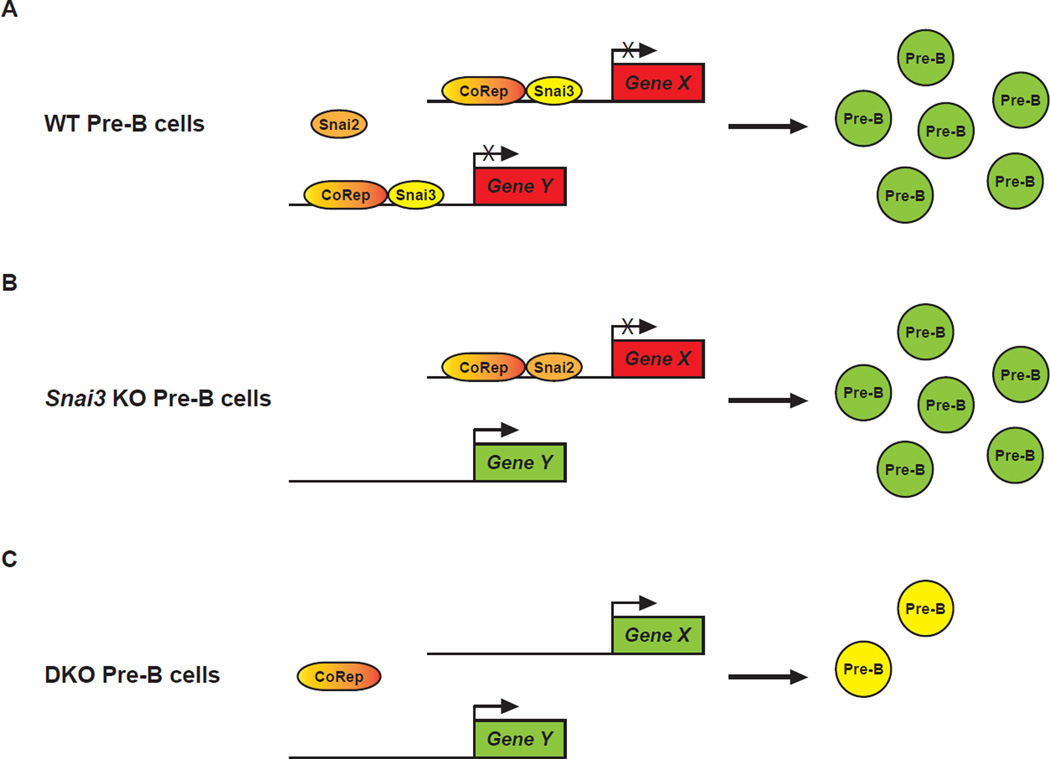
Once a functional redundancy is validated using compound knockouts, the next question becomes the mechanism. The optimal way to assess this phenomenon would be to determine the transcriptional footprint of each factor involved through the combined use of ChIP- and RNA-Seq. ChIP-Seq will identify direct transcriptional targets while RNA-Seq of knockout animals will additionally identify indirect genetic interactions. In the case of transcriptional repressors, genes downregulated in a knockout would most likely represent an indirect regulation (i.e. derepression of another repressor). While seemingly arduous, this approach is much more globally informative and unbiased compared to the examination of predetermined gene targets. For both forms of analyses, WT and knockout animals are required. Wildtype animals become extremely valuable in their ability to tell us what happens normally (e.g. Snai2 versus Snai3 bound genes). Analysis of single and compound knockouts will determine the genetic extent of redundancy and may even narrow the candidate list of “essential” targets. Consider the following hypothetical (and over-simplified) example, Snai3 binds the Gene X and Gene Y promoters in WT Pre-B cells (Figure 1A). Upon loss of Snai3, Snai2 now binds only the Gene X promoter. As such Gene X is still repressed but now expression of Gene Y is enhanced with no resultant phenotype (Figure 1B). Upon deletion of both Snai2 and Snai3, Gene X is now also transcriptionally enhanced and a B cell phenotype is now observable (Figure 1C). This result would identify two things: 1) common and specific targets for each Snail member and 2) genes potentially responsible for phenotypes observed in the compound knockout. Of course, these scenarios become more complex as the family being studied and their list of in vivo targets grow. One factor not being discussed is how dysregulated expression of known post-translational modifiers (PTM) may augment the system being studied. This may be most relevant for factors under the influence of a multitude of PTM modalities.
Figure 1. Redundancy model illustrating potential Snail compensation in lymphocyte development.
(A) In a WT developing Pre-B cell, Snai3 inhibits transcription of its specific hypothetical target genes (X and Y) (red boxes) along with a Co-repressor protein (CoRep) allowing for normal development of Pre-B cells. (B) In the absence of Snai3, the Snai2 protein can facilitate the transcriptional repression of Gene X; however, it lacks the specificity to similarly repress Gene Y transcription (green box). The presence of the Gene Y protein product does not appreciably influence the development of the Pre-B cells. (C) The absence of both Snai2 and Snai3 allows for the transcription of both Gene X and Gene Y that results in the inhibition of the development of functional Pre-B cells (yellow cells).
Conclusions
To summarize, the Snail family consists of three evolutionarily conserved members. These factors have been most extensively studied in the fields of developmental and cancer biology. The examination of how this family regulates various aspects of immunology has been minimal at best. It is the opinion of the authors that the role of the Snail family in hematopoiesis will prove to be much more expansive than initially realized. Part of this reasoning is based on the recent discovery that demonstrated a functional redundancy between Snai2 and Snai3 during lymphocyte development. This data also points to the evolutionary conservation of all three Snail members as a potential fail-safe mechanism allowing the fidelity of hematopoiesis in the face of single gene loss (i.e. germline or somatic mutation). An even more provocative question now becomes: how many functions of a particular gene have not been discovered due to redundancy with another family member? To completely unveil the functions of a specific gene product may ultimately require the generation and analysis of various single and compound knockout combinations.
Acknowledgment
J.H.W. was supported by funds from the Weber Presidential Endowed Chair and from the Department of Pathology, University of Utah. P.D.P. was supported as a predoctoral trainee by the NIH Hematology Research Training Grant T-32DK007115-38.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contribution: P.D.P and J.H.W performed the relevant review of the literature. P.D.P. performed unpublished analysis cited in the article. Both authors wrote the article.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
References
- 1.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nature reviews Molecular cell biology. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 2.Manzanares M, Blanco MJ, Nieto MA. Snail3 orthologues in vertebrates: divergent members of the Snail zinc-finger gene family. Development genes and evolution. 2004;214:47–53. doi: 10.1007/s00427-003-0373-1. [DOI] [PubMed] [Google Scholar]
- 3.Mingot JM, Vega S, Maestro B, Sanz JM, Nieto MA. Characterization of Snail nuclear import pathways as representatives of C2H2 zinc finger transcription factors. Journal of cell science. 2009;122:1452–1460. doi: 10.1242/jcs.041749. [DOI] [PubMed] [Google Scholar]
- 4.Kataoka H, Murayama T, Yokode M, et al. A novel snail-related transcription factor Smuc regulates basic helix-loop-helix transcription factor activities via specific E-box motifs. Nucleic acids research. 2000;28:626–633. doi: 10.1093/nar/28.2.626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Wu Y, Li J, et al. The SNAG domain of Snail1 functions as a molecular hook for recruiting lysine-specific demethylase 1. The EMBO journal. 2010;29:1803–1816. doi: 10.1038/emboj.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peinado H, Ballestar E, Esteller M, Cano A. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Molecular and cellular biology. 2004;24:306–319. doi: 10.1128/MCB.24.1.306-319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tong ZT, Cai MY, Wang XG, et al. EZH2 supports nasopharyngeal carcinoma cell aggressiveness by forming a co-repressor complex with HDAC1/HDAC2 and Snail to inhibit E-cadherin. Oncogene. 2012;31:583–594. doi: 10.1038/onc.2011.254. [DOI] [PubMed] [Google Scholar]
- 8.Bolos V, Peinado H, Perez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. Journal of cell science. 2003;116:499–511. doi: 10.1242/jcs.00224. [DOI] [PubMed] [Google Scholar]
- 9.Oram KF, Gridley T. Mutations in snail family genes enhance craniosynostosis of Twist1 haplo-insufficient mice: implications for Saethre-Chotzen Syndrome. Genetics. 2005;170:971–974. doi: 10.1534/genetics.105.041277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulay JL, Dennefeld C, Alberga A. The Drosophila developmental gene snail encodes a protein with nucleic acid binding fingers. Nature. 1987;330:395–398. doi: 10.1038/330395a0. [DOI] [PubMed] [Google Scholar]
- 11.Alberga A, Boulay JL, Kempe E, Dennefeld C, Haenlin M. The snail gene required for mesoderm formation in Drosophila is expressed dynamically in derivatives of all three germ layers. Development. 1991;111:983–992. doi: 10.1242/dev.111.4.983. [DOI] [PubMed] [Google Scholar]
- 12.Carver EA, Jiang R, Lan Y, Oram KF, Gridley T. The mouse snail gene encodes a key regulator of the epithelial-mesenchymal transition. Molecular and cellular biology. 2001;21:8184–8188. doi: 10.1128/MCB.21.23.8184-8188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lomeli H, Starling C, Gridley T. Epiblast-specific Snai1 deletion results in embryonic lethality due to multiple vascular defects. BMC research notes. 2009;2:22. doi: 10.1186/1756-0500-2-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perez-Losada J, Sanchez-Martin M, Rodriguez-Garcia A, et al. Zinc-finger transcription factor Slug contributes to the function of the stem cell factor c-kit signaling pathway. Blood. 2002;100:1274–1286. [PubMed] [Google Scholar]
- 15.Parent AE, Newkirk KM, Kusewitt DF. Slug (Snai2) expression during skin and hair follicle development. The Journal of investigative dermatology. 2010;130:1737–1739. doi: 10.1038/jid.2010.22. [DOI] [PubMed] [Google Scholar]
- 16.Sanchez-Martin M, Rodriguez-Garcia A, Perez-Losada J, Sagrera A, Read AP, Sanchez-Garcia I. SLUG (SNAI2) deletions in patients with Waardenburg disease. Human molecular genetics. 2002;11:3231–3236. doi: 10.1093/hmg/11.25.3231. [DOI] [PubMed] [Google Scholar]
- 17.Murray SA, Oram KF, Gridley T. Multiple functions of Snail family genes during palate development in mice. Development. 2007;134:1789–1797. doi: 10.1242/dev.02837. [DOI] [PubMed] [Google Scholar]
- 18.Chen Y, Gridley T. Compensatory regulation of the Snai1 and Snai2 genes during chondrogenesis. Journal of bone and mineral research : the official journal of the American Society for Bone and Mineral Research. 2013;28:1412–1421. doi: 10.1002/jbmr.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bradley CK, Norton CR, Chen Y, et al. The snail family gene snai3 is not essential for embryogenesis in mice. PloS one. 2013;8:e65344. doi: 10.1371/journal.pone.0065344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pioli PD, Dahlem TJ, Weis JJ, Weis JH. Deletion of Snai2 and Snai3 results in impaired physical development compounded by lymphocyte deficiency. PloS one. 2013;8:e69216. doi: 10.1371/journal.pone.0069216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inukai T, Inoue A, Kurosawa H, et al. SLUG, a ces-1-related zinc finger transcription factor gene with antiapoptotic activity, is a downstream target of the E2A-HLF oncoprotein. Molecular cell. 1999;4:343–352. doi: 10.1016/s1097-2765(00)80336-6. [DOI] [PubMed] [Google Scholar]
- 22.Seigneurin D, Champelovier P, Mouchiroud G, et al. Human chronic myeloid leukemic cell line with positive Philadelphia chromosome exhibits megakaryocytic and erythroid characteristics. Experimental hematology. 1987;15:822–832. [PubMed] [Google Scholar]
- 23.Cascavilla N, Musto P, D'Arena G, et al. CD117 (c-kit) is a restricted antigen of acute myeloid leukemia and characterizes early differentiative levels of M5 FAB subtype. Haematologica. 1998;83:392–397. [PubMed] [Google Scholar]
- 24.Wakita S, Yamaguchi H, Miyake K, et al. Importance of c-kit mutation detection method sensitivity in prognostic analyses of t(8;21)(q22;q22) acute myeloid leukemia. Leukemia. 2011;25:1423–1432. doi: 10.1038/leu.2011.104. [DOI] [PubMed] [Google Scholar]
- 25.Inoue A, Seidel MG, Wu W, et al. Slug, a highly conserved zinc finger transcriptional repressor, protects hematopoietic progenitor cells from radiation-induced apoptosis in vivo. Cancer cell. 2002;2:279–288. doi: 10.1016/s1535-6108(02)00155-1. [DOI] [PubMed] [Google Scholar]
- 26.Wu WS, Heinrichs S, Xu D, et al. Slug antagonizes p53-mediated apoptosis of hematopoietic progenitors by repressing puma. Cell. 2005;123:641–653. doi: 10.1016/j.cell.2005.09.029. [DOI] [PubMed] [Google Scholar]
- 27.Sun Y, Shao L, Bai H, Wang ZZ, Wu WS. Slug deficiency enhances self-renewal of hematopoietic stem cells during hematopoietic regeneration. Blood. 2010;115:1709–1717. doi: 10.1182/blood-2009-07-232934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. Snail blocks the cell cycle and confers resistance to cell death. Genes & development. 2004;18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu CT, Chang TY, Cheng CC, et al. Snail associates with EGR-1 and SP-1 to upregulate transcriptional activation of p15INK4b. The FEBS journal. 2010;277:1202–1218. doi: 10.1111/j.1742-4658.2009.07553.x. [DOI] [PubMed] [Google Scholar]
- 30.Hale JS, Dahlem TJ, Margraf RL, Debnath I, Weis JJ, Weis JH. Transcriptional control of Pactolus: evidence of a negative control region and comparison with its evolutionary paralogue, CD18 (beta2 integrin) Journal of leukocyte biology. 2006;80:383–398. doi: 10.1189/jlb.0705390. [DOI] [PubMed] [Google Scholar]
- 31.Dahlem T, Cho S, Spangrude GJ, Weis JJ, Weis JH. Overexpression of Snai3 suppresses lymphoid- and enhances myeloid-cell differentiation. European journal of immunology. 2012;42:1038–1043. doi: 10.1002/eji.201142193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagasawa T. Microenvironmental niches in the bone marrow required for B-cell development. Nature reviews Immunology. 2006;6:107–116. doi: 10.1038/nri1780. [DOI] [PubMed] [Google Scholar]
- 33.Gossens K, Naus S, Corbel SY, et al. Thymic progenitor homing and lymphocyte homeostasis are linked via S1P-controlled expression of thymic P selectin/CCL25. The Journal of experimental medicine. 2009;206:761–778. doi: 10.1084/jem.20082502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svensson M, Marsal J, Uronen-Hansson H, et al. Involvement of CCR9 at multiple stages of adult T lymphopoiesis. Journal of leukocyte biology. 2008;83:156–164. doi: 10.1189/jlb.0607423. [DOI] [PubMed] [Google Scholar]
- 35.Shitara S, Hara T, Liang B, et al. IL-7 produced by thymic epithelial cells plays a major role in the development of thymocytes and TCRgammadelta+ intraepithelial lymphocytes. Journal of immunology. 2013;190:6173–6179. doi: 10.4049/jimmunol.1202573. [DOI] [PubMed] [Google Scholar]
- 36.Chen Y, Gridley T. The SNAI1 and SNAI2 proteins occupy their own and each other's promoter during chondrogenesis. Biochemical and biophysical research communications. 2013;435:356–360. doi: 10.1016/j.bbrc.2013.04.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujiwara Y, Chang AN, Williams AM, Orkin SH. Functional overlap of GATA-1 and GATA-2 in primitive hematopoietic development. Blood. 2004;103:583–585. doi: 10.1182/blood-2003-08-2870. [DOI] [PubMed] [Google Scholar]
- 38.Wang CQ, Jacob B, Nah GS, Osato M. Runx family genes, niche, and stem cell quiescence. Blood Cells Mol Dis. 2010;44:275–286. doi: 10.1016/j.bcmd.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 39.Hollenhorst PC, Shah AA, Hopkins C, Graves BJ. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes & development. 2007;21:1882–1894. doi: 10.1101/gad.1561707. [DOI] [PMC free article] [PubMed] [Google Scholar]